Abstract
We have previously demonstrated that double-strand breaks (DSBs) in regions near telomeres are much more likely to result in large deletions, gross chromosome rearrangements, and chromosome instability than DSBs at interstitial sites within chromosomes. In the present study, we investigated whether this response of subtelomeric regions to DSBs is a result of a deficiency in DSB repair by comparing the frequency of homologous recombination repair (HRR) and nonhomologous end joining (NHEJ) at interstitial and telomeric sites following the introduction of DSBs by I-SceI endonuclease. We also monitored the frequency of small deletions, which have been shown to be the most common mutation at I-SceI-induced DSBs at interstitial sites. We observed no difference in the frequency of small deletions or HRR at interstitial and subtelomeric DSBs. However, the frequency of NHEJ was significantly lower at DSBs near telomeres compared to interstitial sites. The frequency of NHEJ was also lower at DSBs occurring at interstitial sites containing telomeric repeat sequences. We propose that regions near telomeres are deficient in classical NHEJ as a result of the presence of cis-acting telomere-binding proteins that cause DSBs to be processed as though they were telomeres, resulting in excessive resection, telomere loss, and eventual chromosome rearrangements by alternative NHEJ.
Keywords: Chromosome instability, double-strand break, nonhomologous end joining, telomere
1. Introduction
DNA double-strand breaks (DSBs) are a critical DNA lesion, responsible for both the toxic effects of ionizing radiation and radiation-induced chromosome rearrangements leading to cancer [1]. The repair of DSBs occurs through either homologous recombination repair (HRR) or nonhomologous end joining (NHEJ). HRR in mammalian cells primarily uses the sister chromatid as a template, and is therefore limited to DSBs that occur after DNA replication [2]. NHEJ involves the joining of broken ends, and therefore can occur at anytime during the cell cycle. There are two forms of NHEJ, classical (C-NHEJ) and alternative (A-NHEJ). C-NHEJ has been extensively studied and many of the proteins that are involved are known, whereas much less is known about A-NHEJ [3, 4]. A-NHEJ has primarily been observed in cells deficient in C-NHEJ, and has therefore been proposed to serve as a backup mechanism for repair of DSBs. Both C-NHEJ and A-NHEJ produce mutations at the site of a DSB caused by ionizing radiation, however, A-NHEJ is commonly associated with large deletions [5, 6] and chromosome rearrangements [5, 7–10]. Another characteristic of A-NHEJ is that repair commonly occurs at sites with microhomology [6, 8, 11–13].
Not all DSBs are repaired equally well. Most DSBs generated by ionizing radiation are repaired within a few hours; however, some DSBs require many hours to be repaired [14, 15]. One factor that can influence the efficiency of repair of DSBs is their location in the genome. Goodarzi et al. [16] found that DSBs within heterochromatin are repaired much more slowly than DSBs occurring in euchromatin. Moreover, they also found that unlike DSBs occurring in euchromatin, the repair of DSBs in heterochromatin is dependent on ATM.
The ends of chromosomes, called telomeres, are another structural feature that can influence DSB repair. Telomeres are composed of the TTAGGG repeat sequence and associated proteins that together form the T-loop structure that keep the ends of chromosomes from appearing as DSBs and prevent chromosome fusion [17, 18]. Telomeres are maintained in human germ line cells by telomerase, but shorten during cell division in somatic cells due to insufficient telomerase expression [19]. The gradual telomere shortening that occurs during growth of somatic cells that do not express sufficient telomerase normally results in telomere-associated DSB repair foci and cell senescence [20, 21]. However, in cells that are unable to senesce, telomeres continue to shorten, eventually resulting in extensive chromosome fusion [22].
The role of the telomere in protecting chromosome ends and preventing chromosome fusion can influence the response to DNA damage within subtelomeric regioins. Richetti et al. [23] found that I-SceI-induced DSBs near telomeres in yeast are much more likely to result in gross chromosome rearrangements (GCRs) than DSBs occurring at interstitial locations, and concluded that this difference in response to DSBs near telomeres was due to differences in DSB repair. We have also reported that I-SceI-induced DSBs occurring near telomeres in a human tumor cell line are much more likely to result in large deletions, GCRs, and chromosome instability than DSBs occurring at interstitial locations within a chromosome [24]. This increased likelihood of telomere loss and GCRs as a result of DSBs within subtelomeric regions is not limited to human cancer cells, because I-SceI-induced DSBs near telomeres in mouse ES cells results in similar types of rearrangements and chromosome instability [25, 26]. The likelihood of telomere loss and GCRs as a result of DSBs within subtelomeric regions has important implications for the chromosome instability associated with cancer. We have previously proposed that the sensitivity of subtelomeric regions to DSBs plays an important role in the high rate of spontaneous telomere loss commonly observed human cancer cells [24, 27, 28]. The chromosome fusions resulting from telomere loss initiate chromosome instability through breakage/fusion/bridge (B/F/B) cycles [29], which can generate many of the chromosome rearrangements leading to cancer [30]. The importance of B/F/B cycles resulting from telomere loss in cancer was demonstrated by the high rate of carcinomas in mice that are deficient in both telomerase and p53 [31]. The sensitivity of subtelomeric regions to DSBs could also be important in ionizing radiation-induced carcinogenesis, because we have demonstrated that the sensitive region extends at least 100 kb from a telomere and therefore poses a large target for DSB-induced chromosome instability [32].
In the current study, we have investigated whether the increased frequency of large deletions and GCRs that result from DSBs within subtelomeric regions in mammalian cells is due to a deficiency in DSB repair. These studies involve the analysis of the frequency of DSB repair by HRR and NHEJ through the activation of the gene for green fluorescent protein (GFP) following the introduction of DSBs with the I-SceI endonuclease. The repair of I-SceI-induced-DSBs through activation of GFP has been used extensively in mammalian cells to study the mechanisms of DSB repair and DSB-induced chromosome rearrangements [12, 33–37]. We compared the frequency of repair of I-SceI-induced DSBs in a GFP gene integrated at telomeric sites, interstitial sites, and interstitial sites containing telomeric repeat sequences, to determine whether telomeres or telomeric repeat sequences influence the efficiency DSB repair.
2. Materials and Methods
2.1. Plasmids
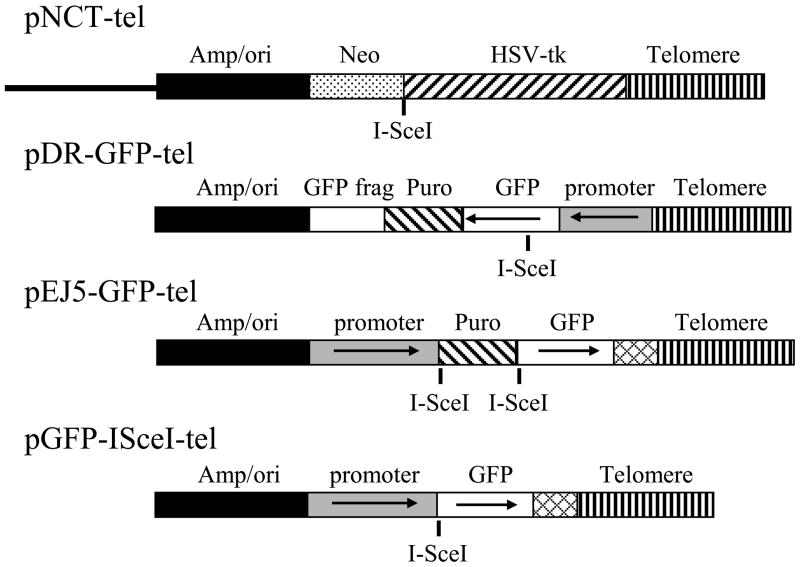
The pNCT-tel plasmid has been previously described [38]. pNCT-tel contains a neomycin-resistance (Neo) gene for positive selection with G418, a herpes simplex virus thymidine kinase (HSV-tk) gene for negative selection with ganciclovir, an I-SceI recognition site for introducing DSBs with the I-SceI endonuclease, and 0.8 kb of telomeric repeat sequences for seeding the formation of new telomeres (Fig. 1). The pQCXIH-ISceI retroviral vector that was used for expression of the I-SceI gene was constructed as previously described [24].
Fig. 1.
The structure of the plasmids used in this study. The pNCT-tel plasmid is located adjacent to the telomere on chromosome 16p in clone B3 of the EJ-30 tumor cell line, and was previously used to demonstrate the sensitivity of subtelomeric regions to DSBs. The pNCT-tel plasmid contains a β-lactamase gene for resistance to ampicillin and bacterial origin of replication (Amp/ori), a Neo gene for resistance to G418, and a gene for Herpes simplex virus thymidine kinase (HSV-tk). The pEJ5-GFP-tel and pDR-GFP-tel plasmids were generated from the pEJ5-GFP and pDR-GFP plasmids by insertion of telomeric repeat sequences. The telomeric repeat sequences were added to seed the formation of a new telomere following targeted integration through the shared homology in the Amp/ori sequences. The pDR-GFP-tel plasmid was used to monitor the frequency of HRR and contains a GFP gene that is defective due to an I-SceI site in the coding sequence, a puro gene for selection with puromycin, and a complimentary fragment of the GFP gene for repair of the I-SceI-induced DSB. The pEJ5-GFP-tel plasmid was used to monitor the frequency of NHEJ and contains a puro gene flanked by I-SceI sites, which is located between the GFP coding sequence and promoter. NHEJ between the two I-SceI-induced DSBs results in activation of the GFP gene. The pGFP-ISceI-tel plasmid was used to determine the frequency of large deletions (>50 bp) and GCRs. The cell clones containing the pGFP-ISceI-tel plasmid were generated from the clones containing the pEJ5-GFP-tel plasmid by I-SceI-mediated deletion of the puro gene, and contain an active GFP gene with an I-SceI site located between the coding sequence and promoter.
The pDR-GFP-tel plasmid was generated from the pDR-GFP plasmid [12, 39] by the insertion of telomeric repeat sequences (Fig. 1). The first step in construction of pDR-GFP-ISceI was to remove the unique NotI restriction site in pDR-GFP. This was accomplished by first digesting the plasmid with NotI, and using the Klenow fragment of E. coli DNA polymerase (Invitrogen) to fill in the single-stranded NotI overhang prior to ligation. The next step was to insert a linker containing the NotI and XhoI restriction sites into the unique SspI restriction site located between the β-lactamase gene and chicken β-actin promoter for the GFP gene. A NotI/XhoI restriction fragment containing 0.8 kb of telomeric repeat sequences from the pNTP-tel plasmid [40] was then inserted into the NotI/XhoI sites in the pDR-GFP plasmid.
The pEJ5-GFP-tel plasmid was generated from the pEJ5-GFP plasmid [12] by the insertion of telomeric repeat sequences (Fig. 1). The first step was to eliminate two NotI restriction sites flanking a 300 bp fragment containing the polyA addition sequences on the GFP gene. This was accomplished by first digesting the plasmid with NotI, then using the Klenow fragment of E. coli DNA polymerase (Invitrogen) to fill in the single-stranded NotI overhang prior to ligation. We next eliminated a unique SacI restriction site, by digesting the plasmid with SacI, then using the T4 DNA polymerase (Invitrogen) to remove the single-stranded SacI overhang prior to ligation. We then inserted a linker containing the NotI and SacI restriction sites between two PvuII restriction sites located between the beta lactamase gene and the GFP gene. The 0.8kb NotI/SacI fragment containing telomeric repeat sequences in the pNTP-tel plasmid [40] was then inserted into the NotI/SacI sites in the pEJ5-GFP plasmid. We then inserted a SmaI/XbaI restriction fragment containing the PolyA addition sequences from the pPGKpuro plasmid into complementary FseI/NheI restriction sites at the end of the GFP gene.
2.2. Cell lines
All of the cell lines used in this study were derived from clone B3 of the EJ-30 human bladder cell carcinoma cell line. EJ-30 is a subclone of the EJ-30 cell line, which is also called MGH-U1 [41]. The cells were grown in αMEM media (UCSF Cell Culture Facility) supplemented with 5% fetal calf serum (Invitrogen-Gibco), 5% newborn calf serum with iron (Invitrogen-Gibco), 1 mM L-glutamine (Invitrogen-Gibco), and were propagated at 37°C in humidified incubators.
Cell clone B3 was derived from EJ-30 following transfection with the linearized pNCT-tel plasmid [38], which seeded the formation of a new telomere 4.3 Mb from the original telomere on the short arm of chromosome 16 (Fig. 1). Rescue of the integrated pNCT-tel plasmid and sequencing of adjacent cellular DNA demonstrated that the plasmid integrated in the first intron at the 5′ end of the sarcalumenin gene (Genbank Accession number GQ475285) [26].
The targeting of the pDR-GFP and pEJ5-GFP plasmids to a telomere was accomplished through the addition of telomeric repeat sequences to generate the plasmids, pDR-GFP-tel and pEJ5-GFP-tel, respectively (Fig. 1). Targeting by homologous recombination into the pNCT-tel plasmid at the telomere on chromosome 16 in clone B3 [30, 38, 42] was facilitated by the presence of an identical β-lactamase and replication origin (Amp/ori) in the pNCT-tel plasmid and transfected plasmids. Following integration, the telomeric repeat sequences in the transfected plasmids are in the proper orientation to seed the formation of a new telomere. Screening for successful targeting was performed using positive selection for the puro gene in the pDR-GFP-tel or pEJ5-GFP-tel plasmids, and selection with ganciclovir for loss of the HSV-tk gene in the pNCT-tel plasmid. The puror/Ganr clones were then selected and their genomic DNA analyzed by Southern blot analysis using a variety of restriction enzymes to identify clones in which the transfected plasmid had replaced the pNCT-tel plasmid (data not shown). Successful targeting of the plasmids was then confirmed by the rescue and sequencing of the integrated plasmid sequences and adjacent cellular DNA (data not shown), as was used previously [32].
A similar protocol was also used to identify cell clones that contain the pEJ5-GFP-tel plasmid integrated at interstitial sites. Southern blot analysis using a variety of restriction enzymes was then used to identify clones that contained a single copy of the plasmid (data not shown). PCR was used to determine whether the integrated pEJ5-GFP plasmid had retained telomeric repeat sequences, using the TRP primer (5′-AACCCTAACCCTAACCCT-3′) within the telomeric repeat sequences and the PIM-2R primer (5′-TCCTCCAGAGTGGATTCG-3′) within the adjacent plasmid DNA (data not shown). PCR involved 94°C for 2 minutes, then 40 cycles of 94°C for 30 seconds, 60°C for 30 seconds, and 72°C for 30 seconds.
Cell clones containing the plasmids integrated at interstitial sites without telomeric repeat sequences were obtained by transfection of clone B3 with the pDR-GFP-tel and pEJ5-GFP-tel plasmid sequences after removal of the telomeric repeat sequences with either NotI and XhoI, or NotI and SacI, respectively. The linearized plasmids were trasfected into clone B3 using Lipofectamine 2000, as recommended by the manufacturer (Invitrogen). The transfected cells were grown in the presence of 2 μg/ml puromycin (Sigma) to select for cells containing the integrated plasmids. Southern blot analysis was then used to identify clones that contained a single copy of the plasmid (data not shown).
Cell clones containing active GFP with an I-SceI site between the GFP coding sequence and promoter (Fig. 1) were generated from clones containing the EJ5-GFP-tel plasmid integrated at a telomeric site (EJ5-6D), interstitial site without telomeric repeat sequences (EJ5-7F), or interstitial site with telomeric repeat sequences (EJ5-7C). Following transient transfection with the pCMV-ISceI expression vector, the cells were plated at low density. After two weeks, the plates were observed using a GFP fluorescent flashlight with GFP filter eyeglasses (NightSea) to detect GFP-positive colonies. The GFP-positive colonies were then selected, expanded in culture, and genomic DNA was isolated. As described below for the analysis of small deletions, the genomic DNA was then amplified by PCR using the GFP-1 and GFP-3 primers, and the PCR product analyzed by digestion with I-SceI to identify clones with an intact I-SceI site.
2.3. Virus preparation and selection for virally infected cells
Packaging of the pQCXIH-I-SceI retroviral vectors and infection of cell cultures was performed as previously described [24]. The selection for cells infected with pQCXIH-ISceI was achieved by growth in medium containing 50 μg/ml hygromycin (Sigma) for 10 days with medium changes every 2 days, to allow for expression of I-SceI endonuclease and the generation of DSBs. After 10 days, the cells were trypsinized, pooled together, and either analyzed for the frequency of GFP-positive cells, or replated for preparation of genomic DNA. For analysis of the frequency of GFP-positive cells, the cells were aliquoted into a counting chamber slide, and total cells and GFP-positive cells counted using a Cellometer Vision (Nexelcom). Two counting chambers were used for each sample, with each chamber being counted two times. All experiments were performed in triplicate, with a minimum of 1000 cells counted for each sample. The results with the Cellometer Vison were confirmed for some of the samples using a fluorescent microscope and manual counting.
2.4. PCR and analysis of small deletions
The presence of small deletions, either at a single I-SceI-induced DSB, or when joining two I-SceI-induced DSBs, was analyzed by first generating PCR products spanning an I-SceI site in the integrated pEJ5-GFP plasmid, and then digesting the PCR products with I-SceI endonuclease. PCR was performed on genomic DNA isolated from the pooled hygromycin-resistant cell cultures following infection with the pQCXIH-ISceI retroviral vector. PCR was performed using Taq 2X Master Mix (New England Biolabs) and primers GFP-1 (5′-GCGGGGTTCGGCTTCTGG-3′) and GFP-3 (5′-CGCTTCCATTGCTCAGCGG-3′) for small deletions resulting from a single I-SceI-induced DSBs, and GFP-1 and GFP-2 (5′-TCTTTCATGGCGGACTTG-3′) for small deletions resulting from joining two I-SceI-induced DSBs. PCR involved 94°C for 2 minutes, then 40 or 41 cycles (single or two I-SceI-induced DSBs, respectively) of 94°C for 30 seconds, 62°C for 30 seconds, and 72°C for 30 seconds. 25 μl of the PCR product was then digested with 20 units of I-SceI endonuclease at 37°C overnight, and the products were run on 4% agarose gels. After staining with ethidium bromide, digital images were analyzed using free ImageJ software (http://www.versiontracker.com/dyn/moreinfo/macosx/37303) to calculate the intensity of the bands. The percent of cells containing small deletions at the I-SceI site was determined by dividing the intensity of the uncut band by the combined intensity of the cut and uncut bands. The values for small deletions at a single I-SceI site were corrected for the fraction of cells that had experienced inactivation of the GFP gene (see Fig. 2), because these cells would not produce a PCR product due to large deletions, GCRs or failure to repair the DSB, and would therefore cause an overestimation of the fraction of cells containing small deletions. This correction involved multiplying by 0.6 (1–0.4) for clones containing subtelomeric integration sites, and 0.95 (1–0.05) for clones containing interstitial integration sites. The validity of this correction was previously demonstrated by the analysis of the frequency of small deletions in 100 individual subclones selected at random [24].
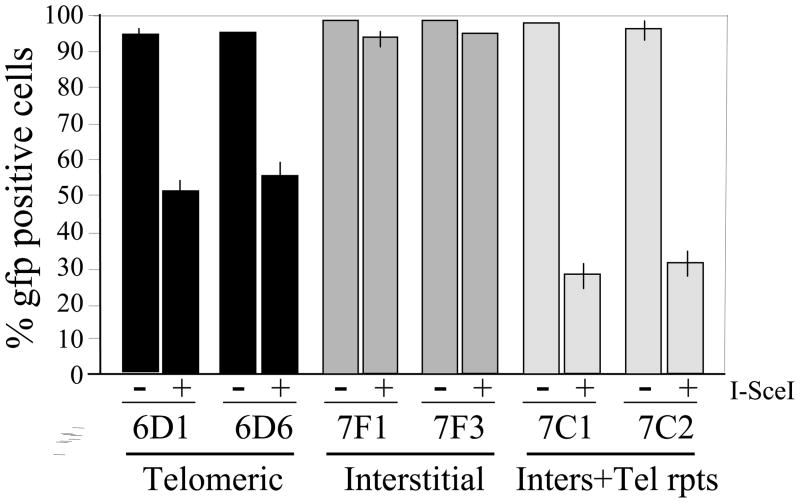
Fig. 2. Increased frequency of loss of the GP gene as a result of DSBs.
near telomeres and interstitial sites containing telomeric repeat sequences. The percentage of GFP-positive cells with (+) or without (-) constitutive expression of I-SceI endonuclease was used to monitor large deletions and GCRs in cell clones containing the pGFP-ISceI plasmid adjacent to a telomere (6D1 and 6D6), at an interstitial site (7F1 and 7F3), and an interstitial site containing telomeric repeat sequences (7C1 and 7C2). Experiments were performed in triplicate and standard deviations are shown.
3.0. Results
3.1. Generation of cell lines used for monitoring DSB repair
DSB repair at interstitial and telomeric sites was compared using the pDR-GFP and pEJ5-GFP plasmids (Fig. 1), which were previously used to monitor the frequency of repair of DSBs generated with the I-SceI endonuclease [12, 39]. The frequency of HRR was determined using the pDR-GFP plasmid, which contains a GFP gene that is inactive due to the presence of an I-SceI recognition site inserted in the coding sequence. The activation of GFP in pDR-GFP results from the repair of the I-SceI-induced DSB by HRR using a complimentary fragment of the GFP gene that is also present in the plasmid. The frequency of NHEJ was determined using the pEJ5-GFP plasmid, in which the GFP gene is inactive due to the presence of a puro gene located between the GFP coding sequence and promoter. The puro gene is flanked at either end by I-SceI recognition sites in the same orientation, so that NHEJ between the two I-SceI sites results in deletion of the puro gene and activation of GFP.
Clone B3 of the EJ-30 human tumor cell line was used to isolate cell clones that contain a single copy of the pDR-GFP and pEJ5-GFP plasmids stably integrated at interstitial sites or at a telomere on the end of the short arm of chromosome 16 (see Materials and Methods). The telomere on the short arm of chromosome 16 in clone B3 demonstrated sensitivity to DSBs, similar to other telomeres that were analyzed in this cell line [24, 32]. Following transfection of the plasmids, we identified three clones containing the interstitial pDR-GFP plasmid, DR-3, DR-4, and DR-8, and six clones that contain the interstitial pEJ5-GFP plasmid, EJ5-7F, EJ5-AB, EJ5-AD, EJ5-AE, EJ5-AI, and EJ5-AL. The targeting of the pDR-GFP and pEJ5-GFP plasmids to a telomeric site was achieved by inserting telomeric repeat sequences into the plasmids in the proper orientation to seed the formation of a new telomere (see Materials and Methods). The plasmids containing the telomeric repeat sequences, pDR-GFP-tel and pEJ5-GFP-tel, were then transfected into clone B3 to target and replace the pNCT-tel plasmid located at a telomere (Fig. 1). We identified three clones containing the telomeric pDR-GFP-tel plasmid, DR-1, DR-2, and DR-7, and three clones that contain the telomeric pEJ5-GFP-tel plasmid, EJ5-6D, Ej5-6F, and EJ5-6J. Using a similar protocol, we also identified six cell clones that contain a single copy of the pEJ5-GFP-tel plasmid containing telomeric repeat sequences integrated at interstitial sites, EJ5-6B, EJ5-7C, EJ5-7G, EJ5-AC, EJ5-AF, and EJ5-AJ.
We also isolated cell clones containing the plasmid, pGFP-ISceI, in which the GFP gene is active, with the I-SceI site located between the coding sequence and the promoter (Fig. 1). The sequences for the translational start site and start codon for the GFP gene are located 20 bps from the I-SceI site, and therefore the inactivation of GFP requires deletions of at least 20 bps. This construct therefore allowed us to determine the frequency of deletions greater than 20 bps as a result of DSBs at telomeric and interstitial sites. These cell clones were generated from cell clones containing the pEJ5-GFP plasmid following transient expression of I-SceI and selection for GFP-positive colonies. Using this protocol, we identified two clones containing the active GFP gene integrated at a telomeric site, GFP-6D1 and GFP-6D6, at an interstitial site, GFP-7F1 and GFP-7F3, and at an interstitial site containing telomeric repeat sequences, GFP-7C1 and GFP-7C2.
3.2. Comparison of large deletions at telomeric and interstitial sites
Our previous studies demonstrated that DSBs near telomeres were much more likely to result in large deletions and GCRs (Del/GCR) than DSBs at interstitial sites [24, 32]. Specifically, these Del/GCR events involve the complete loss of the integrated plasmid sequences that constitute the subtelomeric region immediately adjacent to the telomere. These previous studies were performed using multiple clones and two different assay systems, both with and without selection. To confirm whether the telomeres containing the GFP gene also showed this increase in large deletions in response to DSBs, we compared the frequency of inactivation of the GFP gene at telomeric and interstitial integration sites using cell clones containing the pGFP-ISceI plasmid. DSBs were generated by infecting the cells with the pQCXIH-ISceI retrovirus, followed by growth in medium containing hygromycin for 10 days to allow time to generate DSBs and allow for turnover of the GFP protein. The results demonstrated a 40% decrease in GFP-positive cells as a result of DSBs near telomeres, while DSBs at interstitial sites were found to result in only a 5% decrease in GFP-positive cells (Fig. 2). These results are consistent with earlier studies showing that Del/GCR events are rare at I-SceI-induced DSBs at interstitial sites [33–35], and that DSBs near telomeres are much more likely to result in Del/GCR events than DSBs at interstitial sites [24, 32]. Interestingly, the clones that contain the pGFP-ISceI plasmid located at interstitial sites with telomeric repeat sequences showed a greater decrease in GFP-positive cells than the clones containing the pGFP-ISceI plasmid located at telomeric sites. However, without further analysis, whether the frequency of Del/GCRs as a result of DSBs near interstitial telomeric repeat sequences is consistently greater then at DSBs near telomeres is not known. Regardless, these results demonstrate that the increase in large deletions resulting from DSBs within subtelomeric regions is due to the presence of the telomeric repeat sequences.
3.3. The frequency of small deletions at subtelomeric and interstitial DSBs
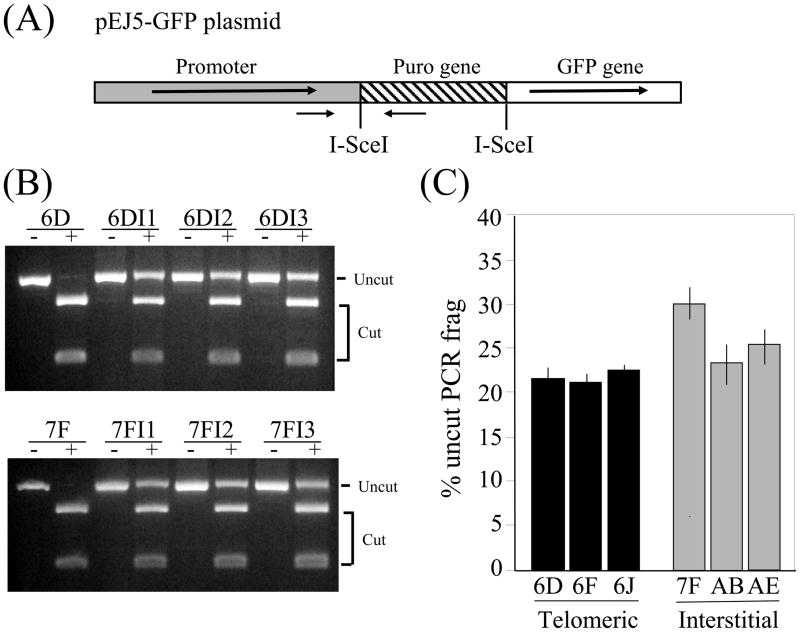
We previously found that unlike the frequency of Del/GCR events, the frequency of small deletions was similar at subtelomeric and interstitial DSBs [24, 32]. To determine whether this is also true for clones containing the pEJ5-GFP plasmid, we compared the frequency of small deletions in clones with interstitial and subtelomeric DSBs. Following infection with the pQCXIH-ISceI retrovirus and selection with hygromycin for 10 days, genomic DNA was isolated and PCR was performed to amplify a DNA fragment spanning one of the two I-SceI sites (Fig. 3A). The percentage of cells in the population that contain small deletions at the I-SceI site was then determined by digesting the PCR fragment with I-SceI. The percentage of the PCR fragment that is not cut by I-SceI is then used to determine the percentage of cells in the population that have small deletions (Fig. 3B). This number must first be corrected for the fraction of cells in the population that have undergone inactivation of the GFP gene (see Fig. 2), because the DNA from these cells would not generate a PCR product, and therefore would cause an overestimation of the fraction of cells containing small deletions. As in our earlier studies, we again saw little if any difference in the frequency of small deletions at the interstitial and subtelomeric DSBs (Fig. 3C). Therefore, the analysis of small deletions at single I-SceI-induced DSBs, which are preferentially generated by A-NHEJ [6, 8, 12], does not indicate a defect in A-NHEJ near telomeres.
Fig. 3.
No difference in small deletions due to DSBs at interstitial and subtelomeric sites. (A) Cell clones with the pEJ5-GFP plasmid integrated at telomeric (EJ5-6D, EJ5-6F, EJ5-6J) or interstitial (EJ5-7F, EJ5-AB, EJ5-AE) sites were used to determine the percentage of cells in the population that contain small deletions resulting from I-SceI-induced DSBs. Genomic DNA from pooled populations of cells infected with the pQCXIH-ISceI retrovirus and selected with hygromycin for 10 days was amplified by PCR using oligonucleotide primers that span one of the I-SceI sites in pEJ5-GFP (small black arrows). (B) The PCR products were then digested with I-SceI endonuclease, and the percentage of cut and uncut DNA was determined from the intensity of the bands using Image J. (C) The percentage of cells in the population containing small deletions was then determined by correcting the fraction of uncut DNA for the number of cells in the population that contained large deletions or GCRs (see Fig. 1), since the DNA from these cells would not amplify and would therefore cause an overestimation of the percentage of cells containing small deletions (see Materials and Methods). Experiments were performed in triplicate and standard deviations are shown.
3.4. Comparison of DSB repair at telomeric and interstitial sites
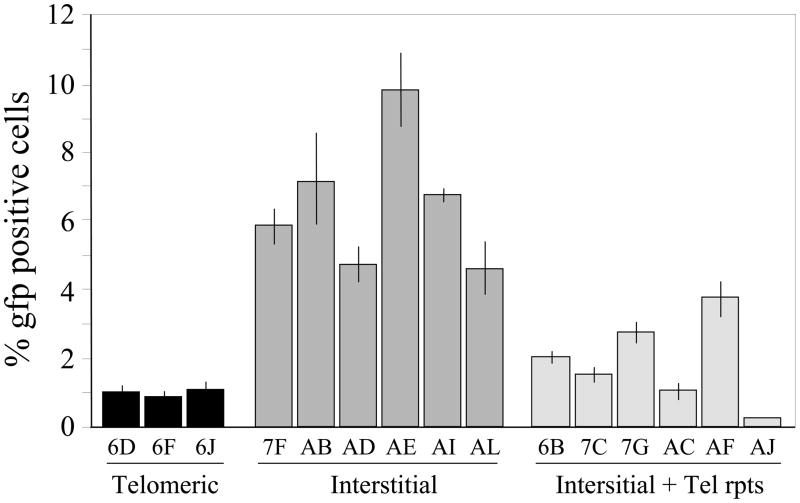
The mechanism responsible for the high frequency of large deletions and GCRs in response to DSBs in subtelomeric regions was investigated by comparing the frequency of repair of I-SceI-induced DSBs at interstitial and telomeric sites. DSBs were generated by infection with the pQCXIH-ISceI retrovirus vector and selection with hygromycin for 10 days. The frequency of DSB repair by NHEJ at interstitial and telomeric sites was analyzed using the clones containing the pEJ5-GFP plasmid (Fig. 4). Only 0.8 to 1.2 % (average 1.0%) of the cells expressing I-SceI were GFP-positive in the clones with subtelomeric DSBs. In contrast, 4.2 to 9.9 % (average 6.8%) of the cells expressing I-SceI were GFP-positive in clones with interstitial DSBs. Therefore, despite the large increase in inactivation of the GFP gene resulting from I-SceI-induced DSBs near telomeres (Fig. 2), the frequency of NHEJ-mediated rejoining of two different I-SceI-induced DSBs is decreased near telomeres. Importantly, the cell clones with interstitial plasmids containing telomeric repeat sequences also showed a much lower frequency of rejoining the two I-SceI-induced DSBs than clones with interstitial DSBs without telomeric repeat sequences, consistent with our observation that DSBs occurring at interstitial sites containing telomeric repeat sequences also have a high frequency of inactivation of the GFP gene compared to interstitial sites without telomeric repeat sequences (see Fig. 2). Despite increase in inactivation of GFP gene, see decrease
Fig. 4.
Decreased NHEJ in repair of DSBs near telomeres and interstitial sites containing telomeric repeat sequences. The appearance of GFP-positive cells following infection with the pQCXIH-ISceI retrovirus and selection with hygromycin for 10 days was used to monitor the frequency of NHEJ in cell clones containing the pEJ5-GFP plasmid at telomeric sites (EJ5-6D, EJ5-6F, EJ5-6J), interstitial sites (EJ5-7F, EJ5-AB, EJ5-AD, EJ5-AE, EJ5-AI, EJ5-AL) or interstitial sites containing telomeric repeat sequences (EJ5-6B, EJ5-7C, EJ5-7G, EJ5-AC, EJ5-AF, EJ5-AJ). Experiments were performed in triplicate and standard deviations are shown.
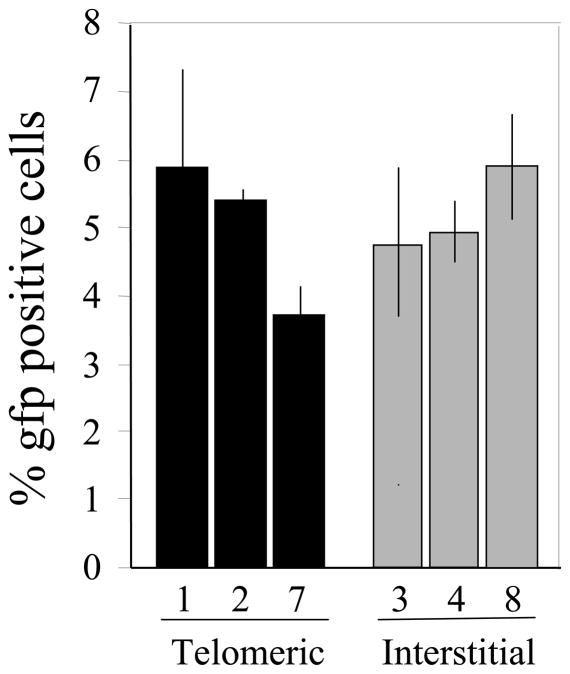
We next compared the frequency of HRR at I-SceI-induced DSBs using the clones containing the DR-GFP plasmid (Fig. 5). The frequency of GFP-positive cells was similar in all of the clones, demonstrating that similar to small deletions, but unlike NHEJ between two different I-SceI sites, there is no difference in the frequency of HRR detected at I-SceI-induced DSBs at interstitial and subtelomeric sites.
Fig. 5.
No difference in HRR in repair of DSBs at interstitial and subtelomeric sites. The appearance of GFP-positive cells following infection with the pQCXIH-ISceI retrovirus and selection with hygromycin for 10 days was used to monitor the frequency of HRR in cell clones containing the pDR-GFP plasmid at telomeric (DR-1, DR-2, DR-7) or interstitial (DR-3, DR-4, DR-8) sites. Experiments were performed in triplicate and standard deviations are shown.
3.5. The frequency of loss of the I-SceI site during DSB repair near telomeres
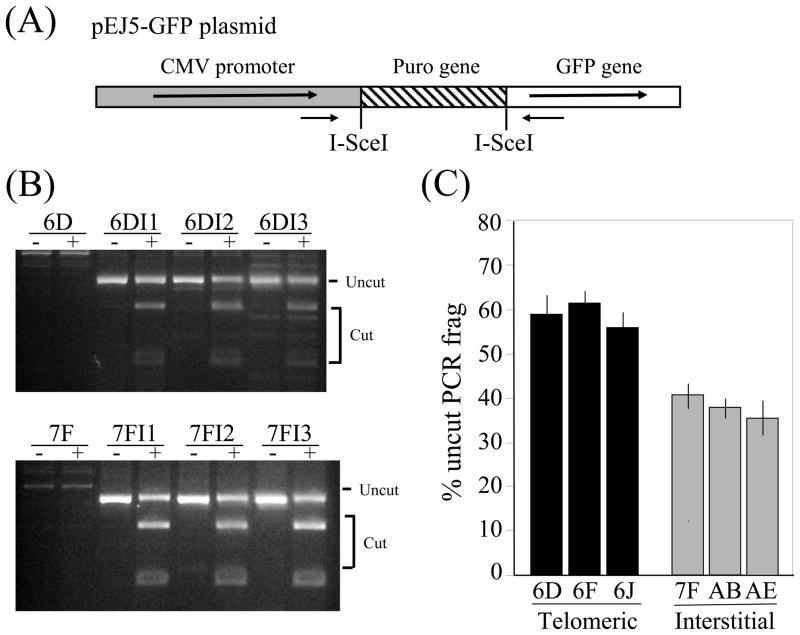
Mammalian cells that are deficient in C-NHEJ have been shown to repair DSBs by A-NHEJ, which is typically associated with large deletions and GCRs at I-SceI-induced DSBs [6, 12, 43, 44], similar to what we have observed as a result of DSBs near telomeres. Several studies have reported that rodent and human cells that are deficient in C-NHEJ are much more likely to experience the loss of the I-SceI site during DSB repair [6, 8, 12, 13, 45]. We therefore compared the relative frequency of the loss of the I-SceI site during NHEJ at interstitial and subtelomeric DSBs to determine whether there was a difference in the likelihood of deletion of the I-SceI site. This analysis was performed using primers distal to the two I-SceI sites, so that only repair events involving the joining of the two I-SceI sites would be amplified (Fig. 6A). Although most cells would not have undergone NHEJ between the two I-SceI sites, the genomic DNA from these cells will not generate a PCR product. Approximately 40% of the PCR product spanning the interstitial DSBs was not cut with I-SceI, i.e., had small deletions when joining the two different I-SceI sites (Figs. 6B and 6C). In contrast, approximately 60% of the PCR product spanning the subtelomeric DSB was not cut with I-SceI. In view of the earlier studies demonstrating an increase in the frequency of loss of the I-SceI site in cells deficient in C-NHEJ [6, 8, 12, 13, 45], this increased frequency of loss of the I-SceI site suggests that the residual NHEJ occurring in subtelomeric regions preferentially involves A-NHEJ.
Fig. 6.
Increased loss of the I-SceI site during NHEJ of DSBs near telomeres. The frequency of loss of the I-SceI site during NHEJ between the two I-SceI sites in the pEJ5-GFP plasmid was compared for cell clones containing interstitial and telomeric integration sites. (A) The pEJ5-GFP plasmid integrated at telomeric (EJ5-6D, EJ5-6F, EJ5-6J) or interstitial (EJ5-7F, EJ5-AB, EJ5-AE) sites were used to determine the relative percentage of loss and restoration of the I-SceI site. Genomic DNA from pooled populations of cells infected with the pQCXIH-ISceI retrovirus and selected with hygromycin for 10 days was amplified by PCR using oligonucleotide primers distal to the two I-SceI sites in pEJ5-GFP (small black arrows). (B) The PCR products were then digested with I-SceI endonuclease. (C) The percentage of cut and uncut DNA was determined from the intensity of the bands using Image J. Experiments were performed in triplicate and standard deviations are shown.
4.0. Discussion
The results presented here demonstrate that the joining of two different I-SceI-induced DSBs by NHEJ is decreased near telomeres compared to interstitial sites in the EJ-30 human tumor cell line. This decrease in NHEJ near telomeres does not appear to be a result of a decrease in the frequency of formation of DSBs by I-SceI at telomeric sites. Although we were not able to determine the frequency of DSBs directly due to the fact that there is only a single DSB that is not present in all of the cells at the same time, the comparison of other types of events served as important controls. We did not observe a difference in the frequency of small deletions (Fig. 3) or HRR (Fig. 5) at subtelomeric and interstitial sites. These observations alone are not conclusive, because we cannot rule out that an increase in small deletions and HRR might compensate for a decrease in the frequency of DSBs near telomeres. However, a decreased frequency of I-SceI-induced DSBs near telomeres would be entirely inconsistent with the increased inactivation of the GFP gene (Fig. 2) and increased frequency of Del/GCR events [24, 32] that we have observed near telomeres. This increased frequency of large deletions and GCRs in response to DSBs in subtelomeric regions is not limited to the EJ-30 human cancer cell line, because the same types of rearrangements i.e., large deletions, GCRs, and chromosome healing, were also observed in the SCC61 human squamous cell carcinoma cell line (unpublished results) and mouse ES cells as a result of DSBs near telomeres [25, 26, 40]. Moreover, the increase in large deletions, GCRs, and chromosome instability in response to DSBs in subtelomeric regions is typical of most telomeres, since it has been observed at multiple telomeres in the EJ-30 human tumor cell line [24, 38] and mouse ES cells [25, 26, 40].
In view of earlier studies demonstrating that a deficiency in C-NHEJ results in an increased frequency of loss of the I-SceI site during DSB repair [6, 8, 12, 13, 45], the increased frequency of loss of the I-SceI site near telomeres compared to interstitial sites suggests that A-NHEJ is the preferred mechanism of NHEJ near telomeres. A-NHEJ has previously been proposed as the mechanism of chromosome fusion following telomere loss in mammalian cells [24, 26, 50, 51]. The predominance of A-NHEJ near telomeres would be consistent with the increase in large deletions, GCRs, and use of microhomology that we have observed as a result of DSBs within subtelomeric regions [24, 26, 32], since these events are all typically associated with A-NHEJ [5–13, 43, 44]. However, it is important to point out that other studies have found that A-NHEJ, not C-NHEJ, is more likely to preserve overhangs generated by restriction enzymes, both in mycobacterium [46] and in an extra-chromosomal system in mammalian cells [47]. Thus, while the restoration of I-SceI sites has been shown to be promoted by Ku and XRCC4, precise rejoining of overhangs can be independent of these factors under some circumstances. In addition, the role of ATM and BRCA1 in the regulation of processing of DSBs by MRE11 is also important in restoration of I-SceI sites during DSB repair [48, 49]. Proof that subtelomeric regions rely predominantly on A-NHEJ will therefore require a more detailed genetic approach. This is an important goal, because the predominance of A-NHEJ for DSB repair near telomeres would have dramatic consequences for chromosome stability due to the increase in Del/GCR events that are associated with A-NHEJ [6, 9, 10, 12, 43, 44].
The deficiency in NHEJ within subtelomeric regions may be a result of the role of telomeric proteins in preventing chromosome fusion. Our results show that the presence of telomeric repeat sequences near interstitial DSBs is sufficient to promote large deletions (Fig. 2) and inhibit NHEJ (Fig. 4). An important function of telomeric proteins is to prevent the telomere from appearing as a DSB and thereby prevent chromosome fusion. TRF2 is a key protein that is involved in telomere maintenance and cap formation [18, 52–54]. TRF2 binds to and inhibits ATM, a key protein in the cellular response to DSBs [55, 56]. Consistent with this observation, a study in yeast demonstrated that the presence of telomeric repeat sequences near DSBs could prevent the initiation of MEC1-mediated cell cycle checkpoints [57]. The inhibition of ATM could be directly involved in the deficiency in C-NHEJ near telomeres, because ATM is required for NHEJ within heterochromatin [14], and subtelomeric regions are composed of heterochromatin [58, 59]. In addition, the role of ATM in limiting MRE11-mediated processing of DSBs is also important in promoting C-NHEJ [48, 49, 60]. Alternatively, the deficiency in NHEJ near telomeres could be a result of the role of TRF2 in regulating DNA resection. Following DNA replication, the leading strand on the end of the chromosome is blunt ended, and therefore must be processed to generate the 3′ single-stranded overhang required for T-loop formation (Fig. 7A). TRF2 has been proposed to facilitate the formation of the 3′ single-stranded overhang by stimulating the 5′-3′ nuclease activity of MRE11 and inhibiting the 3′-5′ nuclease activity of MRE11 [61]. The TPP1 and POT1 proteins then bind to the single strand telomeric DNA and facilitate T-loop formation, preventing further resection of the 5′ strand. Deficiencies in TPP1 or POT1 therefore result in large single-stranded tails due to excessive resection [62, 63]. As a result, chromosome fusions resulting from the loss of TRF2 occur by ATM/53BP1-dependent C-NHEJ, while chromosome fusions resulting from the loss of TPP1, POT1, or naturally shortened telomeres occur by ATM/53BP1-independent A-NHEJ [51]. Importantly, chromosome fusions resulting from loss of TRF2, TPP1, or POT1 are largely eliminated in cells deficient in MRE11 nuclease activity [51, 61]. Based on these results, Rai et al. [51] proposed that TRF2 prevents chromosome fusion by C-NHEJ, and that the loss of telomere function in the presence of TRF2 results in extensive resection/degradation and chromosome fusion by A-NHEJ. As they point out, there are several mechanisms by which TRF2 could inhibit C-NHEJ, one possibility being that TRF2 binds to and inhibits ATM [55], and a second possibility being the role of TRF2 in regulating MRE11 to promote the formation of 3′ overhangs [61], thereby creating single-stranded substrates that are not amenable to C-NHEJ.
Fig. 7.
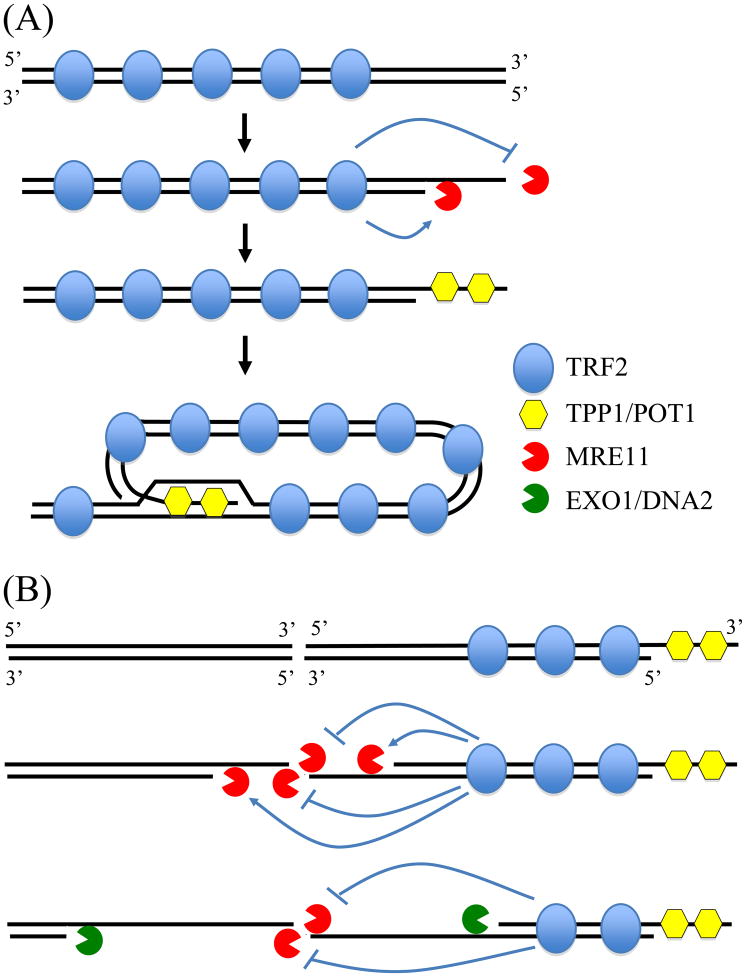
A model to explain the deficiency in C-NHEJ near telomeres. (A) The processing of the leading strand of the telomere following DNA replication is regulated by the telomeric protein TRF2, which inhibits the 3′-5′ nuclease activity of MRE11, and stimulates the 5′-3′ nuclease activity of MRE11. Further resection is prevented by TPP1 and POT1, which bind to the single-stranded telomeric repeat sequences and promote T-loop formation. (B) The processing of DSBs near telomeres is also regulated by TRF2 and involves the resection of the 5′ strand by MRE11 to generate the 3′ single strand overhang. However, because the DNA is not composed of telomeric repeat sequence, TPP1 and POT1 cannot bind and prevent further resection, resulting in large single stranded regions, as are found in cells deficient in TPP1 and POT1 [62]. Further resection then occurs through EXO1 and DNA2, similar to HRR. However, cells that have not undergone DNA replication, and therefore do not contain a template for HRR, cannot repair the break cannot repair the break and experience large deletions, telomere loss, and GCRs.
Unlike the MRE11-mediated resection of DNA ends containing telomeric repeat sequences, which results in the binding of TPP1/POT1 and T-loop formation that limits resection [62, 63], we propose that the resection of DSBs in subtelomeric DNA will continue unabated due to the inability of TPP1/POT1 to bind and form a T-loop (Fig. 7B). Although these long single-stranded 3′ ends would facilitate homologous recombination, this would not be possible for cells experiencing DSBs prior to DNA replication when the sister chromatid is not available for HRR. DSBs in subtelomeric regions would therefore behave like telomeres in TPP1/POT1-deficient cells, which undergo extensive resection [62] that promotes homologous recombination [63] and chromosome fusions involving A-NHEJ [51]. Although earlier studies demonstrated that the inhibition of C-NHEJ by TRF2 was specific for telomeric repeat sequences [64], it is important to point out that these studies were conducted in vitro, and did not involve the processing of DNA ends. As a result, these studies could not rule out the possibility that TRF2 may influence the processing of DSBs within subtelomeric regions as proposed in our model. Similarly, in vitro studies would not rule out the possibility that the inhibition of ATM by TRF2 would prevent the chromatin modification that might be required for repair of DSBs within subtelomeric DNA.
The increased likelihood of large deletions and GCRs due to DSBs within subtelomeric regions has important implications with regard to chromosome instability and cancer. As we have previously proposed, the sensitivity of subtelomeric regions to DSBs is likely to be an important factor in the high frequency of spontaneous telomere loss in cancer cells [65], which leads to many of the types of chromosome rearrangements associated with human cancer [66–68]. Cancer cells often experience a high frequency of DSBs due to oncogene-mediated cell stress, which generates DSBs at regions that are difficult to replicate, called fragile sites [69]. Telomeric regions are fragile sites [70] and therefore would be one of the regions that would experience an increased frequency of DSBs in cancer cells. However, unlike DSBs at most fragile sites, which are repaired, DSBs near telomeres result in telomere loss and chromosome instability. The increased frequency of large deletions and GCRs in response to DSBs in subtelomeric regions may also be important in ionizing radiation-induced carcinogenesis. Ionizing radiation is now known to initiate chromosome instability, which has been proposed to play an important role in radiation-induced carcinogenesis [71]. The region that is sensitive to DSBs extends at least 100 kb from the telomere [24]. Therefore, in view of the fact that there are 96 telomeres in a diploid human cell, the combined size of the sensitive regions (9.6 Mbps), such that one in every 625 DSBs would cause telomere loss and chromosome instability.
Acknowledgments
This work was supported by National Institutes of Health grants CA120205 for J.P.M. and CA120954 for J.M.S.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Jeggo P, Lavin MF. Cellular radiosensitivity: how much better do we understand it? Int J Radiat Biol. 2009;85:1061–1081. doi: 10.3109/09553000903261263. [DOI] [PubMed] [Google Scholar]
- 2.Moynahan ME, Jasin M. Mitotic homologous recombination maintains genomic stability and suppresses tumorigenesis. Nat Rev Mol Cell Biol. 2010;11:196–207. doi: 10.1038/nrm2851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nussenzweig A, Nussenzweig MC. A backup DNA repair pathway moves to the forefront. Cell. 2007;131:223–225. doi: 10.1016/j.cell.2007.10.005. [DOI] [PubMed] [Google Scholar]
- 4.Zha S, Boboila C, Alt FW. Mre11: roles in DNA repair beyond homologous recombination. Nat Struct Mol Biol. 2009;16:798–800. doi: 10.1038/nsmb0809-798. [DOI] [PubMed] [Google Scholar]
- 5.Guirouilh-Barbat J, Huck S, Bertrand P, Pirzio L, Desmaze C, Sabatier L, Lopez BS. Impact of the KU80 pathway on NHEJ-induced genome rearrangements in mammalian cells. Mol Cell. 2004;14:611–623. doi: 10.1016/j.molcel.2004.05.008. [DOI] [PubMed] [Google Scholar]
- 6.Xie A, Kwok A, Scully R. Role of mammalian Mre11 in classical and alternative nonhomologous end joining. Nat Struct Mol Biol. 2009;16:814–818. doi: 10.1038/nsmb.1640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhu C, Mills KD, Fergusion DO, Lee C, Manis J, Fleming J, Gao Y, Morton CC, Alt FW. Unrepaired DNA breaks in p53-deficient cells lead to oncogenic gene amplification subsequent to translocations. Cell. 2002;109:811–821. doi: 10.1016/s0092-8674(02)00770-5. [DOI] [PubMed] [Google Scholar]
- 8.Guirouilh-Barbat J, Rass E, Plo I, Bertrand P, Lopez BS. Defects in XRCC4 and KU80 differentially affect the joining of distal nonhomologous ends. Proc Natl Acad Sci U S A. 2007;104:20902–20907. doi: 10.1073/pnas.0708541104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Weinstock DM, Brunet E, Jasin M. Formation of NHEJ-derived reciprocal chromosomal translocations does not require Ku70. Nat Cell Biol. 2007;9:978–981. doi: 10.1038/ncb1624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Simsek D, Jasin M. Alternative end-joining is suppressed by the canonical NHEJ component Xrcc4-ligase IV during chromosomal translocation formation. Nat Struct Mol Biol. 2010;17:410–416. doi: 10.1038/nsmb.1773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yan CT, Boboila C, Souza EK, Franco S, Hickernell TR, Murphy M, Gumaste S, Geyer M, Zarrin AA, Manis JP, Rajewsky K, Alt FW. IgH class switching and translocations use a robust non-classical end-joining pathway. Nature. 2007;449:478–482. doi: 10.1038/nature06020. [DOI] [PubMed] [Google Scholar]
- 12.Bennardo N, Cheng A, Huang N, Stark JM. Alternative-NHEJ is a mechanistically distinct pathway of mammalian chromosome break repair. PLoS Genet. 2008;4 :e1000110. doi: 10.1371/journal.pgen.1000110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rass E, Grabarz A, Plo I, Gautier J, Bertrand P, Lopez BS. Role of Mre11 in chromosomal nonhomologous end joining in mammalian cells. Nat Struct Mol Biol. 2009;16:819–824. doi: 10.1038/nsmb.1641. [DOI] [PubMed] [Google Scholar]
- 14.Goodarzi AA, Noon AT, Deckbar D, Ziv Y, Shiloh Y, Lobrich M, Jeggo PA. ATM signaling facilitates repair of DNA double-strand breaks associated with heterochromatin. Mol Cell. 2008;31:167–177. doi: 10.1016/j.molcel.2008.05.017. [DOI] [PubMed] [Google Scholar]
- 15.Kato TA, Okayasu R, Bedford JS. Comparison of the induction and disappearance of DNA double strand breaks and gamma-H2AX foci after irradiation of chromosomes in G1-phase or in condensed metaphase cells. Mutat Res. 2008;639:108–112. doi: 10.1016/j.mrfmmm.2007.11.006. [DOI] [PubMed] [Google Scholar]
- 16.Goodarzi AA, Noon AT, Jeggo PA. The impact of heterochromatin on DSB repair. Biochem Soc Trans. 2009;37:569–576. doi: 10.1042/BST0370569. [DOI] [PubMed] [Google Scholar]
- 17.Chan SR, Blackburn EH. Telomeres and telomerase. Philos Trans R Soc Lond B Biol Sci. 2004;359:109–121. doi: 10.1098/rstb.2003.1370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Palm W, de Lange T. How shelterin protects mammalian telomeres. Annu Rev Genet. 2008;42:301–334. doi: 10.1146/annurev.genet.41.110306.130350. [DOI] [PubMed] [Google Scholar]
- 19.De Lange T. Telomere-related genome instability in cancer. Cold Spring Harb Symp Quant Biol. 2005;70:197–204. doi: 10.1101/sqb.2005.70.032. [DOI] [PubMed] [Google Scholar]
- 20.d’Adda di Fagagna FdAd, Reaper PM, Clay-Farrace L, Fiegler H, Carr P, von Zglinicki T, Saretzki G, Carter NP, Jackson SP. A DNA damage checkpoint response in telomere-initiated senescence. Nature. 2003;426:194–198. doi: 10.1038/nature02118. [DOI] [PubMed] [Google Scholar]
- 21.Nakamura AJ, Chiang YJ, Hathcock KS, Horikawa I, Sedelnikova OA, Hodes RJ, Bonner WM. Both telomeric and non-telomeric DNA damage are determinants of mammalian cellular senescence. Epigenetics Chromatin. 2008;1:6. doi: 10.1186/1756-8935-1-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Counter CM, Avilion AA, LeFeuvre CE, Stewart NG, Greider CW, Harley CB, Bacchetti S. Telomere shortening associated with chromosome instability is arrested in immortal cells which express telomerase activity. EMBO J. 1992;11:1921–1929. doi: 10.1002/j.1460-2075.1992.tb05245.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ricchetti M, Dujon B, Fairhead C. Distance from the chromosome end determines the efficiency of double-strand break repair in subtelomeres of haploid yeast. J Mol Biol. 2003;328:847–862. doi: 10.1016/s0022-2836(03)00315-2. [DOI] [PubMed] [Google Scholar]
- 24.Zschenker O, Kulkarni A, Miller D, Reynolds GE, Granger-Locatelli M, Pottier G, Sabatier L, Murnane JP. Increased sensitivity of subtelomeric regions to DNA double-strand breaks in a human tumor cell line. DNA Repair. 2009;8:886–900. doi: 10.1016/j.dnarep.2009.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gao Q, Reynolds GE, Wilcox A, Miller D, Cheung P, Artandi SE, Murnane JP. Telomerase-dependent and -independent chromosome healing in mouse embryonic stem cells. DNA Repair. 2008;7:1233–1249. doi: 10.1016/j.dnarep.2008.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lo AWI, Sprung CN, Fouladi B, Pedram M, Sabatier L, Ricoul M, Reynolds GE, Murnane JP. Chromosome instability as a result of double-strand breaks near telomeres in mouse embryonic stem cells. Mol Cell Biol. 2002;22:4836–4850. doi: 10.1128/MCB.22.13.4836-4850.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gisselsson D, Jonson T, Petersen A, Strombeck B, Dal Cin P, Hoglund M, Mitelman F, Mertens F, Mandahl N. Telomere dysfunction triggers extensive DNA fragmentation and evolution of complex chromosome abnormalities in human malignant tumors. Proc Natl Acad Sci USA. 2001;98:12683–12688. doi: 10.1073/pnas.211357798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nakamura AJ, Redon CE, Bonner WM, Sedelnikova OA. Telomere-dependent and telomere-independent origins of endogenous DNA damage in tumor cells. Aging (Albany NY) 2009;1:212–218. doi: 10.18632/aging.100019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Murnane JP. Telomeres and chromosome instability. DNA Repair (Amst) 2006;5:1082–1092. doi: 10.1016/j.dnarep.2006.05.030. [DOI] [PubMed] [Google Scholar]
- 30.Sabatier L, Ricoul M, Pottier G, Murnane JP. The loss of a single telomere can result in genomic instability involving multiple chromosomes in a human tumor cell line. Mol Cancer Res. 2005;3:139–150. doi: 10.1158/1541-7786.MCR-04-0194. [DOI] [PubMed] [Google Scholar]
- 31.Artandi SE, Chang S, Lee S-L, Alson S, Gottlieb GJ, Chin L, DePinho RA. Telomere dysfunction promotes non-reciprocal translocations and epithelial cancers in mice. Nature. 2000;406:641–645. doi: 10.1038/35020592. [DOI] [PubMed] [Google Scholar]
- 32.Kulkarni A, Zschenker O, Reynolds G, Miller D, Murnane JP. The effect of telomere proximity on telomere position effect, chromosome healing and sensitivity to DNA double-strand breaks in a human tumor cell line. Mol Cell Biol. 2010 doi: 10.1128/MCB.01137-09. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Honma M, Sakuraba M, Koizumi T, Takashima Y, Sakamoto H, Hayashi M. Non-homologous end-joining for repairing I-SceI-induced DNA double strand breaks in human cells. DNA Repair (Amst) 2007;6:781–788. doi: 10.1016/j.dnarep.2007.01.004. [DOI] [PubMed] [Google Scholar]
- 34.Varga T, Aplan PD. Chromosomal aberrations induced by double strand DNA breaks. DNA Repair (Amst) 2005;4:1038–1046. doi: 10.1016/j.dnarep.2005.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rebuzzini P, Khoriauli L, Azzalin CM, Magnani E, Mondello C, Giulotto E. New mammalian cellular systems to study mutations introduced at the break site by non-homologous end-joining. DNA Repair (Amst) 2005;4:546–555. doi: 10.1016/j.dnarep.2004.12.011. [DOI] [PubMed] [Google Scholar]
- 36.Richardson C, Jasin M. Frequent chromosomal translocations induced by DNA double-strand breaks. Nature. 2000;405:697–700. doi: 10.1038/35015097. [DOI] [PubMed] [Google Scholar]
- 37.Richardson C, Moynahan ME, Jasin M. Double-strand break repair by interchromosomal recombination: suppression of chromosomal translocations. Genes Dev. 1998;12:3831–3842. doi: 10.1101/gad.12.24.3831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fouladi B, Miller D, Sabatier L, Murnane JP. The relationship between spontaneous telomere loss and chromosome instability in a human tumor cell line. Neoplasia. 2000;2:540–554. doi: 10.1038/sj.neo.7900107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Moynahan ME, Cui TY, Jasin M. Homology-directed dna repair, mitomycin-c resistance, and chromosome stability is restored with correction of a Brca1 mutation. Cancer Res. 2001;61:4842–4850. [PubMed] [Google Scholar]
- 40.Sprung CN, Reynolds GE, Jasin M, Murnane JP. Chromosome healing in mouse embryonic stem cells. Proc Natl Acad Sci USA. 1999;96:6781–6786. doi: 10.1073/pnas.96.12.6781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.O’Toole CM, Povey S, Hepburn P, Franks LM. Identity of some human bladder cancer cell lines. Nature. 1983;301:429–430. doi: 10.1038/301429a0. [DOI] [PubMed] [Google Scholar]
- 42.Lo AWI, Sabatier L, Fouladi B, Pottier G, Ricoul M, Murnane JP. DNA amplification by breakage/fusion/bridge cycles initiated by spontaneous telomere loss in a human cancer cell line. Neoplasia. 2002;6:531–538. doi: 10.1038/sj.neo.7900267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Richardson C, Jasin M. Coupled homologous and nonhomologous repair of a double-strand break preserves genomic integrity in mammalian cells. Mol Cell Biol. 2000;20:9068–9075. doi: 10.1128/mcb.20.23.9068-9075.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Johnson RD, Jasin M. Double-strand-break-induced homologous recombination in mammalian cells. Biochem Soc Trans. 2001;29:196–201. doi: 10.1042/0300-5127:0290196. [DOI] [PubMed] [Google Scholar]
- 45.So S, Adachi N, Lieber MR, Koyama H. Genetic interactions between BLM and DNA ligase IV in human cells. J Biol Chem. 2004;279:55433–55442. doi: 10.1074/jbc.M409827200. [DOI] [PubMed] [Google Scholar]
- 46.Aniukwu J, Glickman MS, Shuman S. The pathways and outcomes of mycobacterial NHEJ depend on the structure of the broken DNA ends. Genes Dev. 2008;22:512–527. doi: 10.1101/gad.1631908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Fattah F, Lee EH, Weisensel N, Wang Y, Lichter N, Hendrickson EA. Ku regulates the non-homologous end joining pathway choice of DNA double-strand break repair in human somatic cells. PLoS Genet. 2010;6:e1000855. doi: 10.1371/journal.pgen.1000855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bennardo N, Stark JM. ATM limits incorrect end utilization during non-homologous end joining of multiple chromosome breaks. PLoS Genet. 2010;6:e1001194. doi: 10.1371/journal.pgen.1001194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wang HC, Chou WC, Shieh SY, Shen CY. Ataxia telangiectasia mutated and checkpoint kinase 2 regulate BRCA1 to promote the fidelity of DNA end-joining. Cancer Res. 2006;66:1391–1400. doi: 10.1158/0008-5472.CAN-05-3270. [DOI] [PubMed] [Google Scholar]
- 50.Capper R, Britt-Compton B, Tankimanova M, Rowson J, Letsolo B, Man S, Haughton M, Baird DM. The nature of telomere fusion and a definition of the critical telomere length in human cells. Genes Dev. 2007;21:2495–2508. doi: 10.1101/gad.439107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rai R, Zheng H, He H, Luo Y, Multani A, Carpenter PB, Chang S. The function of classical and alternative non-homologous end-joining pathways in the fusion of dysfunctional telomeres. Embo J. 2010;29:2598–2610. doi: 10.1038/emboj.2010.142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Denchi EL, de Lange T. Protection of telomeres through independent control of ATM and ATR by TRF2 and POT1. Nature. 2007;448:1068–1071. doi: 10.1038/nature06065. [DOI] [PubMed] [Google Scholar]
- 53.Smogorzewska A, van Steensel B, Bianchi A, Oelmann S, Schaefer MR, Schnapp G, de Lange T. Control of human telomere length by TRF1 and TRF2. Mol Cell Biol. 2000;20:1659–1668. doi: 10.1128/mcb.20.5.1659-1668.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.van Steensel B, Smogorzewska A, de Lange T. TRF2 protects human telomeres from end-to-end fusions. Cell. 1998;92:401–413. doi: 10.1016/s0092-8674(00)80932-0. [DOI] [PubMed] [Google Scholar]
- 55.Karlseder J, Hoke K, Mirzoeva OK, Bakkenist C, Kastan MB, Petrini JH, de Lange T. The telomeric protein TRF2 binds the ATM kinase and can inhibit the ATM-dependent DNA damage response. PLoS Biol. 2004;2:E240. doi: 10.1371/journal.pbio.0020240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zhu X-D, Kuster B, Mann M, Petrini JHJ, de Lange T. Cell-cycle-regulated association of RAD50/MRE11/NBS1 with TRF2 and human telomeres. Nat Genet. 2000;25:347–352. doi: 10.1038/77139. [DOI] [PubMed] [Google Scholar]
- 57.Michelson RJ, Rosenstein S, Weinert T. A telomeric repeat sequence adjacent to a DNA double-stranded break produces an anticheckpoint. Genes Dev. 2005;19:2546–2559. doi: 10.1101/gad.1293805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Garcia-Cao M, O’Sullivan R, Peters AH, Jenuwein T, Blasco MA. Epigenetic regulation of telomere length in mammalian cells by the Suv39h1 and Suv39h2 histone methyltransferases. Nat Genet. 2004;36:94–99. doi: 10.1038/ng1278. [DOI] [PubMed] [Google Scholar]
- 59.Gonzalo S, Jaco I, Fraga MF, Chen T, Li E, Esteller M, Blasco MA. DNA methyltransferases control telomere length and telomere recombination in mammalian cells. Nat Cell Biol. 2006;8:416–424. doi: 10.1038/ncb1386. [DOI] [PubMed] [Google Scholar]
- 60.Zha S, Guo C, Boboila C, Oksenych V, Cheng HL, Zhang Y, Wesemann DR, Yuen G, Patel H, Goff PH, Dubois RL, Alt FW. ATM damage response and XLF repair factor are functionally redundant in joining DNA breaks. Nature. 2010 doi: 10.1038/nature09604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Deng Y, Guo X, Ferguson DO, Chang S. Multiple roles for MRE11 at uncapped telomeres. Nature. 2009;460:914–918. doi: 10.1038/nature08196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Palm W, Hockemeyer D, Kibe T, de Lange T. Functional dissection of human and mouse POT1 proteins. Mol Cell Biol. 2009;29:471–482. doi: 10.1128/MCB.01352-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wu L, Multani AS, He H, Cosme-Blanco W, Deng Y, Deng JM, Bachilo O, Pathak S, Tahara H, Bailey SM, Behringer RR, Chang S. Pot1 deficiency initiates DNA damage checkpoint activation and aberrant homologous recombination at telomeres. Cell. 2006;126:49–62. doi: 10.1016/j.cell.2006.05.037. [DOI] [PubMed] [Google Scholar]
- 64.Bae NS, Baumann P. A RAP1/TRF2 Complex Inhibits Nonhomologous End-Joining at Human Telomeric DNA Ends. Mol Cell. 2007;26:323–334. doi: 10.1016/j.molcel.2007.03.023. [DOI] [PubMed] [Google Scholar]
- 65.Murnane JP. Telomere loss as a mechanism for chromosomal instability in human cancer. Cancer Res. 2010;70:4255–4259. doi: 10.1158/0008-5472.CAN-09-4357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Brodeur GM, Hogarety MD. Gene amplification in human cancers: biological and clinical significance. In: Vogelstein B, Kinzler KW, editors. The genetic basis of human cancer. McGraw-Hill; New York: 1998. pp. 161–172. [Google Scholar]
- 67.Gollin SM. Chromosomal alterations in squamous cell carcinomas of the head and neck: window to the biology of disease. Head Neck. 2001;23:238–253. doi: 10.1002/1097-0347(200103)23:3<238::aid-hed1025>3.0.co;2-h. [DOI] [PubMed] [Google Scholar]
- 68.Schwab M. Oncogene amplification in solid tumors. Sem Cancer Biol. 1999;9:319–325. doi: 10.1006/scbi.1999.0126. [DOI] [PubMed] [Google Scholar]
- 69.Tsantoulis PK, Kotsinas A, Sfikakis PP, Evangelou K, Sideridou M, Levy B, Mo L, Kittas C, Wu XR, Papavassiliou AG, Gorgoulis VG. Oncogene-induced replication stress preferentially targets common fragile sites in preneoplastic lesions. A genome-wide study. Oncogene. 2008;27:3256–3264. doi: 10.1038/sj.onc.1210989. [DOI] [PubMed] [Google Scholar]
- 70.Sfeir A, Kosiyatrakul ST, Hockemeyer D, MacRae SL, Karlseder J, Schildkraut CL, de Lange T. Mammalian telomeres resemble fragile sites and require TRF1 for efficient replication. Cell. 2009;138:90–103. doi: 10.1016/j.cell.2009.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Huang L, Snyder AR, Morgan WF. Radiation-induced genomic instability and its implications for radiation carcinogenesis. Oncogene. 2003;22:5848–5854. doi: 10.1038/sj.onc.1206697. [DOI] [PubMed] [Google Scholar]