Abstract
Laboratory evolution has generated many biomolecules with desired properties, but a single round of mutation, gene expression, screening or selection, and replication typically requires days or longer with frequent human intervention.1 Since evolutionary success is dependent on the total number of rounds performed,2 a means of performing laboratory evolution continuously and rapidly could dramatically enhance its effectiveness.3 While researchers have accelerated individual steps in the evolutionary cycle,4–9 the only previous example of continuous directed evolution was the landmark study of Joyce,10 who continuously evolved RNA ligase ribozymes with an in vitro replication cycle that unfortunately cannot be easily adapted to other biomolecules. Here we describe a system that enables the continuous directed evolution of gene-encoded molecules that can be linked to protein production in E. coli. During phage-assisted continuous evolution (PACE), evolving genes are transferred from host cell to host cell through a modified bacteriophage life cycle in a manner that is dependent on the activity of interest. Dozens of rounds of evolution can occur in a single day of PACE without human intervention. Using PACE, we evolved T7 RNA polymerases that recognize a distinct promoter, initiate transcripts with A instead of G, and initiate transcripts with C. In one example, PACE executed 200 rounds of protein evolution over the course of eight days. Starting from undetectable activity levels in two of these cases, enzymes with each of the three target activities emerged in less than one week of PACE. In all three cases, PACE-evolved polymerase activities exceeded or were comparable to that of the wild-type T7 RNAP on its wild-type promoter, representing improvements of up to several hundred-fold. By greatly accelerating laboratory evolution, PACE may provide solutions to otherwise intractable directed evolution problems and address novel questions about molecular evolution.
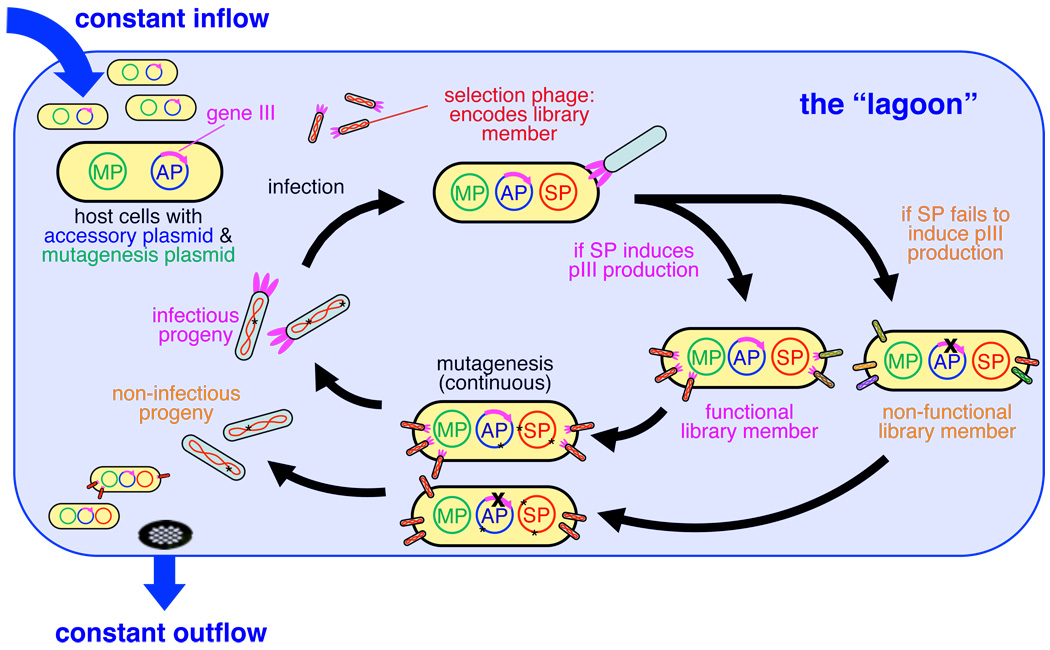
We devised a system that exploits the continuous culture and selection of the M13 filamentous bacteriophage11 (commonly used in phage display12) to enable the continuous directed evolution of proteins or nucleic acids. In phage-assisted continuous evolution (PACE), E. coli host cells continuously flow through a fixed-volume vessel (the “lagoon”) containing a replicating population of phage DNA vectors (“selection phage”, SP) encoding the gene(s) of interest (Supplementary Fig. 1).
The average residence time of host cells in the lagoon is less than the time required for E. coli replication. As a result, mutations accumulate only in the evolving SP population, the only DNA that can replicate faster than the rate of lagoon dilution. The mutation of host cells in the lagoon should therefore have minimal impact on the outcome of the selection over many rounds of phage replication, and mutagenesis conditions are not limited to those that preserve E. coli viability.
PACE achieves continuous selection by linking the desired activity to the production of infectious progeny phage containing the evolving gene(s). Phage infection requires protein III (pIII; encoded by gene III), which mediates F pilus binding and host cell entry.13 Phage lacking pIII are ~108-fold less infectious than wild-type phage.14 Crucially, the production of infectious phage scales with increasing levels of pIII over concentrations spanning two orders of magnitude.15
To couple pIII production to the activity of interest, we deleted gene III from the phage vector and inserted it into an “accessory plasmid” (AP) present in the E. coli host cells (see Supplementary Fig. 2 for plasmid maps). The production of pIII from the AP is dependent on the activity of the evolving gene(s) on the SP. Only phage vectors able to induce sufficient pIII production from the AP will propagate and persist in the lagoon (Fig. 1). Because pIII expression level determines the rate of infectious phage production,15 phage encoding genes that result in a higher level of pIII production will infect more host cells than phage encoding less active genes.
Figure 1.
Overview of the PACE system. PACE in a single lagoon. Host cells continuously flow through a lagoon, where they are infected with selection phage (SP) encoding library members. Functional library members induce production of pIII from the accessory plasmid (AP) and release progeny capable of infecting new host cells, while non-functional library members do not. Increased mutagenesis is triggered through induction of the mutagenesis plasmid (MP). Host cells flow out of the lagoon on average faster than they can replicate, confining the accumulation of mutations to replicating phage.
Due to the speed of the phage life cycle (progeny phage production begins ~10 minutes post-infection),16 PACE can mediate many generations of selective phage replication in a single day. We observed activity-dependent phage vectors that tolerate lagoon flow rates up to 3.2 volumes per hour (Supplementary Fig. 3), corresponding to ~115 population doublings and an average of ~38 phage generations per 24 hours (see the Supplementary Information for an analysis). More conservative flow rates of 2.0–2.5 volumes per hour allow 24–30 generations per day and reduce the risk of complete phage loss (washout) during selections. Multiple lagoons can evolve genes in parallel, with each 100 mL lagoon containing ~5×1010 host cells selectively replicating active phage variants. Importantly, PACE requires no intervention during evolution and obviates the need to create DNA libraries, transform cells, extract genes, or perform DNA cloning steps during each round.
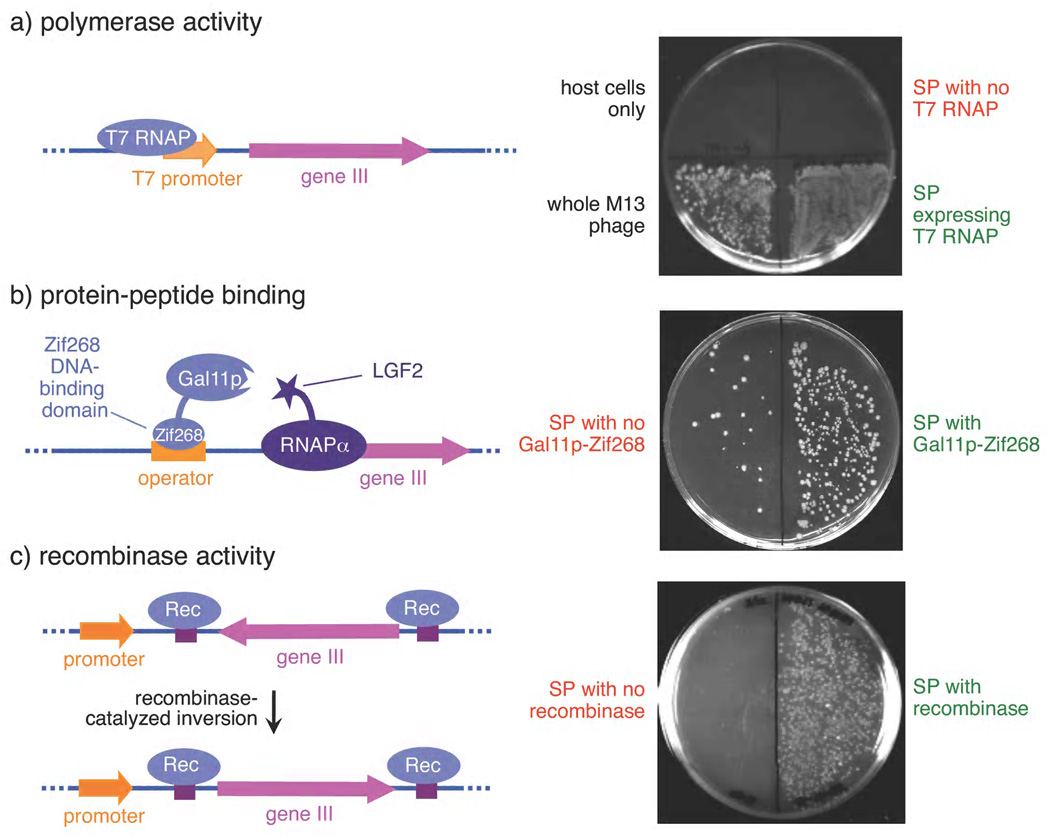
In principle, PACE is capable of evolving any gene that can be linked to pIII production in E. coli. Because a wide variety of functions including DNA binding, RNA binding, protein binding, bond-forming catalysis, and a variety of enzyme activities have been linked to the expression of a reporter protein,17,18 PACE can be applied to the evolution of many different activities of interest. As examples, we successfully linked protein-protein binding, recombinase activity, and RNA polymerase activity to phage infectivity in discrete infection assays by creating variants of the AP that associate each of these activities with pIII production (Fig. 2).
Figure 2.
Linkage of three protein activities to pIII production and phage infectivity using three distinct APs. E. coli cells containing APs encoding conditionally expressed gene III (left) and selection phage were combined with recipient cells. Phage production resulted in colonies with antibiotic resistances conferred by the phage and the recipient cells (right). See Methods for details. (a) RNA polymerase activity leads to gene III expression and infection comparable to wild-type phage, while SP lacking T7 RNAP do not infect. (b) Protein-protein interaction between a Gal11p domain tethered to a Zif268 DNA-binding domain and an LGF2a domain fused to RNA polymerase leads to gene III expression and infection. (c) Recombinase-catalysed gene inversion induces gene III expression and infection.
PACE applies optimal evolutionary pressure when pIII levels are above the minimal threshold required to prevent phage washout, but below the amount needed to maximize infectious phage production. This window can be shifted by varying the copy number of the AP, or by altering the ribosome-binding site (RBS) sequence of gene III to modulate the efficiency with which gene III is transcribed or translated (Supplementary Fig. 4).
We constructed an arabinose-inducible mutagenesis plasmid (MP) that elevates the error rate during DNA replication in the lagoon by suppressing proofreading19 and enhancing error-prone lesion bypass (Supplementary Information).20 Full induction increased the observed mutagenesis rate by ~100-fold, inducing all possible transitions and transversions (Supplementary Fig. 5). This enhanced mutation rate is sufficient to sample all possible single and double mutants of a given sequence each generation (Supplementary Information), in principle enabling single-mutation fitness valleys to be traversed during PACE.
Bacteriophage T7 RNA polymerase (T7 RNAP) is widely used to transcribe RNA in vitro and in cells. T7 RNAP is highly specific for its promoter sequence (TAATACGACTCACTATA), and exhibits virtually no detectable activity on the consensus promoter of the related bacteriophage T3 (AATTAACCCTCACTAAA, differences underlined).21,22 Despite decades of study and several attempts to engineer the specificity of T7 RNAP towards other promoters22,23 including that of T3, a mutant T7 RNAP capable of recognizing the T3 promoter has not been previously reported.
To remove potential interference from evolutionary improvements to the phage vector rather than to T7 RNAP, we propagated an SP expressing wild-type T7 RNAP for three days on host cells containing an AP with the wild-type T7 promoter driving gIII expression. A single plaque presumed to represent vector-optimized SP contained a single mutation (P314T) in T7 RNAP. We confirmed that the activity of the P314T mutant does not significantly differ from that of wild-type T7 RNAP (Supplementary Fig. 6).
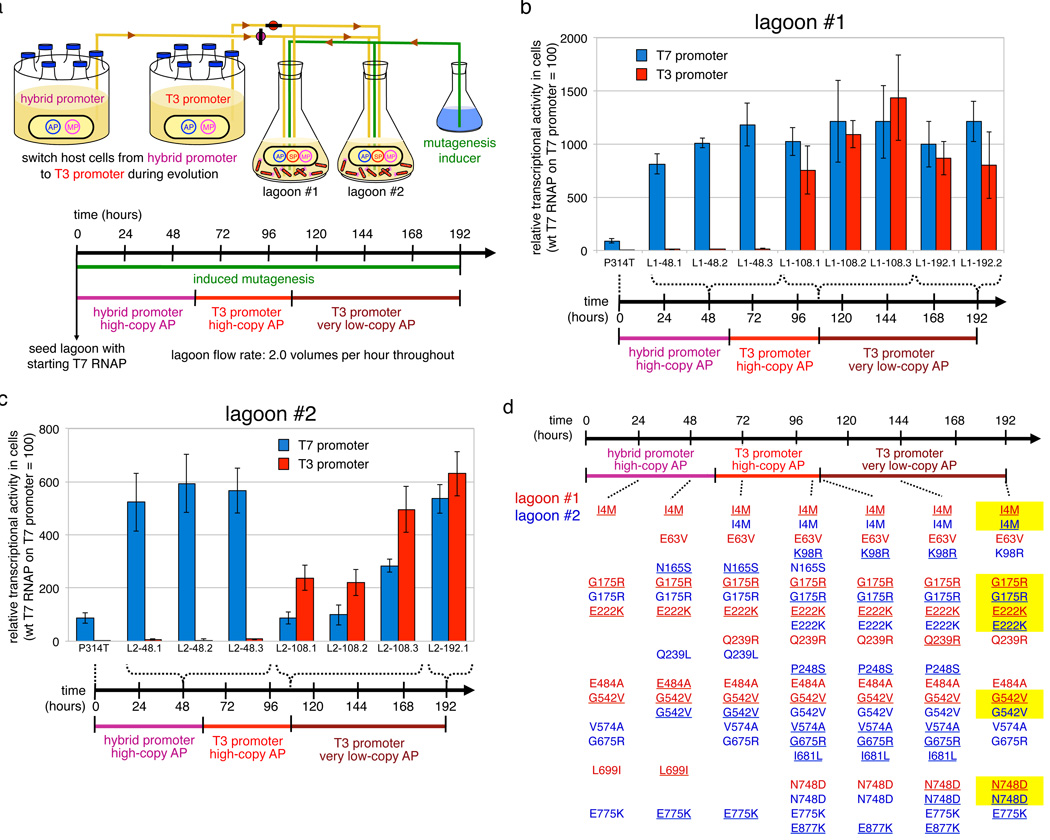
This starting SP failed to propagate on host cells containing the T3 promoter AP. We therefore propagated the phage on cells containing a hybrid T7/T3 promoter AP with the T7 promoter base at the important -11 position21 but all other positions changed to their T3 counterparts. Two initially identical lagoons were evolved in parallel on the hybrid promoter AP for 60 hours, then on the complete T3 promoter AP for 48 hours, and finally on a high-stringency, very low-copy T3 promoter AP for 84 hours (Fig. 3a).
Figure 3.
Continuous evolution of T7 RNAP variants that recognize the T3 promoter. (a) PACE schedule. (b) Activity in cells of T7 RNAP variants isolated from lagoon 1 at 48, 108, and 192 hours on the T7 and T3 promoters. Transcriptional activity was measured spectrophotometrically by subcloning the protein-encoding regions of the T7 RNAP genes into a construct in which the T7 or T3 promoter drives lacZ expression. (c) Activity in cells of T7 RNAP variants isolated from lagoon 2. Error bars in (b) and (c) represent the standard deviation of at least three independent assays. (d) Mutations identified in T7 RNAP clones from lagoons 1 and 2 are shown in red and blue, respectively. Underlined mutations were predominant in that lagoon based on whole-pool sequencing of lagoon aliquots. Mutations highlighted in yellow independently evolved to predominance in both lagoons.
In both lagoons phage persisted after 8 days of PACE, surviving a net dilution of 10167 fold, the equivalent of 555 phage population doublings and ~200 rounds of evolution by the average phage. We isolated, sequenced, and characterized phage vectors from each lagoon after 48, 108, and 192 hours, observing up to eight, ten, and 11 non-silent mutations in single T7 RNAP genes at each time point.
Protein-encoding regions (without upstream promoter sequences) of evolved mutant T7 RNAP genes were subcloned into assay plasmids that quantitatively link transcriptional activity to beta-galactosidase expression in cells.24 We defined the activity of wild-type T7 RNAP on the T7 promoter to be 100%. The starting T7 RNAP exhibited undetectable (< 3%) levels of activity on the T3 promoter in these cell-based assays. The assayed mutants exhibited > 200% activity after 108 hours of PACE, and > 600% activity following high-stringency PACE at 192 hours, improvements of more than 200-fold (Fig. 3b and 3c). These results collectively establish the ability of PACE to very rapidly evolve large changes in enzyme activity and specificity with minimal intervention by the researcher.
Several evolved T7 RNAP mutants were also purified and assayed in vitro using radioactive nucleotide incorporation assays. Purified T7 RNAP mutants exhibited activity levels on the T3 promoter in vitro exceeding that of wild-type T7 RNAP on the T7 promoter, representing improvements of up to 89-fold compared with the starting enzyme (Supplementary Fig. 7), These results indicate that PACE resulted in large improvements in substrate binding or catalytic rate. Evolved activity improvements were higher in cells than in vitro, suggesting that these enzymes also evolved improvements in features such as expression level, polymerase folding, or stability that are specific to the context of the cytoplasm.
Interestingly, the evolutionary dynamics of the two initially identical lagoons differed significantly (Fig. 3d and Supplementary Results). Within 24 hours, lagoon 1 acquired a predominant suite of mutations consisting of I4M, G175R, E222K, and G542V and changed little thereafter beyond acquiring N748D, a mutation known to enable recognition of the T3 base at the -11 position,21 following exposure to the full T3 promoter. In contrast, lagoon 2 accessed these mutations more slowly before a different suite of mutations also including N748D became predominant at 108 hours, only to be displaced by the same suite of mutations observed in lagoon 1. The presence of several mutations unique to lagoon 2 throughout the experiment suggests that lagoon cross-contamination did not occur. The distinct evolutionary trajectories of the two lagoons prior to their ultimate convergence upon a common set of mutations highlight the ability to PACE to rapidly discover multiple viable pathways to a target activity in parallel experiments. This capability may enable a more in-depth experimental study of protein evolutionary dynamics than can be achieved with conventional directed evolution methods that cannot complete so many rounds of evolution on a practical time scale.
T7 RNAP is highly specific for initiation with GTP,25,26 significantly limiting its usefulness for the in vitro transcription of RNAs that begin with other nucleotides. As initiation has been described as a mechanistically challenging step in transcription,27 we next used PACE to evolve T7 RNAP variants capable of initiating transcription with other nucleotides in a template-directed manner. T7 RNAP is known to preferentially initiate with GTP up to several bases downstream of the +1 position if the template is devoid of early guanines in the coding strand.26 We therefore constructed accessory plasmids in which positions +1 through +6 of the gene III transcript were AAAAAA (iA6) or CCCCCC (iC6).
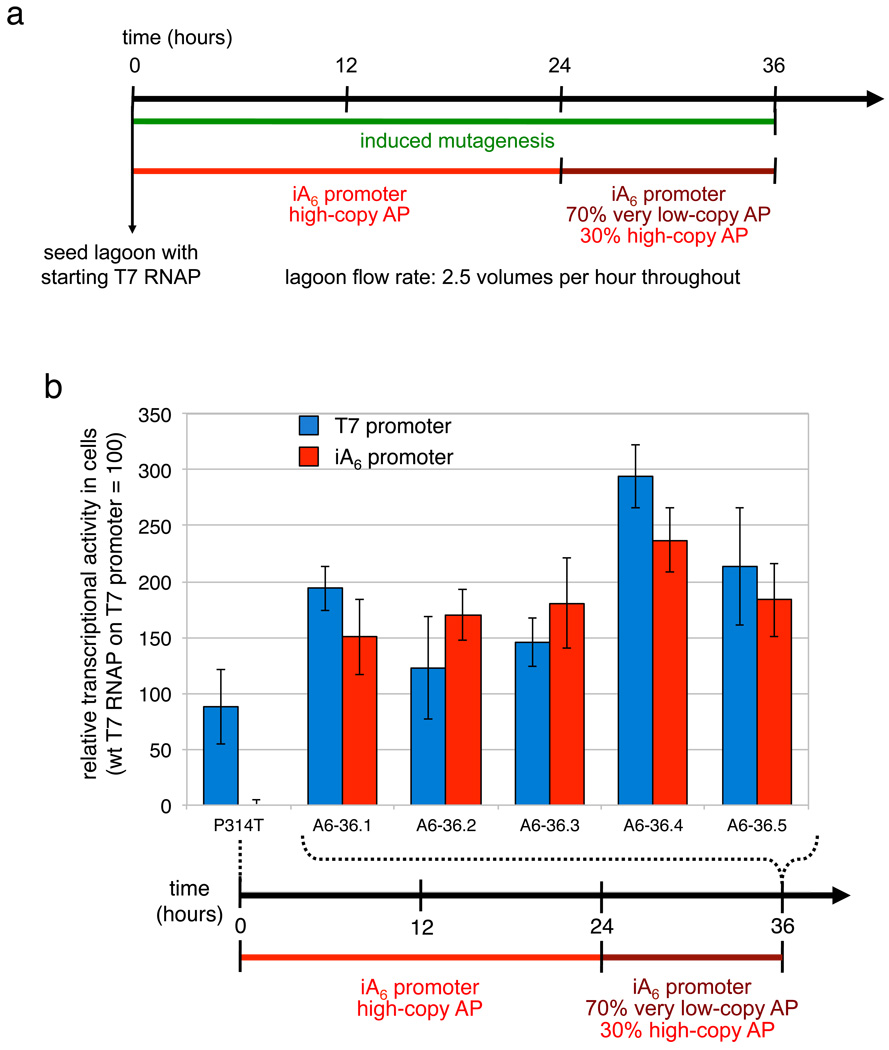
We used PACE to rapidly evolve variants of T7 RNAP capable of initiating with ATP. In light of previous reports indicating varying degrees of initiation of T7 RNAP with ATP,25,26 we propagated starting phage in host cells with a high-copy iA6 AP for 24 hours, followed by a 30:70 high-copy:very low-copy mixture of host cells for 12 hours (Fig. 4a). At a dilution rate of 2.5 volumes per hour, phage survived a total dilution of 1039-fold and experienced an average of ~45 rounds of evolution.
Figure 4.
Continuous evolution of T7 RNAP variants that initiate transcription with A. (a) PACE schedule. (b) Activity in cells of T7 RNAP variants on the T7 and iA6 promoters isolated after 36 hours of PACE. Assays were performed as described in Fig. 3b. Error bars represent the standard deviation of at least three independent assays.
The wild-type enzyme exhibited undetectable initial activity in cells (< 3%) on the iA6 promoter. All six clones isolated after only 36 hours of PACE exhibited at least 170% activity on the iA6 promoter in cell-based assays, while retaining at least 120% activity on the wild-type promoter (Fig. 4b). Purified variants assayed in vitro exhibited activities on the iA6 promoter matching that of wild-type T7 RNAP on the T7 promoter (Supplementary Fig. 8). RACE analysis of transcripts produced by the most active clone (A6-36.4) confirmed that this enzyme begins transcripts with the template-directed bases on the iA6, iC6, and wild-type promoters (Supplementary Fig. 9a). All six characterized clones contained K93T, S397R, and S684Y mutations, while three of the six also contained S228A (Supplementary Table 2). Residue 397 directly contacts the nascent RNA strand,28 suggesting a role for S397R in allowing efficient initiation of iA6 transcripts.
We concurrently evolved T7 RNAP to initiate transcripts with CTP (Supplementary Fig. 10a). We observed that wild-type T7 RNAP retains significant activity on the iC6 promoter (~50%) both in cells and in vitro (Supplementary Fig. 10b and 10c), a surprising observation in light of reports that the enzyme initiates with G at the +2 position if the +1 position is C.25 While the high starting activity precluded large improvements, the most active PACE-evolved variants nevertheless exceeded 100% activity on the iC6 promoter both in cells and in vitro (Supplementary Fig. 10b and 10c and Supplementary Results). RACE analysis of transcripts produced by the most active clone (C6-80.9) confirmed that this enzyme begins transcripts with the template-directed bases (Supplementary Fig. 9b).
The three PACE experiments executed 45 to 200 rounds of evolution in 1.5 to 8 days and yielded T7 RNAP variants with activities on their target promoters or templates that exceed or match the activity of the wild-type enzyme transcribing the wild-type T7 promoter both in cells and in vitro. This degree of improvement is especially significant given that for two of the evolved activities, the starting polymerase exhibited virtually no detectable activity.
The evolved A6-36.4 variant of T7 RNAP can initiate transcription from iC6, iA6, and wild-type templates in a template-directed manner with efficiencies comparable to that of wild-type T7 RNAP initiating with the wild-type template (Supplementary Fig. 11) and sequence fidelity sufficient to mediate the production of functional pIII and LacZ enzyme. These findings suggest that this enzyme, and possibly other PACE-evolved variants, may represent improved, more general T7 RNA polymerases for routine in vitro and in vivo transcription.
The PACE system can be assembled entirely from a modest collection of commercially available equipment (listed in Supplementary Table 3) and does not require the manufacture of any specialized components. The ability to perform dozens of rounds of evolution each day with minimal researcher involvement implies that PACE is particularly well suited to address problems or questions in molecular evolution that require hundreds to thousands of generations, or the execution of many evolution experiments in parallel. More generally, PACE represents the integration and manipulation of many protein and nucleic acid components in a living system to enable the rapid generation of biomolecules with new activities, a significant example and goal of synthetic biology.
Methods Summary
Discrete infection assays
2 µL of donor culture containing AP and SP were mixed with 198 of µL F+ recipient cells and incubated for 1.5 hours at 37 °C. Dilutions were plated to select for recipient cells containing SP.
PACE
E. coli host cells with AP and MP (see Methods) maintained at 5 × 108 cells/mL were pumped at 2.0, 2.5, or 3.2 volumes/hour into a fixed-volume lagoon seeded with SP. Aliquots were taken regularly. Evolved mutants were isolated as individual plaques and subcloned for cell-based or in vitro activity assays.
Transcription assays
Activity in cells was measured using a standard beta-galactosidase expression assay.24 In vitro activity was assessed using a standard radioactive nucleotide incorporation assay29 with His-tag-purified variants.
Methods
General methods
All equipment, reagents, suppliers, and relevant catalogue numbers are detailed in Supplementary Table 3. All PCR reactions were performed with HotStart Phusion II polymerase. Water was purified using a MilliQ water purification system (Millipore, Billerica MA).
DNA cloning
All vectors were constructed by isothermal assembly cloning.30 5x isothermal assembly buffer contained 3 mL 1 M Tris-HCl pH 7.5, 300 µL 1 M MgCl2, 600 µL 10 mM dNTPs, 300 µL 1 M dithiothreitol, 1.5 g PEG-8000, 20 mg NAD, and H2O to 6 mL. Individual 320 µL aliquots were frozen at −20 °C. Isothermal assembly master mix was prepared by mixing 320 µL 5x buffer with 1 µL T5 Exonuclease, 20 µL Phusion polymerase, 160 µL Taq DNA ligase, and H2O to 700 µL. Individual 15 µL aliquots in PCR tubes were frozen at −20 °C. DNA fragments to be assembled were PCR-amplified using oligonucleotide primers designed to ensure between 30 and 40 base pairs of overlap homology with each adjacent fragment. DpnI was added directly to the PCR reactions to remove template DNA, followed by PCR cleanup with MinElute columns according to the manufacturer’s protocol. Fragments were assembled by mixing equimolar amounts totalling 5 µL with 15 µL isothermal assembly master mix and incubating at 50 °C for 1 hr. Assembly mixtures were directly transformed into NEB Turbo competent cells by heat shock or purified by MinElute columns as described prior to electroporation.
Plasmids
T7 RNAP-dependent accessory plasmids (APs) contained, in order, a strong rrnB terminator, the promoter of interest, a desired ribosome binding site, gene III, the bla gene conferring carbenicillin resistance, and either the pUC or SC101 origin of replication. For selection stringency assays, RBS A = 5’-AAGGAGGTAACTCATAGTG-3’, RBS B = 5’-AAGGAAATAACTCATAGTG-3’, and RBS C = 5’-AAGAAAATAACTCATAGTG-3’, where underlined bases represent the start codon of gene III. Reporter plasmids were identical to SC101 accessory plasmids except for the replacement of gene III by full-length lacZ. T7 RNAP selection phage (SP) was constructed by replacing all but the last 180 bp of gene III with the gene encoding T7 RNAP in VCSM13 helper phage. The mutagenesis plasmid (MP) consisted of dnaQ926, umuD’, umuC, and recA730 under control of the araC operon. Expression plasmids used for quantification assays consisted of the cloD13 origin of replication, aadA, and the wild-type gene III promoter and RBS driving expression of the evolved T7 RNAP variant. All plasmids used in this work are described in Supplementary Table 1. Vector maps of representative plasmids are shown in Supplementary Fig. 2.
Bacterial strains
All DNA cloning was performed with Mach1 cells or NEB Turbo cells. Early discrete infection assays and PACE experiments were performed with PirPlus DH10βF'DOT cells. Plaque assays and PACE experiments with T7 RNAP were performed with using E. coli S109 cells derived from DH10B by replacement of the proBA locus with the pir116 allele, as previously described.31 To our knowledge, this modification was not required for PACE experiments with T7 RNAP. Similarly, the lacI cassette was deleted from the F plasmid and from the chromosome to enable mutagenesis assays. S109 cells were rendered F+ by conjugation with ER2738. The complete genotype of the resulting strain is F�proA+B+ Δ(lacIZY) zzf::Tn10(TetR)/ endA1 recA1 galE15 galK16 nupGrpsLΔlacIZYA araD139 Δ(ara,leu)7697 mcrA Δ(mrr-hsdRMS-mcrBC) proBA::pir116 λ−.
Discrete infection assays
Discrete (non-continuous) infectivity assays were performed by co-transforming phage bearing antibiotic resistance genes with the appropriate accessory plasmid into competent cells to generate phage donors. For two-hybrid and recombinase experiments, phage-producing cells contained gIII-deleted helper phage, accessory plasmid, and selection phagemid. The recombinase used was Hin, a member of the serine resolvase family. Colonies were picked and grown overnight in 2xYT media with both antibiotics. 2 µL donor cells were mixed with 198 µL F+ recipient cells in mid-exponential phase containing an antibiotic resistance gene not found in the donor. Mixtures were incubated at 37 °C for 1.5 hours and 20 µL of serial dilutions were spread on plates containing the donor and the recipient antibiotics. Infection was quantified by the number of resulting colonies after incubation at 37 °C overnight. For plaque assays, phage DNA was transformed into electrocompetent cells containing the appropriate accessory plasmid and recovered for 1 hr at 37 °C, or simply isolated from a lagoon. Serial dilutions were mixed with 300 µL F+ recipient cells grown to exponential phase in 14 mL Falcon culture tubes and incubated at 37°C for 15 minutes. 3 mL top agar (7 g/L from LB broth base) at 50 °C was added to each tube, briefly vortexed, and poured onto minimal agar plates incubated at 37 °C for 8 hours or overnight to generate plaques.
Turbidostat assembly
Assembly followed the schematic shown in Fig. 2b. Turbidostats were constructed from BioProbe flasks on magnetic stir plates. Each flask was equipped with a TruCell2 cell density meter held in a GL32 probe holder with compression fitting. GL45 and GL32 septa pierced with needles transferred media to and from the turbidostat via an 8-channel peristaltic pump with Tygon tubing. A needle set at the desired turbidostat volume level pumped excess cells to the waste container. A 0.2 µm filter attached to a 14-gauge needle piercing the septum vented the turbidostat vessel. A two-way valve controlled media flow to the turbidostat, connected such that a closed valve state returned the media to source. The valve opened and closed in response to TruCell2 4–20 mA output processed by a digital panel meter programmed with the desired set point. Panel meters were unlocked according to the instruction manual and programmed to the following settings: Input dc_A, Setup 30_ 10, Config 00000, Filtr 11009, dec.pt ddd.dd, lo in 00.400, lo rd 004.00, hi in 02.000, hi rd 020.00, Alset 00036, deu1h 000.01, deu2h 000.01. The digital panel meter, adapter, and valve were connected with a solderless breadboard according to the diagram shown in Supplementary Fig. 12. Lagoons consisted of 100 mL Pyrex bottles with GL45 septa pierced with needles for fluid delivery, a 0.2 µm filter-terminated vent line, and a magnetic stir bar. Excess lagoon volume was continually pumped to waste via a waste needle set at the desired lagoon volume.
Media preparation
Each 20 L media carboy received 140 g anhydrous potassium phosphate dibasic, 40 g potassium phosphate monobasic, 20 g ammonium sulphate, and 20 mL Tween 80 in 20 L H2O. Carboys were loosely capped with Polyvent filling/venting closures with an autoclavable 0.2 µm filter fastened to the venting port. Media was autoclaved until visibly boiling (typically 120 min at 30 psi, 121 °C) and allowed to cool overnight. Media supplement was prepared from 90 g glucose, 10 g sodium citrate, 0.25 g anhydrous magnesium sulphate, 10 g casamino acids, 0.15 g tetracycline-HCl, 0.6 g carbenicillin, 0.6 g spectinomycin, and 0.5 g (L)-leucine, dissolved in 500 mL H2O, and filtered with a Nalgene 500 mL filtration unit. 500 mL media supplement was added to each carboy under conditions that minimize the risk of media contamination (in the case of the reported experiments, immediately following 1 hour of germicidal UV irradiation).
Sterilization and Cell Culture
The autoclavable components of a turbidostat apparatus include the BioProbe flask, TruCell2 probe, needles, vent filter, and tubing. All such components were autoclaved fully assembled except for tubing, which was connected while hot. Upon equilibrating to ambient temperature, the peristaltic pump responsible for media addition and waste removal was started with the valve opened until the desired volume was reached. The TruCell2 probe was connected to its transmitter and zeroed. Turbidostats were seeded with 100 µL of an overnight culture of host cells. Turbidostats and lagoon cultures were grown at 37 °C.
Cell Density Calibration
Serial dilution plating was used to generate a calibration curve to determine TruCell2 output and panel meter setting corresponding to the desired cell density. For these experiments, both panel meter alarms were programmed to open the valve at 6.80 mA. Cells were pumped from the turbidostat to the lagoons via peristaltic pumps with silicone (platinum) two-stop tubing. Calibration curves relating pump speed in rpm to volumetric flow rate were determined experimentally with a timer and graduated cylinder for each tubing size.
PACE Experiments
Turbidostats and lagoons were assembled as described above. Upon the turbidostat reaching the desired set point of 6.80 mA (corresponding to 5 × 108 cells/mL in our hands), lagoons were connected to turbidostats, waste needles were set at the desired volume, and lagoon pumps were set to a flow rate corresponding to the desired dilution rate. Each lagoon was seeded with 100 µL of an overnight culture producing selection phage. To induced elevated mutagenesis, 10% filter-sterilized (L)-arabinose was delivered by a separate peristaltic pump to each lagoon requiring enhanced mutagenesis to a final concentration of 1%. Lagoon aliquots were taken by sampling lagoon waste lines at the luer lock just after the peristaltic pump. Individual clones were isolated by plaque assay or amplified by PCR, assembled into a T7 RNAP activity assay plasmid, and transformed into cells containing a lacZ reporter plasmid. Active clones were picked by blue/white screening.
Selection phage optimization
T7 RNAP was subcloned into VCSM13 helper phage encoding kanamycin resistance, generating HP-T7RNAP A, used in the discrete infection assay shown in Fig. 3a. To ensure that improvements to the phage genome did not interfere with the evolution of T7 RNAP, HP-T7RNAP A was propagated in a lagoon fed by S109 host cells containing AP-T7 A and DP-QUR and supplemented with arabinose for 72 hours at 2.0 volumes/hour. Individual plaques were isolated and their T7 RNAP genes sequenced. One plaque contained only a single point mutation in T7 RNAP, P314T, and was chosen as the preoptimized selection phage for T7 RNAP evolution. Sequencing of the rest of the selection phage revealed numerous changes relative to the parental VCSM13. Notably, the entire p15a-KanR cassette inserted into the intergenic (IG) region to create VCSM13 had been perfectly deleted to reconstitute the wild-type M13 IG region. Other changes included N79S, F286S, and I360T mutations in gIV, a K249R mutation in gII, three silent mutations back to the corresponding M13 base, two other silent mutations, and the deletion of one thymine residue in the terminator before gIII, possibly increasing the expression of T7 RNAP. These regional patterns of variation parallel those observed by Husimi in more extensive filamentous phage evolution experiments.11 This evolved phage, designated SP-T7RNAP P314T, was used as the starting selection phage for all subsequent PACE experiments.
Mutagenesis assays
The lacI gene was cloned into VCSM13 between the p15a origin and kanr to generate VCSM13-lacI. A turbidostat was grown to a set point equivalent to 5×108 cells/mL with S109 cells containing the mutagenesis plasmid (MP). Lagoons were seeded with 10 µL VCSM13-lacI and run at 2.5 volumes/hour for 3 hours. One lagoon was supplemented with 10% arabinose to a final concentration of 1%, while the other was not. Aliquots were removed after 3 hours and each was used to infect a 100-fold greater volume of recipient cell culture of S109 cells containing a reporter plasmid conferring carbenicillin and spectinomycin resistance with a lacI binding site (lacO) capable of repressing spectinomycin resistance. Mixtures were incubated for 1.5 hr at 37°C, spread on 2xYT plates containing spectinomycin, kanamycin, and carbenicillin, and incubated at 37°C overnight. Colonies were counted for induced and uninduced lagoons to estimate the fold increase in mutagenesis. 72 colonies were sequenced to determine the frequencies of all transitions and transversions within mutated lacI genes. The results are shown in Supplementary Fig. 4. All sequenced colonies contained at least one mutation capable of inactivating repressor function.
Cell-based T7 RNAP activity assays
Overnight cultures of S109 cells grown in 2xYT containing reporter plasmid and expression vector were diluted 4-fold in fresh 2xYT media. 20 µL of the diluted culture was mixed with 80 µL Z buffer (60 mM Na2HPO4, 40 mM NaH2PO4, 10 mM KCl, 1 mM MgSO4, 50 mM beta-mercaptoethanol, pH 7.0) in Falcon Microtest 96-well OptiLux assay plates and the absorbance at 595 nm was measured using a Spectra M5 plate reader. 25 µL of 1 mg/mL methylumbelliferyl-beta-(D)-galactopyranoside was added to each well and the time recorded. Plates were incubated at 30 °C and fluorescence was measured at 360/460nm on a Spectra M5 plate reader. Plates were measured at multiple time points to avoid saturation of the spectrophotometer or consumption of the substrate, depending on the activity level of the T7 RNAP enzyme being assayed. MUG fluorescence units were calculated as previously described.24 The activity level of wild-type T7 RNAP on the T7 promoter was defined as 100%; activities > 3% were considered significantly above the background level of this assay.
T7 RNAP protein purification
T7 RNAP variants were cloned into pT7-911Q, a His-tagged T7 RNAP expression vector.32 Overnight cultures grown at 30°C were diluted 1:500 in LB broth containing 50 µg/mL carbenicillin and 2% glucose. Upon reaching OD600 = ~0.5, cultures were centrifuged at 4000 g for 5 minutes and resuspended in LB broth with 0.4 mM IPTG and 50 µg/mL carbenicillin. Cultures were grown for 4 hours at 30°C, spun at 8000 g for 6 minutes, and the pellet was frozen overnight. Binding buffer consisted of 50 mM Tris, 300 mM NaCl, 5% glycerol, 5 mM beta-mercaptoethanol, and 10 mM imidazole at pH 8.0. Wash buffer consisted of 50 mM Tris, 800 mM NaCl, 5% glycerol, 5 mM beta-mercaptoethanol, and 20 mM imidazole at pH 8.0. Elution buffer was equivalent to wash buffer with 500 mM imidazole. Pellets from 25 mL culture were resuspended in 1 mL wash buffer and cells were lysed by sonication while kept on ice using a Misonix CL4 sonicator at maximal microtip power for 45 seconds in 1-second bursts. Cell debris was spun down at 20,000 g for 15 minutes at 4°C. Ni-NTA spin columns were equilibrated with 500 µL binding buffer and spun at 800 g for 2 minutes. Lysate supernatant was loaded onto each column and spun at 300 g for 5 minutes. Columns were washed twice with 500 µL wash buffer, spinning at 800 g for each, then eluted twice with 250 µL elution buffer. Proteins were dialyzed into 20 mM Tris, 100 mM NaCl, 5% glycerol, 1 mM EDTA, 1 mM DTT, pH 8.0, and concentrated using Amicon Ultra-0.5 30K concentration columns.
In vitro T7 RNAP activity assays
Purified T7 RNAP variant concentrations were determined by Bradford assay and then by Coomassie stain on a 4–12% NuPage gel. Templates were prepared by PCR amplification of 150 bp fragments of the reporter plasmids used for in vivo assays including the promoter and the start of the lacZ gene. Templates were purified by MinElute spin column. Transcription reactions were performed in 1x RNA polymerase buffer consisting of 40 mM Tris-HCl, 6 mM MgCl2, 10 mM dithiothreitol, 2 mM spermidine pH 7.9 with 1 mM rNTPs, 1 ng template DNA, purified polymerase variant, and 2 mCi [α-32P]ATP. Reactions were incubated at 37 °C for 20 minutes, mixed with an equivalent volume of loading dye consisting of 7 M urea, 178 mM Tris-Cl, 178 mM H3BO3, 4 mM EDTA and 0.002% bromophenol blue, then electrophoresed on Criterion 5% or 10% TBE-urea denaturing gels. RNAs were transcribed from double-stranded templates of sequence 5’-TAATACGACTCACTATAGGGAGAGCCACCACCACCACCACCACCA-3’, 5’- TAATACGACTCACTATACCCCCCGCCACCACCACCACCACCACCA -3’, and 5’- TAATACGACTCACTATAAAAAAAGCCACCACCACCACCACCACCA -3’ (the +1 base is underlined in bold). To remove differences in specific radioactivity from [α-32P]-ATP incorporation in iA6, wild-type, and iC6 transcripts arising from the differing number of A nucleotides in their first six bases, transcripts were digested with T1 ribonuclease to remove the first seven nucleotides. This digestion step also enabled all iA6 transcription products to completely enter the gel, which did not always occur for iA6 products of either wild-type or evolved polymerases. Transcripts were electrophoresed on Criterion 15% TBE-urea gels, exposed to phosphor screens, and imaged on a Typhoon Trio phosphorimager. Bands corresponding to transcription products were quantified with ImageJ software.
RACE analysis
In vitro transcription reactions with purified T7 RNAP variants were performed as described above but without the addition of any radioactive nucleotide using polymerase variant C6-80.9 on the iC6 and wild-type templates, and using polymerase variant A4-36.4 on the iA6, iC6, and wild-type templates. DNA oligonucleotides of sequence 5’-TAATACGACTCACTATACCC-3’ and 5’-CCCACCCAAAAAAAAAAAAAAAGGGGGGTATAGTGAGTCGTATTA-3’ formed the iC6 template,5’-TAATACGACTCACTATAAAA-3’ and 5’-CCCACCCAAAAAAAAAAAAAAATTTTTTTATAGTGAGTCGTATTA-3’ formed the iA6 template, and 5’-TAATACGACTCACTATAGGG-3’ and 5’-CCCACCCAAAAAAAAAAAAAAATCTCCCTATAGTGAGTCGTATTA-3’ formed the wild-type template. Each 200 µL transcription reaction was treated with 2 µL calf intestinal phosphatase (CIP) for 1 hour at 37°C, extracted with phenol-chloroform twice to remove enzymes, and precipitated with ethanol. Pellets were resuspended in 1x DNase Turbo buffer with 5 µL DNase in a total volume of 100 µL, incubated for 2 hours at 37°C, extracted with phenol-chloroform twice, and precipitated with ethanol. 5 µL of purified transcript was mixed with 1 µL T4 polynucleotide kinase (PNK) in 1x PNK buffer and incubated at 37°C for 1 hour. 1 µL of treated RNA was ligated to 30 ng RNA adapter of sequence 5’-GCUGAUGGCGAUGAAUGAACACUGCGUUUGCUGGCUUUGAUGAAA-3’ with T4 RNA Ligase in 1x RNA Ligase Buffer at 37°C for 1 hour. 1 µL of ligated RNA was reverse transcribed by mixing with 1 mM dNTPs and a complementary DNA primer of sequence 5’-CCCCCAAACCCCCAAAAAAAAAACCCACCCAAAAAAAAAAAA-3’ at a final concentration of 5 µM at 65 °C for 15 minutes followed by 10 U/µL SuperScript III reverse transcriptase in 1x RT buffer, 5 mM MgCl2, and 10 mM dithiothreitol at 50°C for 1 hour. Enzymes were denatured at 85°C for 5 minutes followed by DNA amplification with sequential PCR reactions using primers 5’-CGATCCGAACGCAGCATTTACGCTGATGGCGATGAATGAACACTG-3’ and 5’-CCCCCAAACCCCCAAAAAAAAAACCCACCCAAAAAAAAAAAA-3’, digestion with MlyI and HinfI in NEBuffer 4 to cleave sequences containing the promoter, and PCR amplification with 5’-GCTAGTTATTGCTCAGCGGAATAACGATCCGAACGCAGCATTTAC-3’ and 5’-GCTAGTTATTGCTCAGCGGAAAAAAAAAAAAACCCCCAAACCCCC-3’. PCR products were cloned into the backbone of plasmid AP-T7 P amplified with primers 5’-CGGATCGTTATTCCGCTGAGCAATAACTAGCAGAGCAAAAGGCCAGC-3’ and 5’-GGGGGTTTGGGGGTTTTTTTTTTTTTCCGGCCTTGTCGGCCTTAC-3’ via isothermal assembly cloning. Individual colonies were picked and sequenced using a primer of sequence 5’-CAGGAAGGCAAAATGCCG-3’.
Supplementary Material
Acknowledgements
This work was supported by NIH/NIGMS R01 GM065400 and by HHMI. KME gratefully acknowledges graduate research fellowships from the Hertz Foundation and NSF. JCC was supported by the Harvard Chemical Biology Graduate Program. We thank Brent Dorr for assistance with phage generation modeling, Dr. Edward Curtis for helpful suggestions, and Professor Victoria D’Souza for plasmid pT7-911Q.
Footnotes
Supplementary Information is linked to the online version of the paper at www.nature.com/nature.
Author Contributions
K.M.E., J.C.C., and D.R.L. designed the experiments; K.M.E. designed and built the PACE apparatus; K.M.E. and J.C.C. performed the experiments. All authors analysed the data and wrote the manuscript.
Author Information
Reprints and permissions information is available at www.nature.com/reprints. The authors declare no competing financial interests.
References Cited
- 1.Yuan L, Kurek I, English J, Keenan R. Laboratory-directed protein evolution. Microbiology and Molecular Biology Reviews: MMBR. 2005;69:373–392. doi: 10.1128/MMBR.69.3.373-392.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Voigt CA, Kauffman S, Wang ZG. Rational evolutionary design: the theory of in vitro protein evolution. Advances in Protein Chemistry. 2000;55:79–160. doi: 10.1016/s0065-3233(01)55003-2. [DOI] [PubMed] [Google Scholar]
- 3.Mills DR, Peterson RL, Spiegelman S. An extracellular Darwinian experiment with a self-duplicating nucleic acid molecule. Proc Natl Acad Sci USA. 1967;58:217–224. doi: 10.1073/pnas.58.1.217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wang L, Jackson WC, Steinbach PA, Tsien RY. Evolution of new nonantibody proteins via iterative somatic hypermutation. Proc Natl Acad Sci USA. 2004;101:16745–16749. doi: 10.1073/pnas.0407752101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Camps M, Naukkarinen J, Johnson BP, Loeb LA. Targeted gene evolution in Escherichia coli using a highly error-prone DNA polymerase I. Proc Natl Acad Sci USA. 2003;100:9727–9732. doi: 10.1073/pnas.1333928100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Makeyev EV, Bamford DH. Evolutionary potential of an RNA virus. Journal of Virology. 2004;78:2114–2120. doi: 10.1128/JVI.78.4.2114-2120.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Davis JN, van den Pol AN. Viral Mutagenesis as a Means for Generating Novel Proteins. Journal of Virology. 2009;84:1625–1630. doi: 10.1128/JVI.01747-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Das AT, et al. Viral evolution as a tool to improve the tetracycline-regulated gene expression system. The Journal of Biological Chemistry. 2004;279:18776–18782. doi: 10.1074/jbc.M313895200. [DOI] [PubMed] [Google Scholar]
- 9.Wang HH, et al. Programming cells by multiplex genome engineering and accelerated evolution. Nature. 2009;460:894–898. doi: 10.1038/nature08187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wright MC, Joyce GF. Continuous in vitro evolution of catalytic function. Science. 1997;276:614–617. doi: 10.1126/science.276.5312.614. [DOI] [PubMed] [Google Scholar]
- 11.Husimi Y. Selection and evolution of bacteriophages in cellstat. Advances in Biophysics. 1989;25:1–43. doi: 10.1016/0065-227x(89)90003-8. [DOI] [PubMed] [Google Scholar]
- 12.Smith GP. Filamentous fusion phage: novel expression vectors that display cloned antigens on the virion surface. Science. 1985;228:1315–1317. doi: 10.1126/science.4001944. [DOI] [PubMed] [Google Scholar]
- 13.Riechmann L, Holliger P. The C-terminal domain of TolA is the coreceptor for filamentous phage infection of E. coli. Cell. 1997;90:351–360. doi: 10.1016/s0092-8674(00)80342-6. [DOI] [PubMed] [Google Scholar]
- 14.Nelson FK, Friedman SM, Smith GP. Filamentous phage DNA cloning vectors: a noninfective mutant with a nonpolar deletion in gene III. Virology. 1981;108:338–350. doi: 10.1016/0042-6822(81)90442-6. [DOI] [PubMed] [Google Scholar]
- 15.Rakonjac J, Model P. Roles of pIII in filamentous phage assembly. Journal of Molecular Biology. 1998;282:25–41. doi: 10.1006/jmbi.1998.2006. [DOI] [PubMed] [Google Scholar]
- 16.Calendar R. The bacteriophages. Oxford University Press US; 2006. [Google Scholar]
- 17.Vidal M, Legrain P. Yeast forward and reverse 'n'-hybrid systems. Nucleic Acids Research. 1999;27:919–929. doi: 10.1093/nar/27.4.919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Baker K, et al. Chemical complementation: a reaction-independent genetic assay for enzyme catalysis. Proc Natl Acad Sci USA. 2002;99:16537–16542. doi: 10.1073/pnas.262420099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fijalkowska IJ, Schaaper RM. Mutants in the Exo I motif of Escherichia coli dnaQ: defective proofreading and inviability due to error catastrophe. Proc Natl Acad Sci USA. 1996;93:2856–2861. doi: 10.1073/pnas.93.7.2856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Opperman T, Murli S, Smith BT, Walker GC. A model for a umuDC-dependent prokaryotic DNA damage checkpoint. Proc Natl Acad Sci USA. 1999;96:9218–9223. doi: 10.1073/pnas.96.16.9218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Raskin CA, Diaz G, Joho K, McAllister WT. Substitution of a single bacteriophage T3 residue in bacteriophage T7 RNA polymerase at position 748 results in a switch in promoter specificity. Journal of Molecular Biology. 1992;228:506–515. doi: 10.1016/0022-2836(92)90838-b. [DOI] [PubMed] [Google Scholar]
- 22.Ikeda RA, Chang LL, Warshamana GS. Selection and characterization of a mutant T7 RNA polymerase that recognizes an expanded range of T7 promoter-like sequences. Biochemistry. 1993;32:9115–9124. doi: 10.1021/bi00086a016. [DOI] [PubMed] [Google Scholar]
- 23.Raskin CA, Diaz GA, McAllister WT. T7 RNA polymerase mutants with altered promoter specificities. Proc Natl Acad Sci USA. 1993;90:3147–3151. doi: 10.1073/pnas.90.8.3147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Vidal-Aroca F, et al. One-step high-throughput assay for quantitative detection of beta-galactosidase activity in intact gram-negative bacteria, yeast, and mammalian cells. BioTechniques. 2006;40:433–434. doi: 10.2144/000112145. 436, 438 passim. [DOI] [PubMed] [Google Scholar]
- 25.Imburgio D, Rong M, Ma K, McAllister WT. Studies of promoter recognition and start site selection by T7 RNA polymerase using a comprehensive collection of promoter variants. Biochemistry. 2000;39:10419–10430. doi: 10.1021/bi000365w. [DOI] [PubMed] [Google Scholar]
- 26.Brieba LG, Padilla R, Sousa R. Role of T7 RNA polymerase His784 in start site selection and initial transcription. Biochemistry. 2002;41:5144–5149. doi: 10.1021/bi016057v. [DOI] [PubMed] [Google Scholar]
- 27.Kuzmine I, Gottlieb PA, Martin CT. Binding of the priming nucleotide in the initiation of transcription by T7 RNA polymerase. The Journal of Biological Chemistry. 2003;278:2819–2823. doi: 10.1074/jbc.M208405200. [DOI] [PubMed] [Google Scholar]
- 28.Cheetham GM, Jeruzalmi D, Steitz TA. Structural basis for initiation of transcription from an RNA polymerase-promoter complex. Nature. 1999;399:80–83. doi: 10.1038/19999. [DOI] [PubMed] [Google Scholar]
- 29.Martin CT, Coleman JE. Kinetic analysis of T7 RNA polymerase-promoter interactions with small synthetic promoters. Biochemistry. 1987;26:2690–2696. doi: 10.1021/bi00384a006. [DOI] [PubMed] [Google Scholar]
References for the Online-only Methods Section
- 30.Gibson DG, et al. Enzymatic assembly of DNA molecules up to several hundred kilobases. Nature Methods. 2009;6:343–345. doi: 10.1038/nmeth.1318. [DOI] [PubMed] [Google Scholar]
- 31.Datsenko KA, Wanner BL. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci USA. 2000;97:6640–6645. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ichetovkin IE, Abramochkin G, Shrader TE. Substrate recognition by the leucyl/phenylalanyl-tRNA-protein transferase. Conservation within the enzyme family and localization to the trypsin-resistant domain. J Biol Chem. 1997;272:33009–33014. doi: 10.1074/jbc.272.52.33009. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.