Abstract
Several genes have been suggested as dyslexia candidates. Some of these candidate genes have been recently shown to be associated with literacy measures in sample cohorts derived from the general population. Here, we have conducted an association study in a novel sample derived from the Australian population (the Raine cohort) to further investigate the role of dyslexia candidate genes. We analysed markers, previously reported to be associated with dyslexia, located within the MRPL19/C2ORF3, KIAA0319, DCDC2 and DYX1C1 genes in a sample of 520 individuals and tested them for association with reading and spelling measures. Association signals were detected for several single nucleotide polymorphisms (SNPs) within DYX1C1 with both the reading and spelling tests. The high linkage disequilibrium (LD) we observed across the DYX1C1 gene suggests that the association signal might not be refined by further genetic mapping.
Keywords: Association study, dyslexia, DYX1C1, Raine study, reading skills
Dyslexia (reading disability) is a developmental condition with a prevalence ranging from 5% to 17% in school-aged children (Pennington 1990; Shaywitz 1998; Shaywitz et al. 1990). As for other neurodevelopmental disorders, initial reports indicated a higher prevalence in males (Finucci & Childs 1981); however, subsequent studies have reported no gender differences (DeFries & Alarcón 1996; Guerin et al. 1993). Some studies have suggested that the higher male prevalence could be explained by a referral bias (Vogel 1990) or by greater variance of reading skills in males (Hawke et al. 2009).
A strong genetic component for dyslexia has been widely documented (Fisher & DeFries 2002; Habib 2000) and molecular genetic analyses have led to the identification of several candidate genes (Scerri & Schulte-Korne 2010). These include the MRPL19/C2ORF3 locus on chromosome 2, ROBO1 on chromosome 3, KIAA0319 and DCDC2 on chromosome 6 and DYX1C1 on chromosome 15. For all of these genes, with the exception of ROBO1, associations with common single nucleotide polymorphisms (SNPs) have been reported suggesting that the genetic variants conferring susceptibility to dyslexia are found in the general population. ROBO1 was identified by the refinement of a breakpoint translocation in one individual with dyslexia and the identification of a rare haplotype co-segregating with dyslexia in a large Finnish family (Hannula-Jouppi et al. 2005). So far, support for ROBO1 has not been reported in additional samples, suggesting its role in the etiology of dyslexia might be restricted to isolated cases. For the other genes, evidence of associations has been described in at least two independent samples.
DYX1C1 (dyslexia susceptibility 1 candidate 1) was the first candidate gene for dyslexia susceptibility to be identified (Taipale et al. 2003). The identification of the gene was initially led by the breakpoint mapping of a chromosome translocation co-segregating with dyslexia in one family. Association analysis in a larger sample supported these findings and implicated two variants in DYX1C1, with a putative functional effect. The -3A variant (at the -3G>A or rs3743205 SNP) has a putative effect of transcription regulation and the 1249T variant (at the 1249G>T or rs57809907 SNP) introduces a premature stop codon. These initial associations have been followed by the largest number of replication studies conducted so far for dyslexia candidate genes, with the majority of studies targeting these two specific SNPs. Nine independent studies have analysed rs3743205 and rs57809907 producing conflicting results (Scerri & Schulte-Korne 2010), where association was not detected (Bates et al. in press; Bellini et al. 2005; Cope et al. 2005b; Meng et al. 2005a), association was detected with the opposite alleles (-3G or 1249G) (Brkanac et al. 2007; Dahdouh et al. 2009; Scerri et al. 2004; Wigg et al. 2004) or association was detected with the same alleles as in the original study (Marino et al. 2007). Analysis of allele-specific effect on gene regulation has suggested a functional role for rs3743205 as well as for two other DYX1C1 markers (rs16787 and rs12899331) (Tapia-Paez et al. 2008). Associations with other markers have also been reported for rs3743204, rs685935 and rs17819126 using quantitative reading-related measures in a twin-based Australian sample representing the general population (Bates et al. in press). The rs17819126 marker, which was the most significant finding of that study, is a nonsynonymous coding SNP and was associated with three different traits. However, the same SNP, previously called 271G>A, did not yield significant associations in previous studies (Scerri et al. 2004; Taipale et al. 2003). The rs3743204 marker has also been reported to be associated with dyslexia but only as part of different haplotypes (Dahdouh et al. 2009; Wigg et al. 2004).
Taken together, association studies of DYX1C1 have generated contradictory findings based on modest association signals. It has been suggested that differences in population ethnicity or stratification effects could explain conflicting association (Paracchini et al. 2007) (Table S1). Indeed, the first association was reported in a Finnish sample, which is a genetic isolate and expected to have a different linkage disequilibrium (LD) landscape. Replications studies have been carried out in samples that varied not only in ethnic origin but also in structure (nuclear families, trios and cases) and ascertainment criteria. However, associations have been consistently reported in independent samples supporting the role of this gene in the development of dyslexia and suggesting that additional efforts are required to elucidate the meaning of these results.
Greater consensus has been reached on the allelic trend of associations for markers within the KIAA0319 and DCDC2 genes at the chromosome 6 locus; however, negative replications have also been reported. Both genes have been analysed by testing for association markers distributed all along their genomic sequence in several independent samples (Table S1). Most of the positive associations for KIAA0319 cluster around the first intron and regulatory sequences upstream of the 5′ end of the gene (Cope et al. 2005a; Deffenbacher et al. 2004; Dennis et al. 2009; Francks et al. 2004; Harold et al. 2006; Kaplan et al. 2002). In particular, rs4504469 which is a coding SNP showed association in independent studies (Cope et al. 2005a; Francks et al. 2004). This marker is also part of a specific haplotype, effectively tagged by rs2143340, found to be associated in two independent samples selected for severity of phenotype (Francks et al. 2004) as well as with reduction in expression of the KIAA0319 gene (Paracchini et al. 2006). The same haplotype was found to be associated with the reading abilities of the general population in two samples representing the general population either with same trend as in the original reports (Paracchini et al. 2008) or with opposite direction (Luciano et al. 2007). This haplotype has been shown to capture the effect of a common polymorphism, rs9461045, which creates the binding site for a nuclear protein explaining the inhibition of gene expression (Dennis et al. 2009). Lack of association at this locus has been reported by studies that instead identified association with the DCDC2 gene (Meng et al. 2005b; Schumacher et al. 2006). A functional effect has been proposed for an intronic deletion within DCDC2 (Meng et al. 2005b) but replication attempts have provided only minor support (Brkanac et al. 2007; Harold et al. 2006; Wilcke et al. 2009). Associations have also been reported for several markers including rs807701 (Ludwig et al. 2008; Schumacher et al. 2006), rs807724 (Meng et al. 2005b), rs1087266 (Harold et al. 2006; Meng et al. 2005b) and rs793862 (Ludwig et al. 2008; Meng et al. 2005b; Schumacher et al. 2006). DCDC2 has also been investigated in an epidemiological sample, with the most significant association reported for the rs1419228 marker (Lind et al. 2010).
Associations at the MRPL19/C2ORF3 locus on chromosome 2 have been reported in two independent samples (Table S1) with overlapping haplotypes (Anthoni et al. 2007). These associations are yet to be replicated in separate studies.
Follow-up studies in epidemiological samples have represented a valid alternative to replicate the original association with dyslexia candidate genes. All these studies have been based on the quantitative analysis of reading-related measures available for individuals representing the general population, regardless of whether a diagnosis of reading impairment was ever conferred (Bates et al. in press; Lind et al. in press; Luciano et al. 2007; Paracchini et al. 2008). Besides supporting the roles of these genes in contributing to dyslexia, these investigations have corroborated the notion that dyslexia represents the lower tail of reading abilities, which are normally distributed across the population rather than being a distinct disorder.
Here, we have conducted a replication study by analysing markers previously reported to be associated with dyslexia within the DYX1C1, KIAA0319, DCDC2 and MRPL19/C2ORF3 genes in the Western Australian Pregnancy Cohort (Raine) study, which is a longitudinal cohort for which reading-related measures are available. We report association with several markers located within the DYX1C1 gene.
Methods
Participants
The Raine study is a pregnancy cohort that was recruited prior to 18 weeks' gestation from the public antenatal clinic at King Edward Memorial Hospital (KEMH) or surrounding private clinics in Perth, Western Australia (WA) (Newnham et al. 1993). Approximately 100 unselected antenatal patients per month were enrolled during this period from August 1989 to April 1992, with a final sample of 2979 women. The inclusion criteria were (1) English language skills sufficient to understand the study demands, (2) an expectation to deliver at KEMH and (3) an intention to remain in WA to enable future follow-up of their child. Participant recruitment and all follow-ups of the study families were approved by the Human Ethics Committee at KEMH and/or Princess Margaret Hospital for Children in Perth. From this original cohort of women, 2868 of their children have been followed over the last two decades with detailed assessments performed every 2–3 years.
Reading and spelling assessment of children at age 10 years
The Western Australian Literacy and Numeracy Assessment (WALNA) is administered annually to all students across WA in school grades three (age 8), five (age 10) and seven (age 12). The WALNA was developed in consultation with educators to provide information on whether children have reached the minimum standards of reading, writing, spelling and numeracy. The tests have been written to cater for the diverse range of students in Australian schools, and to ensure that there is no systematic bias associated with factors such as gender, culture or geographic location. Every year the WALNA is evaluated by expert judges for content and construct validity and scrutinized by psychometricians for misfitting items, precision and bias. The current study concerned performance on the reading and spelling subtests of the WALNA, completed by the Raine cohort during school grade 5 (between 2000 and 2002).
For the reading test, children were given a magazine and required to answer 33 multiple-choice questions on its contents. A further two questions required a short answer of one to two sentences each. For each item, children were directed to the relevant page and article title (e.g. ‘read Helicopter on page 2 of the magazine and answer questions 1–5’). The spelling subtest consisted of two tasks. In the first task, participants were provided with a written paragraph that included 10 spelling mistakes, each of which were circled. The passage was first read aloud by the teacher from beginning to end. The teacher then read through the passage again, this time pausing at each circled word (spelling mistake), upon which children were required to write down the correct spelling of the word. The second spelling task was similar to the first; however, rather than spelling mistakes, the written passage given to children included 14 blank spaces for missing words. The passage, including the missing words, was then read aloud by the teacher twice: the first time, children were instructed to follow the text with their finger, and the second time, children were required to write down each missing word. For items in both the spelling and reading tests, a score (of 1) was awarded for correct answers only. Raw scores for the reading and spelling tests were summed and then converted via a Rasch measurement model (Doig & Groves 2006) into an interval scale to enable easier interpretation of the results. Scores on both the reading and spelling subscales could range from −100 to 800, with higher scores indicating better performance.
These data, collected by the WA Department of Education and Training, were then linked to the Raine study dataset by the WA Data Linkage System using a probabilistic method of matching based on a full name, date of birth, gender and address (Kelman et al. 2002). Western Australian Literacy and Numeracy Assessment records were linked for 1038 Raine study children who were in grade five and attending government schools at the time of assessment.
Genotyping
In the Raine study, DNA was collected using standardized procedures from 74% of adolescence who attended the 14-year follow-up on and a further 5% at the 16-year follow-up. Genome-wide data were generated using an Illumina 660 Quad array for each individual. Only SNPs that passed quality control (QC) criteria (call rate ≥ 95%, minor allele frequency >0.05 and Hardy–Weinberg disequilibrium P-value >0.01) were retained for genetic analysis.
Sample inclusion criteria
Inclusion criteria for the current study were no known intellectual disability; a nonverbal IQ within normal limits as assessed by the Raven's Colored Progressive Matrices (CPM; i.e. a score ≥16th percentile, corresponding to approximately >−1 SD the population average of the 50th centile) and biological parents who were both of white European origin. Furthermore, because the current study had an interest in literacy development of the birth cohort, only those children who spoke English at home were included for analysis.
Statistical analyses
Our first analysis examined selected SNPs within the DYX1C1, KIAA0319, DCDC2 and MRPL19/C2ORF3 genes that have been previously found to be associated with dyslexia. Fourteen of these SNPs were available for the Raine sample (Table S2). These SNPs were tested for association with quantitative measures of reading and spelling scores (Table 1) using an allelic test of association within PLINK version 1.07 (Purcell et al. 2007). We also tested for association haplotypes derived from the markers at the MRPL19/C2ORF3 locus, as previously reported (Anthoni et al. 2007). Haplotypes were inferred using SimHap version 1.0.2. In all analyses, gender was specified as a covariate. Principal components analysis with Eigenstrat (Price et al. 2006) showed evidence of population stratification and the first two principal components were also included in all analyses.
Table 1.
Descriptive statistics of phenotypic measures
| n | Mean (SD) | Range | |
|---|---|---|---|
| Reading (Rasch scale) | 520 | 391.55 (95.42) | −47, 703 |
| Spelling (Rasch scale) | 520 | 422.79 (118.26) | −99, 717 |
| Raven's Colored Progressive Matrices (CPM) (percentiles) | 520 | 62.58 | 18, 100 |
The reported P-values are not corrected for multiple testing and we show any results with a P-value <0.1. The application of a Bonferroni correction would be too conservative because SNPs within the same locus are highly correlated. Instead, we aimed to limit the number of tests by analysing targeted SNPs previously reported in the literature. Any trend of association, even if not significant, would provide additional support for the role of these genes in contributing to dyslexia or reading skills. The LD among DYX1C1 SNPs in this sample was determined with Haploview version 4.2 (http://www.broadinstitute.org/haploview/haploview) (Barrett et al. 2005). Power calculations were computed using Quanto version 4.02 (Gauderman et al. 2002) (Fig. S1).
Results
Literacy data were available for 520 (272 males and 248 females) of the 895 participants who met inclusion criteria (Table 1). All children were turning 10 years of age during the year of WALNA completion. Independent samples t-tests and chi-square analyses found that these children differed somewhat to the remainder of the cohort. For example, children who participated in the current study were heavier at birth (participated: M = 3394.55 g, SD = 591.62 g; did not participate: M = 3261.46 g, SD = 625.43 g; t(2856) = 4.43, P < 0.01) and had mothers who were older at conception (participated: M = 28.39, SD = 5.83; did not participate: M = 27.09 years, SD = 5.91; t(2865) = 4.55, P < 0.01). Therefore, while there was a modest degree of bias in the current sample favoring more socially advantaged families, the children were not bias toward any pathology.
No significant association was detected for SNPs within KIAA0319, DCDC2 and MRPL19/C2ORF3 (data not shown). Haplotype analysis for the MRPL19/C2ORF3 markers also showed no significant patterns of association. Two SNPs within the DYX1C1 gene showed a trend of association: rs3743205 (reading: df = 4, β = 25.46, P = 0.085) and rs685935 (spelling: df = 4, β = −12.28, P = 0.097). The allelic trend of association for rs3743205, which showed the major allele (or ‘-3G’ allele) associated with poor reading performance, was in agreement with a previous replication study (Wigg et al. 2004) reporting opposite trend compared to the original findings (Taipale et al. 2003). The minor allele of rs685935 was associated with poor spelling performance, showing the same trend as reported previously with a short-term memory measure (Bates et al. in press).
Previous studies of DYX1C1 have reported associations with a wide range of markers and with opposite allelic trend compared to other dyslexia susceptibility candidate genes. Therefore, while our initial analyses fell short of statistical significance at P < 0.05, we followed up the association trend by analysis of all the SNPs available for DYX1C1 in the Raine sample (Table S3). The SNPs that passed our QC criteria were tested for association with the reading and spelling measures (Table 2). The most significant association (df = 4, β = −19.32, P = 0.012) was detected between the rs8043049 marker and spelling. This SNP is adjacent to marker rs685935, which was tested in our initial analysis (df = 4, β = −12.28, P = 0.097) and within a cluster of five SNPs showing a trend of association, including rs8037376 which showed a P-value = 0.027 (df = 4, β = −17.39). The strongest association with the reading measure was detected with the rs8040756 marker (df = 4, β = 18.68, P = 0.026).
Table 2.
Association P-values for SNPs in the DYX1C1 gene
| Reading | Spelling | ||||||
|---|---|---|---|---|---|---|---|
| SNP | Minor allele | Major allele | MAF | β | P | β | P |
| rs8034029 | T | C | 0.09 | — | — | — | — |
| rs12324434 | C | T | 0.42 | — | — | — | — |
| rs12594039 | C | T | 0.06 | — | — | — | — |
| rs7174102 | A | T | 0.34 | — | — | −15.98 | 0.036 |
| rs3759864 | C | T | 0.06 | — | — | — | — |
| rs4774768 | T | G | 0.47 | — | — | 12.22 | 0.096 |
| rs7181226 | C | T | 0.09 | — | — | — | — |
| rs687623 | C | T | 0.41 | — | — | — | — |
| rs622097 | T | C | 0.33 | — | — | 14.35 | 0.067 |
| rs600753 | T | C | 0.48 | — | — | — | — |
| rs2290981 | A | G | 0.06 | — | — | — | — |
| rs7181999 | C | T | 0.09 | — | — | — | — |
| rs692690 | T | C | 0.42 | — | — | — | — |
| rs692691 | T | C | 0.38 | — | — | −12.37 | 0.094 |
| rs7182524 | T | C | 0.06 | — | — | — | — |
| rs692646 | A | G | 0.37 | — | — | — | — |
| rs8037376 | C | T | 0.32 | — | — | −17.39 | 0.027 |
| rs4144134 | T | C | 0.43 | — | — | −12.28 | 0.097 |
| rs685935 | C | T | 0.43 | — | — | −12.28 | 0.097 |
| rs8043049 | C | T | 0.33 | — | — | −19.32 | 0.012 |
| rs6493791 | G | A | 0.47 | — | — | −12.08 | 0.098 |
| rs16976343 | C | T | 0.14 | — | — | — | — |
| rs16976349 | C | T | 0.06 | — | — | — | — |
| rs16976351 | C | G | 0.07 | 19.12 | 0.079 | — | — |
| rs12594443 | T | C | 0.06 | — | — | — | — |
| rs17819126 | T | C | 0.06 | — | — | — | — |
| rs3743204 | T | G | 0.20 | — | — | — | — |
| rs3743205 | A | G | 0.05 | 25.46 | 0.085 | — | — |
| rs8040756 | A | G | 0.15 | 18.68 | 0.026 | — | — |
Bolded rows indicate SNPs that previous studies have identified as conferring risk for dyslexia.
MAF, minor allele frequency.
[Correction added after online publication 19 October 2010: in Table 2, major and minor allele were interchanged.]
Evaluation of intermarker LD showed high LD across the DYX1C1 locus (Fig. 1). The multiple association signals detected at different markers are most likely a reflection of the high LD background.
Figure 1. SNPs location and LD across DYX1C1.
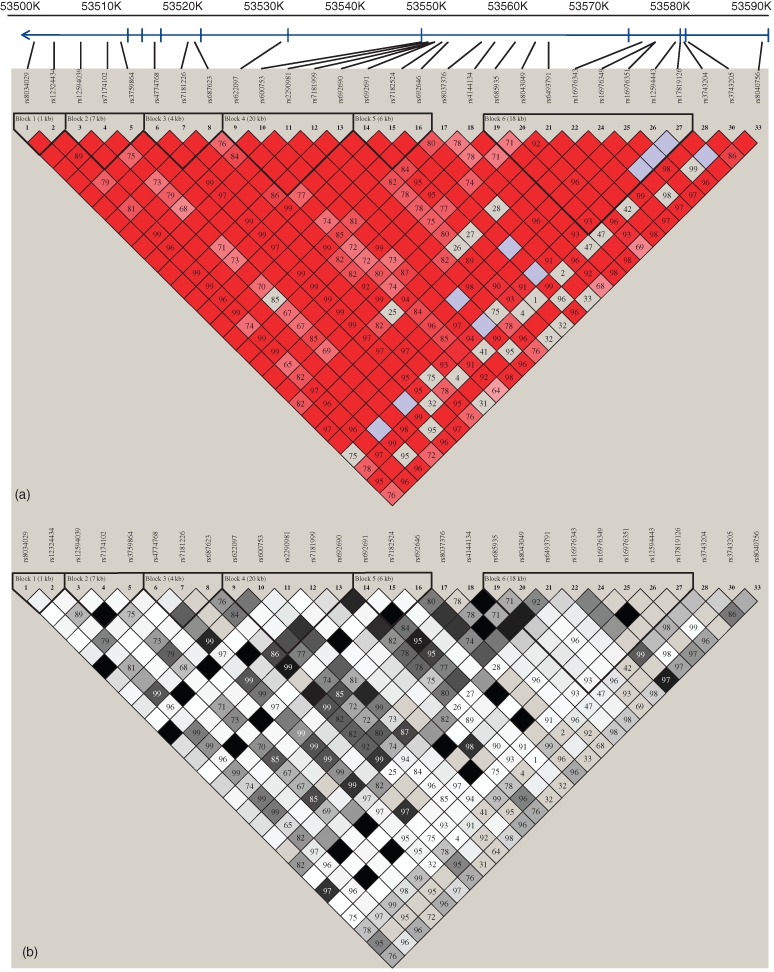
The top of the figure shows the structure of the DYX1C1 gene (blue arrow) indicating the position of exons (blue vertical lines), with an indication of their genomic location on chromosome 15. In total, 29 SNPs were analysed across an 86-kb interval. Black lines indicate the position of each SNP within DYX1C1. Inter-SNP LD was generated with Haploview. (a) D′ values are reported within cells while empty red cells represent full LD and empty blue cells represent lack of LD. (b) r2 values are reported within cells where empty white cells represent lack of LD and darker shadings represent increasingly stronger LD. Haploview identified six LD blocks (black solid lines) using the confidence interval method (Gabriel et al. 2002), but LD is strong all across the examined region.
Discussion
In the present study, we have investigated the effect of dyslexia candidate genes on reading and spelling abilities in the Raine sample representing the general population of WA. This is the first study describing a genetic investigation of cognitive traits in this sample, which has so far been used primarily for epidemiological investigations (Chivers et al. in press; O'Sullivan et al. in press). We detected nominal association signals for several markers within the DYX1C1 gene, further supporting the role of this gene in contributing to dyslexia and reading abilities more generally. Lack of association with the other genes does not rule out their involvement in dyslexia but their effect cannot be detected in the Raine sample. It is important to note that the reading and spelling WALNA tests used here are different from the tests generally used to ascertain dyslexia. Most of the dyslexia studies are based on test of single word reading or, as in the case of German samples, single word spelling (Schumacher et al. 2006). In contrast, the reading test used here is a reading comprehension test and the spelling test is based on the recognition of mistakes. The use of these particular measures combined with a small sample size, yielding limited power to detect genetic associations (Fig. S1), might have prevented the detection of additional genuine associations.
The association P-values reported here are weak. However, the association P-values reported previously in the literature for DYX1C1 have been relatively modest, even in samples selected for dyslexia (Paracchini et al. 2007). The reason could be that the DYX1C1 (as well as the other dyslexia candidate genes) effect size is very small. In addition, the discovery samples employed so far have also been of modest size (usually <1000 individuals). Therefore, if any of these genes would contribute to reading abilities in the Raine sample (which has not been selected for dyslexia), we would expect to observe similar levels of association with weak P-values.
The role of dyslexia candidate genes have not been completely established, and with the exception of the KIAA0319 gene (Dennis et al. 2009), functional molecular mechanisms have not been described to explain genetic associations. Replication analysis is currently the most valid approach to establish whether these genes indeed contribute to dyslexia and even if association do not reach full statistical significance, association trends may show interesting observation and provide additional evidence.
DYX1C1 has received the largest number of positive replications of all dyslexia susceptibility candidate genes together with different negative reports. The positive replications have been reported with different SNPs or with opposite alleles of the same SNPs lacking to provide unanimous consensus on the role of this gene. It is important therefore that the present study is considered in the context of these findings. Our data are consistent with previous studies reporting association within DYX1C1 and, in addition, provide novel elements to interpret of the current body of literature.
We show that in our sample of white European origin, LD is high all across DYX1C1 and may hinder refinement of the association signal by fine genetic mapping. Therefore, it is not possible to indicate a specific role of coding or regulatory SNPs simply on the basis of genetic associations. These markers, showing relatively stronger P-values in some studies (Bates et al. in press; Taipale et al. 2003), could simply reflect an a priori preferential bias in choosing SNPs with potential functional effects but actually acting as proxy of other functional DNA variants in the region. Consistently, our strongest association signals were not observed for markers that have been suggested to have a direct effect on the phenotype, such as rs3743205, rs57809907 (Taipale et al. 2003) or rs17819126 (Bates et al. in press). Sampling effects and missing data can also lead to differences in association patterns when markers are highly correlated and sample sizes are not particularly large. Our interpretation of the data available so far is that they are suggestive of at least one genuine functional variant within the DYX1C1 locus, the effect of which has been picked up by different genetic markers, all in high LD, depending on differences in the samples analysed. Therefore, caution should be applied when interpreting association with markers with a putative functional effect.
DYX1C1 is an attractive candidate for dyslexia, given its proposed role in neuronal migration and brain development (Wang et al. 2006). Neuronal migration has been indicated as a possible biological mechanism underlying dyslexia by neuroanatomical evidences (Galaburda et al. 1985). More recently, DYX1C1 has been shown to interact with estrogen receptors (Massinen et al. 2009). The estrogen pathway has been shown to be important during brain development (Wang et al. 2003).
In conclusion, our findings are consistent with previous reports in supporting the role of DYX1C1 in the etiology of dyslexia and modulating reading abilities. Our data suggest that additional work is needed to identify functional genetic variants relevant to dyslexia and to fully understand the function of this gene. However, the high LD across the gene may prevent further refinement of the association by genetic mapping and alternative strategies should be adopted.
Acknowledgments
The authors would like to acknowledge the National Health and Medical Research Council (NH&MRC) for their long-term contribution to funding the study over the last 20 years. Core Management of the Raine study has been funded by the University of Western Australia (UWA), the UWA Faculty of Medicine, Dentistry and Health Sciences, the Raine Medical Research Foundation, the Telethon Institute for Child Health Research and the Women's and Infants Research Foundation. The genotyping of the Raine cohort was funded by a project grant from the NH&MRC (572613). The authors are extremely grateful to the study participants and their families as well as the Raine study team for cohort co-ordination and data collection. S.P and A.P.M. are supported by the Wellcome Trust (9076566/Z/05/Z) and work in a Wellcome Trust-funded institute (075491/Z/04). We wish to thank Dr. Tom Scerri for useful comments.
Supporting Information
Additional Supporting Information may be found in the online version of this article:
Figure S1: Power analysis. Computed statistical power for the reading (a) and spelling (b) analyses, given a certain β-coefficient and minor allele frequency (MAF) and assuming a sample size of 500 participants.
Table S1: Summary of sample characteristics reported in the literature
Table S2: Descriptive information of SNPs previously reported in the literature to be associated with dyslexia [Correction added after online publication 19 October 2010: Table S2 has been resupplied.]
Table S3: Descriptive information of SNPs in the DYX1C1 gene
As a service to our authors and readers, this journal provides supporting information supplied by the authors. Such materials are peer-reviewed and may be re-organized for online delivery, but are not copy-edited or typeset. Technical support issues arising from supporting information (other than missing files) should be addressed to the authors.
References
- Anthoni H, Zucchelli M, Matsson H, Muller-Myhsok B, Fransson I, Schumacher J, Massinen S, Onkamo P, Warnke A, Griesemann H, Hoffmann P, Nopola-Hemmi J, Lyytinen H, Schulte-Korne G, Kere J, Nothen MM, Peyrard-Janvid M. A locus on 2p12 containing the co-regulated MRPL19 and C2ORF3 genes is associated to dyslexia. Hum Mol Genet. 2007;16:667–677. doi: 10.1093/hmg/ddm009. [DOI] [PubMed] [Google Scholar]
- Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21:263–265. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- Bates TC, Lind PA, Luciano M, Montgomery GW, Martin NG, Wright MJ. Dyslexia and DYX1C1: deficits in reading and spelling associated with a missense mutation. Mol Psychiatry. doi: 10.1038/mp.2009.120. (in press) (in press) [DOI] [PubMed] [Google Scholar]
- Bellini G, Bravaccio C, Calamoneri F, Donatella Cocuzza M, Fiorillo P, Gagliano A, Mazzone D, del Giudice EM, Scuccimarra G, Militerni R, Pascotto A. No evidence for association between dyslexia and DYX1C1 functional variants in a group of children and adolescents from Southern Italy. J Mol Neurosci. 2005;27:311–314. doi: 10.1385/jmn:27:3:311. [DOI] [PubMed] [Google Scholar]
- Brkanac Z, Chapman NH, Matsushita MM, Chun L, Nielsen K, Cochrane E, Berninger VW, Wijsman EM, Raskind WH. Evaluation of candidate genes for DYX1 and DYX2 in families with dyslexia. Am J Med Genet B Neuropsychiatr Genet. 2007;144B:556–560. doi: 10.1002/ajmg.b.30471. [DOI] [PubMed] [Google Scholar]
- Chivers P, Hands B, Parker H, Bulsara M, Beilin LJ, Kendall GE, Oddy WH. Body mass index, adiposity rebound and early feeding in a longitudinal cohort (Raine study) Int J Obes (Lond) doi: 10.1038/ijo.2010.61. (in press) (in press) [DOI] [PubMed] [Google Scholar]
- Cope N, Harold D, Hill G, Moskvina V, Stevenson J, Holmans P, Owen MJ, O'Donovan MC, Williams J. Strong evidence that KIAA0319 on chromosome 6p is a susceptibility gene for developmental dyslexia. Am J Hum Genet. 2005a;76:581–591. doi: 10.1086/429131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cope NA, Hill G, van den Bree M, Harold D, Moskvina V, Green EK, Owen MJ, Williams J, O'Donovan MC. No support for association between dyslexia susceptibility 1 candidate 1 and developmental dyslexia. Mol Psychiatry. 2005b;10:237–238. doi: 10.1038/sj.mp.4001596. [DOI] [PubMed] [Google Scholar]
- Dahdouh F, Anthoni H, Tapia-Paez I, Peyrard-Janvid M, Schulte-Korne G, Warnke A, Remschmidt H, Ziegler A, Kere J, Muller-Myhsok B, Nothen MM, Schumacher J, Zucchelli M. Further evidence for DYX1C1 as a susceptibility factor for dyslexia. Psychiatr Genet. 2009;19:59–63. doi: 10.1097/YPG.0b013e32832080e1. [DOI] [PubMed] [Google Scholar]
- Deffenbacher KE, Kenyon JB, Hoover DM, Olson RK, Pennington BF, DeFries JC, Smith SD. Refinement of the 6p21.3 quantitative trait locus influencing dyslexia: linkage and association analyses. Hum Genet. 2004;115:128–138. doi: 10.1007/s00439-004-1126-6. [DOI] [PubMed] [Google Scholar]
- DeFries JC, Alarcón M. Genetics of specific reading disability. Ment Retard Dev Disabil Res Rev. 1996;2:39–47. [Google Scholar]
- Dennis MY, Paracchini S, Scerri TS, Prokunina-Olsson L, Knight JC, Wade-Martins R, Coggill P, Beck S, Green ED, Monaco AP. A common variant associated with dyslexia reduces expression of the KIAA0319 gene. PLoS Genet. 2009;5:e1000436.. doi: 10.1371/journal.pgen.1000436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doig B, Groves S. Easier analysis and better reporting: modelling ordinal data in mathematics education research. Math Educ Res J. 2006;18:56–76. [Google Scholar]
- Finucci JM, Childs B. Are there really more dyslexic boys than girls? In: Ansara A, Geshwind N, Galaburda AM, Albert M, Gartrell M, editors. Sex Differences in Dyslexia. Towson, MD: Orton Dyslexia Society; 1981. pp. 1–9. [Google Scholar]
- Fisher SE, DeFries JC. Developmental dyslexia: genetic dissection of a complex cognitive trait. Nat Rev Neurosci. 2002;3:767–780. doi: 10.1038/nrn936. [DOI] [PubMed] [Google Scholar]
- Francks C, Paracchini S, Smith SD, Richardson AJ, Scerri TS, Cardon LR, Marlow AJ, MacPhie IL, Walter J, Pennington BF, Fisher SE, Olson RK, DeFries JC, Stein JF, Monaco AP. A 77-kilobase region of chromosome 6p22.2 is associated with dyslexia in families from the United Kingdom and from the United States. Am J Hum Genet. 2004;75:1046–1058. doi: 10.1086/426404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gabriel SB, Schaffner SF, Nguyen H, Moore JM, Roy J, Blumenstiel B, Higgins J, DeFelice M, Lochner A, Faggart M, Liu-Cordero SN, Rotimi C, Adeyemo A, Cooper R, Ward R, Lander ES, Daly MJ, Altshuler D. The structure of haplotype blocks in the human genome. Science. 2002;296:2225–2229. doi: 10.1126/science.1069424. [DOI] [PubMed] [Google Scholar]
- Galaburda AM, Sherman GF, Rosen GD, Aboitiz F, Geschwind N. Developmental dyslexia: four consecutive patients with cortical anomalies. Ann Neurol. 1985;18:222–233. doi: 10.1002/ana.410180210. [DOI] [PubMed] [Google Scholar]
- Guerin DW, Griffin JR, Gottfried AW, Christenson GN. Dyslexic subtypes and severity levels: are there gender differences? Optom Vis Sci. 1993;70:348–351. doi: 10.1097/00006324-199305000-00002. [DOI] [PubMed] [Google Scholar]
- Habib M. The neurological basis of developmental dyslexia: an overview and working hypothesis. Brain. 2000;123(Pt. 12):2373–2399. doi: 10.1093/brain/123.12.2373. [DOI] [PubMed] [Google Scholar]
- Hannula-Jouppi K, Kaminen-Ahola N, Taipale M, Eklund R, Nopola-Hemmi J, Kaariainen H, Kere J. The axon guidance receptor gene ROBO1 is a candidate gene for developmental dyslexia. PLoS Genet. 2005;1:e50.. doi: 10.1371/journal.pgen.0010050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harold D, Paracchini S, Scerri T, Dennis M, Cope N, Hill G, Moskvina V, Walter J, Richardson AJ, Owen MJ, Stein JF, Green ED, O'Donovan MC, Williams J, Monaco AP. Further evidence that the KIAA0319 gene confers susceptibility to developmental dyslexia. Mol Psychiatry. 2006;11:1085–1091. 1061.. doi: 10.1038/sj.mp.4001904. [DOI] [PubMed] [Google Scholar]
- Hawke JL, Olson RK, Willcut EG, Wadsworth SJ, DeFries JC. Gender ratios for reading difficulties. Dyslexia. 2009;15:239–242. doi: 10.1002/dys.389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaplan DE, Gayan J, Ahn J, Won TW, Pauls D, Olson RK, DeFries JC, Wood F, Pennington BF, Page GP, Smith SD, Gruen JR. Evidence for linkage and association with reading disability on 6p21.3-22. Am J Hum Genet. 2002;70:1287–1298. doi: 10.1086/340449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelman CW, Bass AJ, Holman CD. Research use of linked health data – a best practice protocol. Aust N Z J Public Health. 2002;26:251–255. doi: 10.1111/j.1467-842x.2002.tb00682.x. [DOI] [PubMed] [Google Scholar]
- Lind PA, Luciano M, Wright MJ, Montgomery GW, Martin NG, Bates TC. Dyslexia and DCDC2: normal variation in reading and spelling is associated with DCDC2 polymorphisms in an Australian population sample. Eur J Hum Genet. doi: 10.1038/ejhg.2009.237. ((in press)) (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lind PA, Luciano M, Wright MJ, Montgomery GW, Martin NG, Bates TC. Dyslexia and DCDC2: normal variation in reading and spelling is associated with DCDC2 polymorphisms in an Australian population sample. Eur J Hum Genet. 2010;18:668–673. doi: 10.1038/ejhg.2009.237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luciano M, Lind PA, Duffy DL, Castles A, Wright MJ, Montgomery GW, Martin NG, Bates TC. A haplotype spanning KIAA0319 and TTRAP is associated with normal variation in reading and spelling ability. Biol Psychiatry. 2007;62:811–817. doi: 10.1016/j.biopsych.2007.03.007. [DOI] [PubMed] [Google Scholar]
- Ludwig KU, Schumacher J, Schulte-Korne G, Konig IR, Warnke A, Plume E, Anthoni H, Peyrard-Janvid M, Meng H, Ziegler A, Remschmidt H, Kere J, Gruen JR, Muller-Myhsok B, Nothen MM, Hoffmann P. Investigation of the DCDC2 intron 2 deletion/compound short tandem repeat polymorphism in a large German dyslexia sample. Psychiatr Genet. 2008;18:310–312. doi: 10.1097/YPG.0b013e3283063a78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marino C, Citterio A, Giorda R, Facoetti A, Menozzi G, Vanzin L, Lorusso ML, Nobile M, Molteni M. Association of short-term memory with a variant within DYX1C1 in developmental dyslexia. Genes Brain Behav. 2007;6:640–646. doi: 10.1111/j.1601-183X.2006.00291.x. [DOI] [PubMed] [Google Scholar]
- Massinen S, Tammimies K, Tapia-Paez I, Matsson H, Hokkanen ME, Soderberg O, Landegren U, Castren E, Gustafsson JA, Treuter E, Kere J. Functional interaction of DYX1C1 with estrogen receptors suggests involvement of hormonal pathways in dyslexia. Hum Mol Genet. 2009;18:2802–2812. doi: 10.1093/hmg/ddp215. [DOI] [PubMed] [Google Scholar]
- Meng H, Hager K, Held M, Page GP, Olson RK, Pennington BF, DeFries JC, Smith SD, Gruen JR. TDT-association analysis of EKN1 and dyslexia in a Colorado twin cohort. Hum Genet. 2005a;118:87–90. doi: 10.1007/s00439-005-0017-9. [DOI] [PubMed] [Google Scholar]
- Meng H, Smith SD, Hager K, Held M, Liu J, Olson RK, Pennington BF, DeFries JC, Gelernter J, O'Reilly-Pol T, Somlo S, Skudlarski P, Shaywitz SE, Shaywitz BA, Marchione K, Wang Y, Paramasivam M, Loturco JJ, Page GP, Gruen JR. DCDC2 is associated with reading disability and modulates neuronal development in the brain. Proc Natl Acad Sci U S A. 2005b;102:17053–17058. doi: 10.1073/pnas.0508591102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newnham JP, Evans SF, Michael CA, Stanley FJ, Landau LI. Effects of frequent ultrasound during pregnancy: a randomised controlled trial. Lancet. 1993;342:887–891. doi: 10.1016/0140-6736(93)91944-h. [DOI] [PubMed] [Google Scholar]
- O'Sullivan TA, Ambrosini G, Beilin LJ, Mori TA, Oddy WH. Dietary intake and food sources of fatty acids in Australian adolescents. Nutrition. doi: 10.1016/j.nut.2009.11.019. (in press) (in press) [DOI] [PubMed] [Google Scholar]
- Paracchini S, Thomas A, Castro S, Lai C, Paramasivam M, Wang Y, Keating BJ, Taylor JM, Hacking DF, Scerri T, Francks C, Richardson AJ, Wade-Martins R, Stein JF, Knight JC, Copp AJ, Loturco J, Monaco AP. The chromosome 6p22 haplotype associated with dyslexia reduces the expression of KIAA0319, a novel gene involved in neuronal migration. Hum Mol Genet. 2006;15:1659–1666. doi: 10.1093/hmg/ddl089. [DOI] [PubMed] [Google Scholar]
- Paracchini S, Scerri T, Monaco AP. The genetic lexicon of dyslexia. Annu Rev Genomics Hum Genet. 2007;8:57–79. doi: 10.1146/annurev.genom.8.080706.092312. [DOI] [PubMed] [Google Scholar]
- Paracchini S, Steer CD, Buckingham LL, Morris AP, Ring S, Scerri T, Stein J, Pembrey ME, Ragoussis J, Golding J, Monaco AP. Association of the KIAA0319 dyslexia susceptibility gene with reading skills in the general population. Am J Psychiatry. 2008;165:1576–1584. doi: 10.1176/appi.ajp.2008.07121872. [DOI] [PubMed] [Google Scholar]
- Pennington BF. The genetics of dyslexia. J Child Psychol Psychiatry. 1990;31:193–201. doi: 10.1111/j.1469-7610.1990.tb01561.x. [DOI] [PubMed] [Google Scholar]
- Price AL, Patterson NJ, Plenge RM, Weinblatt ME, Shadick NA, Reich D. Principal components analysis corrects for stratification in genome-wide association studies. Nat Genet. 2006;38:904–909. doi: 10.1038/ng1847. [DOI] [PubMed] [Google Scholar]
- Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, Maller J, Sklar P, de Bakker PI, Daly MJ, Sham PC. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scerri TS, Schulte-Korne G. Genetics of developmental dyslexia. Eur Child Adolesc Psychiatry. 2010;19:179–197. doi: 10.1007/s00787-009-0081-0. [DOI] [PubMed] [Google Scholar]
- Scerri TS, Fisher SE, Francks C, MacPhie IL, Paracchini S, Richardson AJ, Stein JF, Monaco AP. Putative functional alleles of DYX1C1 are not associated with dyslexia susceptibility in a large sample of sibling pairs from the UK. J Med Genet. 2004;41:853–857. doi: 10.1136/jmg.2004.018341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schumacher J, Anthoni H, Dahdouh F, Konig IR, Hillmer AM, Kluck N, Manthey M, Plume E, Warnke A, Remschmidt H, Hulsmann J, Cichon S, Lindgren CM, Propping P, Zucchelli M, Ziegler A, Peyrard-Janvid M, Schulte-Korne G, Nothen MM, Kere J. Strong genetic evidence of DCDC2 as a susceptibility gene for dyslexia. Am J Hum Genet. 2006;78:52–62. doi: 10.1086/498992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaywitz SE. Dyslexia. N Engl J Med. 1998;338:307–312. doi: 10.1056/NEJM199801293380507. [DOI] [PubMed] [Google Scholar]
- Shaywitz SE, Shaywitz BA, Fletcher JM, Escobar MD. Prevalence of reading disability in boys and girls. Results of the Connecticut Longitudinal Study. JAMA. 1990;264:998–1002. [PubMed] [Google Scholar]
- Taipale M, Kaminen N, Nopola-Hemmi J, Haltia T, Myllyluoma B, Lyytinen H, Muller K, Kaaranen M, Lindsberg PJ, Hannula-Jouppi K, Kere J. A candidate gene for developmental dyslexia encodes a nuclear tetratricopeptide repeat domain protein dynamically regulated in brain. Proc Natl Acad Sci U S A. 2003;100:11553–11558. doi: 10.1073/pnas.1833911100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tapia-Paez I, Tammimies K, Massinen S, Roy AL, Kere J. The complex of TFII-I, PARP1, and SFPQ proteins regulates the DYX1C1 gene implicated in neuronal migration and dyslexia. Faseb J. 2008;22:3001–3009. doi: 10.1096/fj.07-104455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogel SA. Gender differences in intelligence, language, visual-motor abilities, and academic achievement in students with learning disabilities: a review of the literature. J Learn Disabil. 1990;23:44–52. doi: 10.1177/002221949002300111. [DOI] [PubMed] [Google Scholar]
- Wang L, Andersson S, Warner M, Gustafsson JA. Estrogen receptor (ER)beta knockout mice reveal a role for ERbeta in migration of cortical neurons in the developing brain. Proc Natl Acad Sci U S A. 2003;100:703–708. doi: 10.1073/pnas.242735799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Paramasivam M, Thomas A, Bai J, Kaminen-Ahola N, Kere J, Voskuil J, Rosen GD, Galaburda AM, Loturco JJ. DYX1C1 functions in neuronal migration in developing neocortex. Neuroscience. 2006;143:515–522. doi: 10.1016/j.neuroscience.2006.08.022. [DOI] [PubMed] [Google Scholar]
- Wigg KG, Couto JM, Feng Y, Anderson B, Cate-Carter TD, Macciardi F, Tannock R, Lovett MW, Humphries TW, Barr CL. Support for EKN1 as the susceptibility locus for dyslexia on 15q21. Mol Psychiatry. 2004;9:1111–1121. doi: 10.1038/sj.mp.4001543. [DOI] [PubMed] [Google Scholar]
- Wilcke A, Weissfuss J, Kirsten H, Wolfram G, Boltze J, Ahnert P. The role of gene DCDC2 in German dyslexics. Ann Dyslexia. 2009;59:1–11. doi: 10.1007/s11881-008-0020-7. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


