Abstract
Defining genetic networks underlying animal behavior in a high throughput manner is an important but challenging task that has not yet been achieved for any organism. Using Caenorhabditis elegans, we collected quantitative parametric data related to various aspects of locomotion from wild type and thirty-one mutant worm strains with single mutations in genes functioning in sensory reception, neurotransmission, G-protein signaling, neuromuscular control or other facets of motor regulation. We applied unsupervised and constrained K-means clustering algorithms to the data and found that the genes that clustered together due to the behavioral similarity of their mutants encoded proteins in the same signaling networks. This approach provides a framework to identify genes and genetic networks underlying worm neuromotor function in a high-throughput manner. A publicly accessible database harboring the visual and quantitative behavioral data collected in this study adds valuable information to the rapidly growing C. elegans databanks that can be employed in a similar context.
1. Introduction
Human and animal behaviors are regulated by genes acting in coordinated, often complex, networks. Delineation of these networks holds the key to understanding the genetic mechanisms underlying different behaviors. This goal remains largely unmet, due at least in part to the fact that genes have traditionally been studied individually for their roles in behavioral regulation. Since the data generated by such isolated studies is scattered in various resources, it has been difficult to systematically identify genetic networks regulating animal behaviors with modern data pattern recognition algorithms. Such algorithms have been successfully used to identify relationships between genes through systematic profiling of centralized and standardized data such as genomic sequences, gene expression levels, protein-protein interactions and cellular metabolic activity (Inoue et al. 2005; Lee et al. 2002; Patil and Nielsen 2005; Ren et al. 2000; Sandmann et al. 2007; Shlomi et al. 2008; Wikman et al. 2007). Therefore, the goal of the current study was to compile a behavioral data set to which pattern recognition algorithms could be applied to reveal the relationships between genes involved in regulating the behavior under study (in this case, locomotion).
The nematode C. elegans demonstrates a number of quantifiable behaviors, including locomotion (Rankin 2002). Worm locomotion is directly regulated by the worm neuromotor system and is closely associated with sensory input and experiences (de Bono and Maricq 2005; Giles and Rankin 2008). Aspects of worm locomotion are widely used to study neuronal and genetic mechanisms involved in neurotransmission (Zheng et al. 1999), sensory transduction (Ward et al. 2008), learning and memory (Rose and Rankin 2001), and drug dependence (Feng et al. 2006; Ward et al. 2009), although the specifics of behavioral measurement may differ for these different types of studies. Recently, a number of groups independently developed automated worm behavioral analysis systems that are capable of providing reliable and sensitive behavioral data (Baek et al. 2002; Cronin et al. 2006; Feng et al. 2004; Fontaine et al. 2006; Pierce-Shimomura et al. 2005; Simonetta and Golombek 2007; Tsibidis and Tavernarakis 2007). However, these systems were not designed to reveal the relationships among genes involved in regulation of a particular behavior or predict the signaling pathway of an involved gene.
Here, we hypothesized that mutations in genes regulating worm behaviors that function within the same genetic network would produce tractable behavioral signatures. Use of these signatures to place the variants into clusters in behavioral parametric space would be expected to unveil genetic pathways and functional partners of a given gene regulating the tracked behavior. As ‘proof of principle’, we first developed a behavioral analysis system that provided quantitative measurements of worm locomotion parameters. We then used this system to collect parametric behavior data from ~2000 animals representing 32 different worm genetic strains. The strains tested included wild type (WT) and 31 single gene mutants in which the affected genes are known to function in sensory reception, neurotransmission, G-protein signaling, neuromuscular junction signaling and/or have undefined roles in regulating worm movement. We next applied one unsupervised and two constrained K-means clustering algorithms on the resulting parametric behavioral data to observe natural clustering of these worm variants according to their behavioral signatures. As predicted, we found that genes that clustered together encoded proteins that function in the same signaling pathway. Therefore, this work provides a framework to identify genes and genetic networks underlying worm neuromotor function in a high-throughput manner. In addition, we have placed the visual and parametric behavioral data collected and analyzed in this study in a publicly accessible database which will serve as a useful research and educational resource.
2. Materials and Methods
2.1. Worm strains, culture and methods
Worm strains were cultured by standard methods. About 120 fourth-stage larvae were scored to a Nematode Growth Media (NGM) plate (stock plate) one day before each experiment and cultured overnight at 22°C. Forty young adults were randomly selected from the stock plate and tracked the following morning at 22 °C. The animals were acclimated in the tracking plates for 4 minutes before data acquisition. The tracking plates were prepared and worm visual and motion data acquisition was conducted as previously described (Feng et al. 2006).
2.2. Implementation of constrained K-means clustering algorithms
Measurement of distance between two data points is the fundamental first step for all clustering methods. Euclidean distance (ED) is commonly used in systematic biology including the classification of worm motor behavioral phenotypes (Baek et al. 2002; Geng et al. 2003; Geng et al. 2004). Therefore, we also chose ED. ED between two worm strains S1 and S2 is defined by Equation 1:
| Equation 1 |
where any feature value S’i (k) in S’i is defined as the difference between the same entity and that of the wild type strain (Equation 2):
| Equation 2 |
where N2j(k) is the wild type strain feature values.
We first adopted a constrained K-means clustering algorithm for data pattern recognition (Wagstaff et al. 2001), defined as CKMCA. In this algorithm, must-link constraints specify that two instances have to be in the same cluster and cannot-link constraints specify that two instances (worm strains) must not be placed in the same cluster. We implemented must-link and cannot-link as the following. 1) For strain k and strain j, if the number of strains other than k and j that was closer to k than j by their relative feature distance was no more than three, we added a must-link between k and j. 2) We selected six strains as the seeds of clusters (see main text for details) and stipulated that a cannot-link existed between any pair of the seed strains. Such a rule was applied to any strain s1 that was in the same cluster as seed Sa and strain s2 that was in the same cluster as seed Sb. Thus, any strain could not be grouped into more than one cluster.
During clustering, conflicts of must-links might exist. Specifically, must-links may be found between a non-seed strain si with multiple seeds, and thus require si to be grouped into more than one cluster. We might use the expression pattern of the involved genes to clear the conflicts. If the expression patterns of all involved genes were not available or were not sufficient to clear the conflicts, we used the following rule (Rule of Proximity) to resolve such a conflict.
For any non-seed strain si, si might connect with several seeds with different behavioral similarities. For any pair of seeds Sa and Sb competing for s1, we calculated the distance ranking between si and Sa (top 1 or top 2, etc.), defined as Ia, and the behavioral distance between si and Sb, defined as Ib, in behavioral parametric space according to Equation 1. If la < lb (top 1 < top 2, etc.), s1 was assigned to the cluster seeded with Sa, by reasoning that si displayed more behavioral similarity with Sa than Sb. If this did not resolve the conflict, we counted the number of must-links (Table 2) of si with all possible strain members seeded with Sa (na) or Sb (nb). If na > nb, si was assigned to the cluster seeded with Sa. We reasoned that the Rule of Proximity further measured and compared similarities between si and clusters seeded with Sa or Sb. In the present study, application of the Rule of Proximity resolved all conflicts.
Table 2.
List of Must-links for CKMCA
| Gene 1 | Gene 2 | Gene 1 | Gene 2 |
|---|---|---|---|
| bas-1 | dop-2 | mec-4 | dop-2 |
| bas-1 | mod-1 | mec-4 | mod-1 |
| bas-1 | mod-5 | mec-4 | mod-5 |
| cat-2 | cat-4 | mod-1 | dop-1 |
| cat-2 | dop-3 | mod-1 | mec-3 |
| cat-2 | eat-16 | mod-1 | mod-5 |
| cat-4 | dop-1 | mod-5 | mec-3 |
| cat-4 | eat-16 | mod-5 | mec-4 |
| cat-4 | mod-1 | mod-5 | mod-1 |
| dgk-1 | eat-16 | pde-4 | tdc-1 |
| dgk-1 | eat-4 | pde-4 | tph-1 |
| dgk-1 | gpb-2 | pde-4 | tbh-1 |
| dop-1 | mec-3 | tdc-1 | tbh-1 |
| dop-1 | mod-1 | tdc-1 | unc-13 |
| dop-1 | mod-5 | tdc-1 | unc-29(x29) |
| dop-2 | bas-1 | tph-1 | pde-4 |
| dop-2 | mec-4 | tph-1 | tdc-1 |
| dop-2 | mod-5 | tph-1 | unc-13 |
| dop-3 | bas-1 | tbh-1 | pde-4 |
| dop-3 | cat-2 | tbh-1 | tdc-1 |
| dop-3 | cat-4 | tbh-1 | unc-13 |
| eat-16 | cat-4 | unc-13 | tdc-1 |
| eat-16 | eat-4 | unc-13 | tph-1 |
| eat-16 | gpb-2 | unc-13 | tbh-1 |
| eat-4 | cat-4 | unc-18 | unc-29(e193) |
| eat-4 | dgk-1 | unc-18 | unc-73 |
| eat-4 | eat-16 | unc-18 | unc-2 |
| egl-10 | egl-8 | unc-29(x29) | tdc-1 |
| egl-10 | egl-30(md) | unc-29(x29) | tbh-1 |
| egl-10 | goa-1 | unc-29(x29) | unc-13 |
| egl-8 | goa-1 | unc-29(e193) | tbh-1 |
| egl-8 | mec-3 | unc-29(e193) | unc-36 |
| egl-8 | mod-5 | unc-29(e193) | unc-63 |
| egl-30(js) | eat-16 | unc-36 | tbh-1 |
| egl-30(js) | goa-1 | unc-36 | unc-29e |
| egl-30(js) | gpb-2 | unc-36 | unc-63 |
| egl-30(md) | egl-10 | unc-38 | tph-1 |
| egl-30(md)) | egl-8 | unc-38 | unc-13 |
| egl-30(md) | goa-1 | unc-38 | unc-63 |
| goa-1 | bas-1 | unc-43 | tdc-1 |
| goa-1 | mec-4 | unc-43 | tbh-1 |
| goa-1 | mod-5 | unc-43 | unc-29(x29) |
| gpb-2 | dgk-1 | unc-63 | unc-13 |
| gpb-2 | eat-16 | unc-63 | unc-29(e193) |
| gpb-2 | eat-4 | unc-63 | unc-36 |
| mec-3 | dop-1 | unc-73 | unc-29(e193) |
| mec-3 | mod-1 | unc-73 | unc-36 |
| mec-3 | mod-5 | unc-73 | unc-63 |
| unc-2 | unc-18 | ||
| unc-2 | unc-29(e193) | ||
| unc-2 | unc-36 |
Last, we applied CKMCA to cluster the strains, with the number of clusters set at six.
Alternatively, constraints were applied to a K-means clustering algorithm as described in the following. We first selected seeds for the total K clusters, one seed per cluster, as the centroids (ci) of the clusters, where the values of each behavioral parameter of the seeds are the coordinates of ci. Then, the K-mean algorithm was iteratively repeated to obtain and refine the clustering results. At each iteration, we assigned a non-seed strain si, to a cluster by must-link rules or the closest cluster by ranking its behavioral distances to the centroids of all clusters. Next, we updated the coordinates of ci with the mean behavior parametric values of all the current members in each cluster. The must-link rules were generated with known gene expression and gene functional information for a small proportion of genes (~1/3) (See details in the result section). The algorithm terminated when the clustering results converged. We called this algorithm ACKMCA.
3. Results
3.1. Development of a System for Automated and Quantitative Analysis of Nematode Behaviors
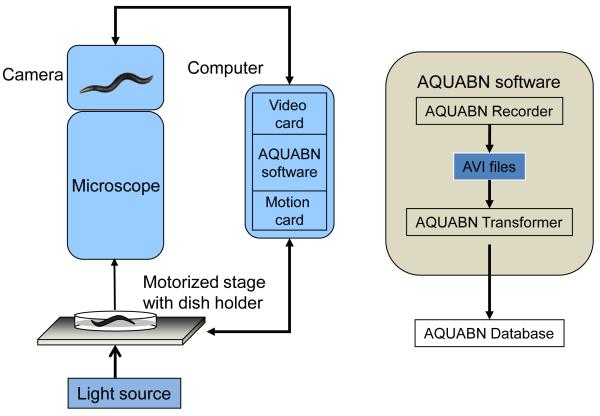
We previously developed a worm tracking system that quantifies various aspects of worm locomotion. This system was used to identify critical genes for proprioception (Li et al. 2006), substance dependence (Feng et al. 2006; Ward et al. 2009), and photoreceptor neuron identification (Ward et al. 2008) and resulted in identification of a behavioral predictor for worm ageing (Hsu et al. 2008). The system was also used in a number of other biomedical studies (Cao et al. 2010; Lee et al. 2005). We developed this system (named the Automated and Quantitative Analysis of Behaviors of Nematode (AQUABN) system, Figure 1) further by using 114 previously published worm behavioral or morphological parameters (Feng et al. 2004) and adding 47 novel behavioral parameters (see Supplemental Materials and Methods) mainly describing detailed aspects of worm reversal or directional changes. Thus, the AQUABN system provides a total of 161 behavioral parameters for quantifying speed, foraging, body waves, body posture, and four classes of worm locomotion behavior in addition to four parameters that measure worm morphology (Table S1).
Figure 1.
Schematic representation of the AQUABN system.
Left panel: hardware and software of the AQUABN system. Right panel: workflow chart of the AQUABN software.
3.2. Collecting and processing quantitative parametric data for worm locomotory behavior
We selected 32 strains including WT and 31 single mutation genetic variants representing mutations in 29 genes, most of which encode proteins from several well defined pathways implicated in neurobiological aspects of animal behavioral regulation: sensory perception, neuronal signal transduction and muscle contraction (Table 1).
Table 1.
Molecular identities of selected strains
| Gene | Allele | Molecular Identity | |
|---|---|---|---|
| bas-1 | ad446 | Aromatic amino acid decarboxylase | Dopamine neurotransmission |
| cat-2 | e1112 | Tyrosine hydroxylase | Dopamine neurotransmission |
| cat-4 | ok-342 | GTP cyclohydrolase | Dopamine and serotonin neurotransmission |
| dgk-1 | sy428 | Diacylglycerol kinase | Go signaling |
| dop-1 | ok398 | D1-like dopamine receptor | Dopamine neurotransmission |
| dop-2 | vs105 | D2-like dopamine receptor | Dopamine neurotransmission |
| dop-3 | vs106 | D2-line dopamine receptor | Dopamine neurotransmission |
| eat-16 | sa609 | A Regulator of G-protein signaling | G protein signaling |
| eat-4 | ad572 | Glutamate transporter | Glutamate neurotransmission |
| egl-10 | md176 | Regulator of G protein Signaling | G protein signaling |
| egl-30 | js126, md186 | Heterotrimeric G-protein (Gq) alpha subunit | Gq signaling |
| egl-8 | md1971 | Phospholipase C beta | Gq signaling |
| goa-1 | m1134 | Heterotrimeric G-protein (Go) alpha subunit | Go signaling |
| gpb-2 | sa603 | Heterotrimeric G protein beta subunit | G protein signaling |
| mec-3 | e1338 | Transcriptional regulator required for mechanosensory neuron | Mechanosensory |
| mec-4 | e1611 | Amiloride sodium channel required for mechanosensory function | Mechanosensory |
| mod-1 | ok103 | Serotonin-gated chloride channel | Serotonin neurotransmission |
| mod-5 | n822 | Serotonin transporter | Serotonin neurotransmission |
| pde-4 | ce268 | Cyclic nucleotide phosphodiesterase | Unknown function |
| tbh-1 | n3247 | Dopamine beta-hydroxylase | Octopamine neurotransmission |
| tdc-1 | ok914 | Aromatic-L-amino acid decarboxylase | Octopamine neurotransmission |
| tph-1 | mg280 | Tryptophan hydroxylase | Serotonin neurotransmission |
| unc-18 | n2813 | Homolog of MUNC-18, regulating synaptic vehicle docking | Synaptic regulation |
| unc-2 | e55 | Homolog of human CACNA1A, a calcium alpha subunit | Voltage-gated calcium channel |
| unc-29 | x29, e193 | Nicotinic acetylcholine receptor non-alpha subunit | Neuromuscular junction |
| unc-36 | ad698 | L-type Calcium channel alpha subunit | Voltage-gated calcium channel |
| unc-38 | x20 | Nicotinic acetylcholine receptor alpha subunit | Neuromuscular junction |
| unc-43 | sa200 | Calcium/calmodulin-dependent protein kinase | Synaptic regulation |
| unc-63 | x13 | Nicotinic acetylcholine receptor alpha subunit | Neuromuscular junction |
| unc-73 | gm33 | Guanine nucleotide exchange factor | Unknown function |
Additional information regarding the roles of these genes in worm locomotion regulation is available in Supplemental Materials.
We collected 1991 video clips of 1991 animals from these 32 strains with the AQUABN Recorder. Each video clip provided visual and motion data from a four minute tracking session of one animal at a 10 Hz frame rate. Each video clip was processed by the AQUABN Transformer to obtain data for 161 behavioral parameters, which were then exported to the AQUABN Behavioral Database as a single data entry (Figure 1) (available online at http://beijing.case.edu/worm/).
3.3. Defining the distance between behavior parameters of different worm strains
We chose Euclidean distance (ED) to measure the effects of genetic mutations on various parameters of worm locomotion. ED has been used in a number of systematic studies to investigate the roles of genes in worm behavior regulation (Baek et al. 2002; Geng et al. 2003; Geng et al. 2004). We first standardized our behavioral parametric dataset to values between 0 and 1 with the min-max method (Theodoridis and Koutroumbas 2009) to avoid the bias in clustering generated by uneven scales of behavioral parameter quantification and to suppress outliers introduced by noise and errors during behavioral parameter quantification. It was previously shown that different standardization methods have little or no effects on pattern recognition of worm behavioral data (Geng et al. 2003). We then selected the median value (Fmedian) of a behavioral parameter (f) calculated by using all animals from a given worm strain (s) to represent f of s in the distance computation. Each s has m (161) Fmedian values: F1, F2, …, Fm. The ED between two worm strains (si and sj) was defined as where is the kth Fmedian value of si. The distance matrix of 31 strains is shown in Table S2.
3.4. Choosing a data mining algorithm
The AQUABN system was initially designed to quantify subtle modifications in worm behavior elicited by various genetic or environmental means. To maximize its sensitivity and reliability, we used several different computational algorithms combined with a statistical approach to quantify the same or similar worm behavioral aspects (Feng et al. 2004). Therefore, a significant proportion of measured behavioral parameters provided by the AQUABN system could be either completely redundant or correlated with other parameters. Defining worm behavioral signatures with a K-means clustering algorithm (Baek et al. 2002; Geng et al. 2004) or classifying worm locomotion phenotypes with principle component analysis (PCA) (Geng et al. 2003), however, obtained similar if not the same results when using the full or a subset of worm locomotion parameters where redundant data was removed. These studies also demonstrated that the variability of worm parametric locomotion data within the same genetic mutant is significantly smaller than that of worms from different genetic background including unc mutants, and that the locomotion difference among various worm strains is not dominated by one or a subgroup of behavioral parameters. Using our dataset and PCA, we reached the same conclusions (data not shown).
We decided to use the K-means clustering algorithm, a useful general clustering method for many applications (Han and Kamber 2006) including defining worm locomotion signature (Baek et al. 2002). In addition, we reasoned that certain biological background information would constitute a valuable input to facilitate or guide data pattern recognition in this study. The recently developed CKMCA was adapted to incorporate such background information into the clustering process. In CKMCA, domain background information undergoes two kinds of restrictions, must-links and cannot-links, representing pairs of data entries that should and should not be clustered together (Wagstaff et al. 2001).
CKMCA was implemented as follows. First, we arbitrarily selected six clusters, each representing one of the six different means that regulate worm locomotion listed: dopamine neurotransmission (cluster 1), Go signaling (cluster 2) and, Gq signaling (cluster 3), sensory input (cluster 4), fundamental synaptic function (cluster 5), and “undefined” (cluster 6). Except for the “undefined” cluster seeded with pde-4 (a gene that impacts locomotion, but has little information available regarding its biological function), we could select a worm strain from each cluster to form 1 set of seeds. One advantage of CKMCA is that it is possible to use different numbers (k values) and/or sets of genes to serve as seeds in the analysis. There are over hundreds of sets of possible seed combinations. We randomly, with some biological input, chose 3 sets of seeds, set 1 (dop-1, dgk-1, egl-30 (md186), mec-3, unc-13 and pde-4), set 2 (dop-2, goa-1, egl-30 (md186), mec-3, unc-18, pde-4) and set 3 (cat-2, dgk-1, egl-8, mec-4, unc-13, pde-4) (Table 1). We obtained a similar data pattern with these different sets of seeds. Hence, only one set of results were presented below.
We further stipulated that no pair of seeds could be clustered together, the latter representing the only cannot-link restriction. We next defined the must-link restrictions. For a strain k, if the relative distance between k and another strain j ranked in the top-three of the shortest in the distance matrix (Table S2), we reasoned that k and j shared sufficient similarity in their behavioral signatures to cluster them together. Hence, we added a must-link between k and j in the must-link table as an entry (Table 2). In the data set of 32 strains, defining must-link tables with top-one, top-two, or top-four ranking in behavioral similarity led to too low (top-one and top-two) or too high (top-four) must-link restriction and a consequent failure of data pattern recognition (data not shown).
3.5. Data pattern recognition
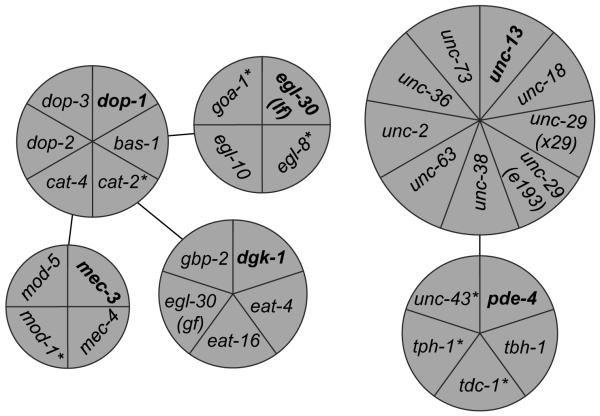
Next, we applied CMKCA, with the six cluster seeds and the restrictions described above, to our standardized behavior parametric dataset. 18 of the 26 non-seed strains unambiguously segregated into one of the six seeded clusters (Figure 2), demonstrating that most strains formed natural clusters according to their behavioral similarities. Each of the remaining 7 non-seed strains could be segregated into two different clusters. In this situation, the expression pattern of representative genes was the first input to guide clustering. If such information was not available or inadequate, we applied the Rule of Proximity (see Supplemental Materials and Methods for details), generated to further measure behavioral similarity. For supervised algorithms of data pattern recognition, human input is essential to facilitate data pattern recognition and generate more accurate and meaningful results (Mitchell 1997).
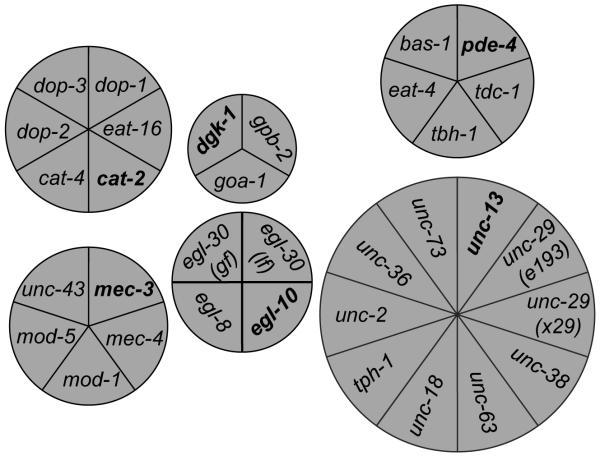
Figure 2.
Clustering of genes according to the behavioral signature of their genetic mutants.
The selected genes segregated into six clusters according to the behavioral signature of their mutants. Seed worm strains are indicated in bold font. While eighteen variants unambiguously fell into a single cluster without supervision, seven variants could fit into two clusters. In these cases, the two involved clusters are connected with lines and the variants are labeled with asterisks. The area of each section and the spatial position of each circular diagram are only for presenting purpose without biological meaning. egl-30 (lf) and egl-30 (gf) are a loss-of-function (md186) or a gain-of-function (js126) allele of egl-30, respectively. We also used two loss-of-function mutations of unc-29 (x29 and e193).
The first cluster, seeded with the dopamine receptor gene dop-1, contained two genes encoding dopamine synthases (bas-1 and cat-4) and two other dopamine receptor genes (dop-2 and dop-3). The loss-of-function mutant of cat-2, encoding another dopamine synthase, shared behavioral similarity with both dop-1- and dgk-1-seeded clusters. We grouped cat-2 into the cluster seeded with dop-1, because it has a lock-and-key relationship with dop-1 in its protein expression pattern (functional partner pair expressed in pre-/post synaptic neurons) (Baruch et al. 2008). The close behavioral similarity between the cat-2 mutant and G-protein signaling mutants (in the dgk-1-seeded cluster) is consistent with the observation that dopamine regulates worm locomotion by activating antagonistic Gq/Go signaling via D1- and D2-like receptors (Chase et al. 2004).
The second cluster, seeded with dgk-1, contained eat-16, which encodes a regulator of G-protein signaling (RGS) that affects locomotion through both Go and Gq signaling, gpb-2, which encodes a G-protein beta-subunit that may interact with both Go and Gq signaling, and a gain-of-function mutant of egl-30, which encodes the alpha-subunit of Gq. It was previously reported that both Go and Gq regulate worm locomotion in a coordinated network (Bastiani and Mendel 2006). The dgk-1-seeded cluster also included eat-4, which encodes a vesicular glutamate transporter (Lee et al. 1999). This suggests that some aspects of worm locomotion regulated by glutamate neurotransmission are performed through the Go/Gq signaling network.
The third cluster, seeded with egl-30 (lf), included egl-10, egl-8 and goa-1. The egl-10 gene encodes an RGS protein that interacts with both Go and Gq signaling to regulate locomotion. Loss-of-function mutants of goa-1 and egl-8 fell into two clusters, dop-1- and egl-30 (md186)-seeded. This is consistent with the observation that goa-1 and elg-8 are involved in behavioral regulation of dopamine neurotransmission (Chase et al. 2004). Because of the Rule of Proximity, goa-1 and egl-8 were placed in the cluster seeded with egl-30 (md186). Segregation of goa-1 into the cluster seeded with egl-30 is consistent with the notion that Go and Gq regulate many aspects of worm locomotion in a highly coordinated genetic network (Bastiani and Mendel 2006) and that the ED algorithm that we used to generate our distance matrix (Table S2) does not consider whether mutations regulate such behavioral parameters negatively or positively (see Supplemental Material and Method for details).
The fourth cluster, seeded with mec-3, included mec-4, mod-1 and mod-5, indicating that mutation of these genes produced similar changes in locomotory behavior. Similar to mec-3, the loss-of-function mutant of mec-4, which encodes an amiloride-sensitive sodium channel protein, exhibits degenerated touch sensory neurons causing defective mechanosensory perception (Bianchi et al. 2004; Way and Chalfie 1988). On the other hand, mod-1 and mod-5 encode a serotonin-gated chloride channel and a Na+, Cl−-dependent serotonin transporter, which is required for serotonin uptake, respectively. Although interaction between serotonin and mechanosensory perception has not been established in worms, serotonin has been reported to suppress the release of neurotransmitters from mechanosensory neurons and modify mechanosensory-related behaviors of medicinal leeches (Gaudry and Kristan 2009). Our data suggests that modulation of mechanosensory neuron output by serotonin neurotransmission might be conserved in C. elegans.
The fifth cluster, seeded with pde-4, contained tbh-1, tdc-1 and tph-1. Since these genes all encode neurotransmitter synthases, this finding suggests that pde-4 plays a role in synaptic function, possibly through affecting neurotransmitter synthesis or packing. This is consistent with a previous report showing that PDE-4 is located in an inactive zone of synapses throughout the worm nervous system (Charlie et al. 2006). Interestingly, unc-43, which encodes the type II calcium/calmodulin-dependent protein kinase (CaMKII) that regulates synaptic strength and maturation (Rongo and Kaplan 1999), also clustered with pde-4. Mutants of tdc-1, tph-1 and unc-43 also shared behavioral similarity with the unc-13-seeded cluster. They were supervised into the pde-4-seeded cluster because of the Rule of Proximity.
The last cluster, seeded with unc-13, which encodes a protein that regulates the release of neurotransmitters at the synapse, contained unc-18, unc-38, unc-63, unc-29, unc-36, unc-2 and unc-73, loss-of-function mutants of which demonstrated severe locomotion defects. These genes encode homologs of mammalian MUNC18 (unc-18), a syntaxin binding protein that enables vesicle docking in synaptic regions (Hata et al. 1993), nicotinic receptor genes (unc-38, unc-29, and unc-63) (Culetto et al. 2004), which are required at neuromuscular junctions (NMJ) for muscle contraction, and excitatory neuronal N-type calcium channels (unc-2 and unc-36) that regulate muscle activity at NMJs (Schafer and Kenyon 1995; Schafer et al. 1996). The unc gene group also included unc-73, which encodes a nucleotide exchange factor with an undefined role in behavioral regulation (Steven et al. 1998). These genes likely clustered together due to their diverse roles in regulating muscle activity as indicated by the sluggish locomotion of their mutants (See Discussion for details).
3.6. Data pattern validation
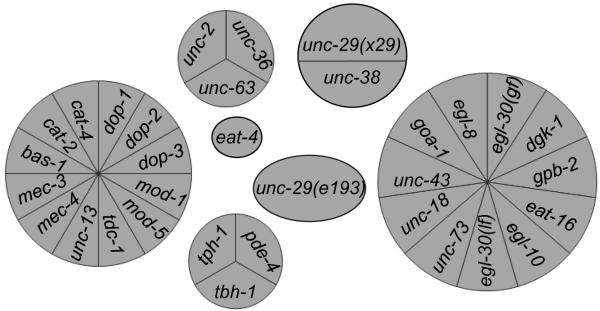
To validate the performance of CMKCA, we first used the unsupervised K-means clustering method that succeeded in defining the worm locomotion phenotype (Geng et al. 2003) (Figure 3). We found that thirty-one strains self-aggregated into 7 clusters according to their locomotion signature: one cluster mainly containing genes of G protein pathways, one cluster consisting of genes related to dopamine/serotonin neurotransmission and touch sensory, a group of pde-4 and two genes encoding neurotransmitters, a group of 3 genes that primarily function at neuromuscular junctions (NMJ) to regulate muscle contraction (unc-63, unc-36, unc-2), a cluster of unc-38 and an allele of unc-29, two genes encoding nAChR receptor subunits, and isolated eat-4 and another allele of unc-29 (Figure S1). Although there is no available “golden rule” to quantify the difference between clustering results generated with the unsupervised method and CMKCA, the latter clearly provides more biologically meaningful data pattern recognition. Specifically, genes regulating worm locomotion through sensory input and several types of neurotransmission were indistinguishable in data pattern recognition with the unsupervised method. Although genes of the Go and Gq pathways were consistently grouped together, unc-13 and unc-18, two genes required to release neurotransmitters at synapses were separated. Moreover, two loss-of-function alleles of unc-29 were not clustered together. However, the unsupervised K-means algorithm grouped unc-43, unc-18 and unc-73 with genes of G protein signaling pathways, consistent with several reports that the G protein signaling pathway interacts with unc-43 (Robatzek and Thomas 2000), unc-18 (Johnson et al. 2009) and unc-73 (Williams et al. 2007) to regulate synaptic transmission in locomotion regulation. This observation indicates that an unsupervised algorithm has some strength over a supervised method, possibly resulting from the fact that no assumption on the number and members of clusters that were made. With an unsupervised clique-based clustering algorithm (Edachery et al. 1999) that was demonstrated to discover intrinsic data patterns in high dimensional data sets with redundant and correlated features (Pei et al. 2005; Yan et al. 2005), we obtained a similar data pattern as the one presented in Figure 3 (data not shown) and reached the same conclusions.
Figure 3.
Self-aggregation of worm strains according to the behavioral signature of their genetic mutants using an unsupervised K-means clustering algorithm.
The selected genes self-aggregated into five clusters and two isolated genes according to the behavioral signature of their mutants. The area of each section and the spatial position of each circular diagram are only for presenting purposes without biological meaning.
We next used ACKMCA to cross validate the results obtained with CKMCA. First, we reasoned that gene expression patterns and functional data of a proportion of strains could be used to supervise clustering, this generated two rules. 1) Strains representing genes that have a lock-and-key relationship (Baruch et al. 2008) (for example, cat-2 and dop-1) or encoding subunits of the same receptor (for example, unc-63, unc-38 and unc-29) must be grouped together. 2). Strains representing genes that have clear functional relationship in behavioral regulation (for example, mec-3 and mec-4) should also be grouped together. We further reasoned that the expression and functional information of a small portion of strains is sufficient to guide the clustering of a larger population of strains based on their behavioral similarity. We thus defined a must-link table (Table 3) to define 1) the lock-and-key relationship of cat-2, a dopamine-specific synthase, and three dopamine receptors; 2) three genes encoding subunits of the same acetylcholine receptor (unc-63, unc-38 and unc-29); 3) two genes related to touch sensory (mec-3 and mec-4) and 4) two strains related to Gq signaling (egl-30 (lf) and egl-8). We chose the same number of clusters (six) but a different set of seeds (dop-1, dgk-1, egl-10, pde-4 and unc-63) to keep the biological significance consistent with the result of Figure 2. We found that the data pattern recognized by CKMCA is largely conserved with the clustering results produced with ACKMCA (Figure 4). The few differences are listed below. 1) bas-1, a gene required for the synthesis of both dopamine and serotonin was grouped with eat-4, encoding a vesicular glutamate transporter, and three neurotransmitter sythases (tbh-1, tdc-1 and tph-1). 2) unc-43, encoding CaMKII, was grouped into the cluster seeded with mec-3, suggesting that unc-43 may regulate some aspects of mechanosensory- or serotonin-regulated locomotion (Donohoe et al. 2008; Robatzek and Thomas 2000). 3) tph-1 was surprisingly grouped together with unc mutants, possibly due to the variety of strains in the unc group.
Table 3.
List of Must-links for ACKMCA.
| Gene 1 | Gene 2 | Biological reason |
|---|---|---|
| cat-2 | dop-1 | lock-and-key expression |
| cat-2 | dop-2 | lock-and-key expression |
| cat-3 | dop-3 | lock-and-key expression |
| mec-3 | mec-4 | regulating mechanosensory |
| egl-30 (gf) | egl-8 | components of Gq signaling |
| unc-63 | unc-29 (e193) | subunits of the same acetylcholine receptor |
| unc-63 | unc-29 (x29) | subunits of the same acetylcholine receptor |
| unc-63 | unc-38 | subunits of the same acetylcholine recptor |
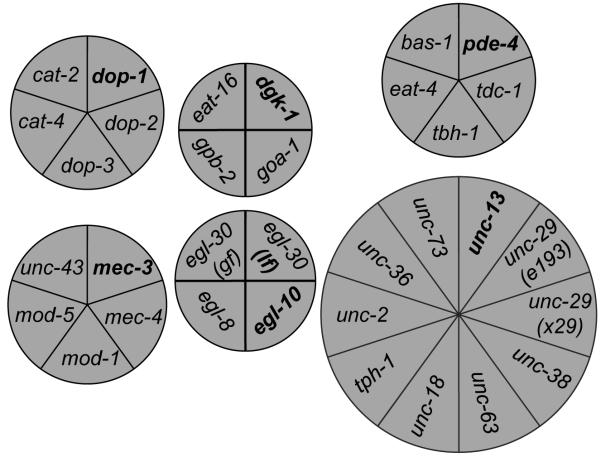
Figure 4.
Clustering of genes according to the behavioral signature of their genetic mutants with ACKMCA.
The selected genes segregated into six clusters with ACKMCA according to the behavioral signature of their mutants. Seed worm strains are indicated in bold font. The area of each section and the spatial position of each circular diagram are only for presenting purposes without biological meaning.
Previously, it was reported that redundant or correlated behavioral features have little effect on the results of defining worm behavioral signatures (Baek et al. 2002; Geng et al. 2004) or classifying worm locomotion phenotypes (Geng et al. 2003) with data mining algorithms. To explore whether redundant or correlated behavioral features distort recognized gene relationship in behavioral space, we removed features 148-158 (Table S1) containing many missing values, and features 48-50, 54, 57, 109, 110, 112-114, which had more than 0.9 Pearson product-moment correlation coefficient with other features (Rodgers and Nicewander 1988). We obtained the same data pattern except that eat-16 was clustered into the dopamine neurotransmission group (Figure 5). This observation is consistent with the report that an eat-16 mutation suppressed dopamine-mediated paralysis (Chase et al. 2004). Therefore, our results further confirmed that redundant/correlated behavioral features have little effect on the result of data mining algorithms.
Figure 5.
Effect of redundant/correlated data on supervised clustering genes according to the behavioral signature of their genetic mutants.
The selected genes segregated into six clusters with ACKMCA according to the behavioral signature of their mutants. Seed worm strains are indicated in bold font. The area of each section and the spatial position of each circular diagram are only for presenting purposes without biological meaning.
To analyze the effects of the ACKMCA algorithm on various data set size (number of behavioral parameters and genes), we generated a new reduced data set by removing redundant/correlated behavioral parameters, as we did above, and unc strains. Next, we applied the ACKMCA algorithm to this data set. By comparing the results on the full data set and the reduced data set, we could gain insights on how the results of the supervised clustering algorithm change with various data set sizes (Figure 6). We found that tph-1 was clustered together with two serotonin receptors (mod-1 and mod-5), supporting that tph-1 was clustered into the unc group, in results shown in Figure 4 and Figure 5, due to the variety of strains in the unc group.
Figure 6.
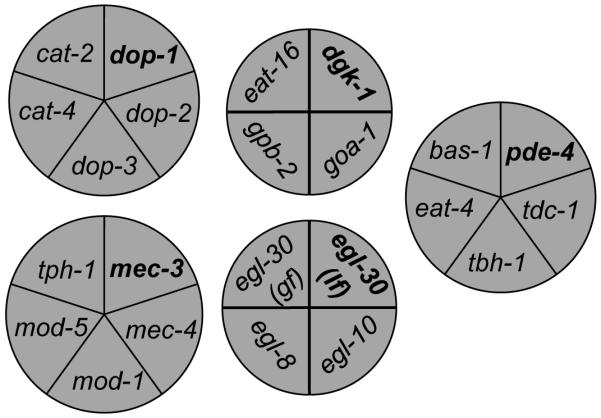
Effect of data size on supervised clustering genes according to the behavioral signature of their genetic mutants.
The selected genes segregated into five clusters with ACKMCA according to the behavioral signature of their mutants. Seed worm strains are indicated in bold font. The area of each section and the spatial position of each circular diagram are only for presenting purposes without biological meaning.
To further address how data size affects clustering result, we used the dataset with redundant/correlated data removed, but all the strains were kept. We randomly selected behavioral data from 50, 40, 30, 20 and 10 animals per strain and applied ACKMCA. We found that the data pattern was largely conserved even when we reduced the number of animals per strain to 30 (Figure S1-S3). Further reduction of sample size did not change aggregation results. This observation indicates that AQUABN reliably quantifies many worm behavioral aspects and that the variation of individuals in a strain is significantly smaller than variation among strains. A similar conclusion was previously reached (Geng et al. 2003; Geng et al. 2004).
4. Discussion
Our results demonstrate that quantitative worm behavioral parameter data provided by an automated system such as AQUABN can be paired with a supervised data pattern recognition algorithm to identify genetic networks regulating worm behavior. We found that a specific set of seeds did not significantly affect data pattern recognition of worm locomotion parameters but biological background knowledge is a critical input to achieve better results. In life science research, it is common that profiling of the relationship of genes/proteins in certain phenotypes is conducted under the scenario where the sequences, gene expression patterns, or protein function/homology of full or a proportion of the genetic or proteomic groups is known. Such domain-specific knowledge was utilized before and/or after running the data mining algorithm to facilitate data analysis or prune data patterns (Nierman et al. 2005; Ross-Macdonald et al. 1999), although this pre-knowledge was not necessarily utilized as an integral part of the data mining algorithms. Albeit supervised clustering methods were demonstrated to provide more meaningful data pattern recognition than traditional unsupervised algorithms (Eric et al. 2004) and could be valuable to biomedical research as this is, to the best of our knowledge, the first time CMKCA has been applied to life science.
The connectivity pattern of all 302 worm neurons has been well characterized at the electron microscopic (EM) level. Moreover, the expression patterns of hundreds of worm neuronal genes have already been identified and efforts are ongoing to provide expression patterns for the whole worm genome in the near future. In addition, knockout mutants of the entire worm genome should be available within the next several years. These advances, together with the collection of a neuromotor behavioral parametric dataset that covers most of the worm genome, will allow systematic analysis of the relationship between genes and neuromotor behavior on a genome-wide scale by applying supervised data mining algorithms to these databanks.
Although human supervision was helpful in recognizing a meaningful data pattern for the dataset presented in this study, where the biological function of most of the genes was partially studied, whether a supervised algorithm is a better method over the unsupervised method is still an open question when the number of mutant strains is increased to cover a significant proportion of the worm genome. In the latter case, little information is known regarding the function of most genes in behavioral regulation. Here, we found that some biological information of a small group of strains can be used to quickly and consistently identify interesting data patterns to unveil the relationship among genes in regulating worm locomotion behaviors. In contrast, an unsupervised data mining algorithm may identify novel relationship which may be missed by supervised methods. This is because unsupervised algorithms do not assume any existing functional relationships among genes, and may be useful in addressing some aspects of behavioral regulation, especially in cases where genes are involved in multiple signaling pathways. It is probably wise to use both methods and explain data patterns as a whole.
In this study, we also found that sample size (the number of animals per strain) has a minor effect on clustering results. This may reflect that the AQUABN system provides reliable worm locomotion quantification and that the behavioral data variation within stains is significantly smaller than the behavioral variation among strains in this particular study. In general, the larger the sample size, the more reliable and consistent the recognized data pattern will be with data mining algorithms.
However, even the approach used here to explore the relationship between genes and neuromotor behavior has some limitations. For example, unc mutant worms are extremely sluggish and frequently coil themselves without demonstrating obvious locomotion. Notably, all of the unc mutants, with the exception of unc-43, clustered together in this study. Thus, quantifying the unc behavioral phenotype represents a challenge (Baek et al. 2002; Geng et al. 2003; Geng et al. 2004; Huang et al. 2006). Previously, behavioral parameters were used to separate some unc phenotypes (Geng et al. 2003), suggesting that it is possible to develop more sophisticated computational and data mining algorithms for more accurate phenotype quantification and better data pattern recognition. It is also worth emphasizing that profiling quantifiable behavioral phenotypes only supplements traditional approaches focused on elucidating genetic mechanisms underlying worm behaviors. This is because experimental conditions in our study were optimized to provide standardized data for data mining algorithms, not to study the genetic mechanism involved in a specific behavior.
Online nematode research databases such as Wormbase (Harris et al. 2010) provide useful research and educational resources. However, these databanks often contain only descriptive information on behavioral phenotypes of worm genetic mutants. Sharing of visual and quantitative parametric worm neuromotor behavior data, as was done in the present study will provide the scientific community with a novel and useful research and educational resource. For instance, clustering methods based on ED computation evaluate the dataset as a whole. It is difficult, if not impossible, to explore which specific behavioral parameter(s) were commonly altered by a specific subgroup of mutants. Our publicly accessible data allows researchers to use other data analysis tools to explore these questions.
Supplementary Material
Acknowledgements
The authors thank Kristopher Kramp, Drs. Shawn X.Z. Xu and Bing Ren for critical reading of this manuscript. This work was supported by a Case Western Reserve University President’s Research Initiative Award (PRI Award) to ZF and JY, and NIH grant R01GM083241 to X.Z.S. Xu and ZF. ZF is a Mt. Sinai Scholar.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Reference
- Baek JH, Cosman P, Feng Z, Silver J, Schafer WR. Using machine vision to analyze and classify Caenorhabditis elegans behavioral phenotypes quantitatively. J Neurosci Methods. 2002;118:9–21. doi: 10.1016/s0165-0270(02)00117-6. [DOI] [PubMed] [Google Scholar]
- Baruch L, Itzkovitz S, Golan-Mashiach M, Shapiro E, Segal E. Using expression profiles of Caenorhabditis elegans neurons to identify genes that mediate synaptic connectivity. PLoS Comput Biol. 2008;4:e1000120. doi: 10.1371/journal.pcbi.1000120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bastiani C, Mendel J. WormBook. 2006. Heterotrimeric G proteins in C. elegans; pp. 1–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bianchi L, Gerstbrein B, Frokjaer-Jensen C, Royal DC, Mukherjee G, et al. The neurotoxic MEC-4(d) DEG/ENaC sodium channel conducts calcium: implications for necrosis initiation. Nat Neurosci. 2004;7:1337–1344. doi: 10.1038/nn1347. [DOI] [PubMed] [Google Scholar]
- Cao P, Yuan Y, Pehek EA, Moise AR, Huang Y, et al. Alpha-synuclein disrupted dopamine homeostasis leads to dopaminergic neuron degeneration in Caenorhabditis elegans. PLoS ONE. 2010;5:e9312. doi: 10.1371/journal.pone.0009312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charlie NK, Thomure AM, Schade MA, Miller KG. The Dunce cAMP phosphodiesterase PDE-4 negatively regulates G alpha(s)-dependent and G alpha(s)-independent cAMP pools in the Caenorhabditis elegans synaptic signaling network. Genetics. 2006;173:111–130. doi: 10.1534/genetics.105.054007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chase DL, Pepper JS, Koelle MR. Mechanism of extrasynaptic dopamine signaling in Caenorhabditis elegans. Nat Neurosci. 2004;7:1096–1103. doi: 10.1038/nn1316. [DOI] [PubMed] [Google Scholar]
- Cronin CJ, Feng Z, Schafer WR. Automated imaging of C. elegans behavior. Methods Mol Biol. 2006;351:241–251. doi: 10.1385/1-59745-151-7:241. [DOI] [PubMed] [Google Scholar]
- Culetto E, Baylis HA, Richmond JE, Jones AK, Fleming JT, et al. The Caenorhabditis elegans unc-63 gene encodes a levamisole-sensitive nicotinic acetylcholine receptor alpha subunit. J Biol Chem. 2004;279:42476–42483. doi: 10.1074/jbc.M404370200. [DOI] [PubMed] [Google Scholar]
- de Bono M, Maricq AV. Neuronal substrates of complex behaviors in C. elegans. Annu Rev Neurosci. 2005;28:451–501. doi: 10.1146/annurev.neuro.27.070203.144259. [DOI] [PubMed] [Google Scholar]
- Donohoe DR, Phan T, Weeks K, Aamodt EJ, Dwyer DS. Antipsychotic drugs up-regulate tryptophan hydroxylase in ADF neurons of Caenorhabditis elegans: role of calcium-calmodulin-dependent protein kinase II and transient receptor potential vanilloid channel. J Neurosci Res. 2008;86:2553–2563. doi: 10.1002/jnr.21684. [DOI] [PubMed] [Google Scholar]
- Edachery J, Sen A, Brandenburg F. Graph Clustering Using distance-K cliques in Process of Graph Drawing. 1999.
- Eric CF, Zeidat N, Zhao Z. Supervised Clustering - Algorithms and Benefits. Proceedings of the 16th IEEE International Conference on Tools with Artificial Interlligence.2004. [Google Scholar]
- Feng Z, Cronin CJ, Wittig JH, Jr., Sternberg PW, Schafer WR. An imaging system for standardized quantitative analysis of C. elegans behavior. BMC Bioinformatics. 2004;5:115. doi: 10.1186/1471-2105-5-115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng Z, Li W, Ward A, Piggott BJ, Larkspur ER, et al. A C. elegans model of nicotine-dependent behavior: regulation by TRP-family channels. Cell. 2006;127:621–633. doi: 10.1016/j.cell.2006.09.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fontaine E, Burdick J, Barr A. Automated tracking of multiple C. Elegans. Conf Proc IEEE Eng Med Biol Soc. 2006;1:3716–3719. doi: 10.1109/IEMBS.2006.260657. [DOI] [PubMed] [Google Scholar]
- Gaudry Q, Kristan WB., Jr. Behavioral choice by presynaptic inhibition of tactile sensory terminals. Nat Neurosci. 2009;12:1450–1457. doi: 10.1038/nn.2400. [DOI] [PubMed] [Google Scholar]
- Geng W, Cosman P, Baek JH, Berry CC, Schafer WR. Quantitative classification and natural clustering of Caenorhabditis elegans behavioral phenotypes. Genetics. 2003;165:1117–1126. doi: 10.1093/genetics/165.3.1117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geng W, Cosman P, Berry CC, Feng Z, Schafer WR. Automatic tracking, feature extraction and classification of C elegans phenotypes. IEEE Trans Biomed Eng. 2004;51:1811–1820. doi: 10.1109/TBME.2004.831532. [DOI] [PubMed] [Google Scholar]
- Giles AC, Rankin CH. Behavioral and genetic characterization of habituation using Caenorhabditis elegans. Neurobiol Learn Mem. 2008 doi: 10.1016/j.nlm.2008.08.004. [DOI] [PubMed] [Google Scholar]
- Han J, Kamber M. Data Mining: Concepts and Techniques. Morgan Kaufmann; 2006. [Google Scholar]
- Harris TW, Antoshechkin I, Bieri T, Blasiar D, Chan J, et al. WormBase: a comprehensive resource for nematode research. Nucleic Acids Res. 2010;38:D463–467. doi: 10.1093/nar/gkp952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hata Y, Slaughter CA, Sudhof TC. Synaptic vesicle fusion complex contains unc-18 homologue bound to syntaxin. Nature. 1993;366:347–351. doi: 10.1038/366347a0. [DOI] [PubMed] [Google Scholar]
- Hsu AL, Feng Z, Hsieh MY, Xu XZ. Identification by machine vision of the rate of motor activity decline as a lifespan predictor in C. elegans. Neurobiol Aging. 2008 doi: 10.1016/j.neurobiolaging.2007.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang KM, Cosman P, Schafer WR. Machine vision based detection of omega bends and reversals in C. elegans. J Neurosci Methods. 2006;158:323–336. doi: 10.1016/j.jneumeth.2006.06.007. [DOI] [PubMed] [Google Scholar]
- Inoue T, Wang M, Ririe TO, Fernandes JS, Sternberg PW. Transcriptional network underlying Caenorhabditis elegans vulval development. Proc Natl Acad Sci U S A. 2005;102:4972–4977. doi: 10.1073/pnas.0408122102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson JR, Ferdek P, Lian LY, Barclay JW, Burgoyne RD, et al. Binding of UNC-18 to the N-terminus of syntaxin is essential for neurotransmission in Caenorhabditis elegans. Biochem J. 2009;418:73–80. doi: 10.1042/BJ20081956. [DOI] [PubMed] [Google Scholar]
- Lee J, Li W, Guan KL. SRC-1 mediates UNC-5 signaling in Caenorhabditis elegans. Mol Cell Biol. 2005;25:6485–6495. doi: 10.1128/MCB.25.15.6485-6495.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee RY, Sawin ER, Chalfie M, Horvitz HR, Avery L. EAT-4, a homolog of a mammalian sodium-dependent inorganic phosphate cotransporter, is necessary for glutamatergic neurotransmission in caenorhabditis elegans. J Neurosci. 1999;19:159–167. doi: 10.1523/JNEUROSCI.19-01-00159.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee TI, Rinaldi NJ, Robert F, Odom DT, Bar-Joseph Z, et al. Transcriptional regulatory networks in Saccharomyces cerevisiae. Science. 2002;298:799–804. doi: 10.1126/science.1075090. [DOI] [PubMed] [Google Scholar]
- Li W, Feng Z, Sternberg PW, Xu XZ. A C. elegans stretch receptor neuron revealed by a mechanosensitive TRP channel homologue. Nature. 2006;440:684–687. doi: 10.1038/nature04538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell T. Machine Learning. McGraw Hill; 1997. [Google Scholar]
- Nierman WC, Pain A, Anderson MJ, Wortman JR, Kim HS, et al. Genomic sequence of the pathogenic and allergenic filamentous fungus Aspergillus fumigatus. Nature. 2005;438:1151–1156. doi: 10.1038/nature04332. [DOI] [PubMed] [Google Scholar]
- Patil KR, Nielsen J. Uncovering transcriptional regulation of metabolism by using metabolic network topology. Proc Natl Acad Sci U S A. 2005;102:2685–2689. doi: 10.1073/pnas.0406811102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pei J, Jiang D, Zhang A. On Mining Cross-graph Quasi-cliques. Proc. of KDD; Chicago, Illinois. 2005. pp. 228–238. [Google Scholar]
- Pierce-Shimomura JT, Dores M, Lockery SR. Analysis of the effects of turning bias on chemotaxis in C. elegans. J Exp Biol. 2005;208:4727–4733. doi: 10.1242/jeb.01933. [DOI] [PubMed] [Google Scholar]
- Rankin CH. From gene to identified neuron to behaviour in Caenorhabditis elegans. Nat Rev Genet. 2002;3:622–630. doi: 10.1038/nrg864. [DOI] [PubMed] [Google Scholar]
- Ren B, Robert F, Wyrick JJ, Aparicio O, Jennings EG, et al. Genome-wide location and function of DNA binding proteins. Science. 2000;290:2306–2309. doi: 10.1126/science.290.5500.2306. [DOI] [PubMed] [Google Scholar]
- Robatzek M, Thomas JH. Calcium/calmodulin-dependent protein kinase II regulates Caenorhabditis elegans locomotion in concert with a G(o)/G(q) signaling network. Genetics. 2000;156:1069–1082. doi: 10.1093/genetics/156.3.1069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodgers JL, Nicewander WA. Thirteen ways to look at the correlation coefficient. The American Statistician. 1988;42:59–66. [Google Scholar]
- Rongo C, Kaplan JM. CaMKII regulates the density of central glutamatergic synapses in vivo. Nature. 1999;402:195–199. doi: 10.1038/46065. [DOI] [PubMed] [Google Scholar]
- Rose JK, Rankin CH. Analyses of habituation in Caenorhabditis elegans. Learn Mem. 2001;8:63–69. doi: 10.1101/lm.37801. [DOI] [PubMed] [Google Scholar]
- Ross-Macdonald P, Coelho PS, Roemer T, Agarwal S, Kumar A, et al. Large-scale analysis of the yeast genome by transposon tagging and gene disruption. Nature. 1999;402:413–418. doi: 10.1038/46558. [DOI] [PubMed] [Google Scholar]
- Sandmann T, Girardot C, Brehme M, Tongprasit W, Stolc V, et al. A core transcriptional network for early mesoderm development in Drosophila melanogaster. Genes Dev. 2007;21:436–449. doi: 10.1101/gad.1509007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schafer WR, Kenyon CJ. A calcium-channel homologue required for adaptation to dopamine and serotonin in Caenorhabditis elegans. Nature. 1995;375:73–78. doi: 10.1038/375073a0. [DOI] [PubMed] [Google Scholar]
- Schafer WR, Sanchez BM, Kenyon CJ. Genes affecting sensitivity to serotonin in Caenorhabditis elegans. Genetics. 1996;143:1219–1230. doi: 10.1093/genetics/143.3.1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shlomi T, Cabili MN, Herrgard MJ, Palsson BO, Ruppin E. Network-based prediction of human tissue-specific metabolism. Nat Biotechnol. 2008;26:1003–1010. doi: 10.1038/nbt.1487. [DOI] [PubMed] [Google Scholar]
- Simonetta SH, Golombek DA. An automated tracking system for Caenorhabditis elegans locomotor behavior and circadian studies application. J Neurosci Methods. 2007;161:273–280. doi: 10.1016/j.jneumeth.2006.11.015. [DOI] [PubMed] [Google Scholar]
- Steven R, Kubiseski TJ, Zheng H, Kulkarni S, Mancillas J, et al. UNC-73 activates the Rac GTPase and is required for cell and growth cone migrations in C. elegans. Cell. 1998;92:785–795. doi: 10.1016/s0092-8674(00)81406-3. [DOI] [PubMed] [Google Scholar]
- Theodoridis S, Koutroumbas K. Pattern Recognition. Academic Press; Burlinton: 2009. [Google Scholar]
- Tsibidis GD, Tavernarakis N. Nemo: a computational tool for analyzing nematode locomotion. BMC Neurosci. 2007;8:86. doi: 10.1186/1471-2202-8-86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagstaff K, Cardie C, Rogers S, Schorodl S. Constrained K-means Clustering with background knowledge. Proceedings of the Eighteenth International Conference on Machine Learning.2001. pp. 577–584. [Google Scholar]
- Ward A, Liu J, Feng Z, Xu XZ. Light-sensitive neurons and channels mediate phototaxis in C. elegans. Nat Neurosci. 2008;11:916–922. doi: 10.1038/nn.2215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward A, Walker VJ, Feng Z, Xu XZ. Cocaine modulates locomotion behavior in C. elegans. PLoS ONE. 2009;4:e5946. doi: 10.1371/journal.pone.0005946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Way JC, Chalfie M. mec-3, a homeobox-containing gene that specifies differentiation of the touch receptor neurons in C. elegans. Cell. 1988;54:5–16. doi: 10.1016/0092-8674(88)90174-2. [DOI] [PubMed] [Google Scholar]
- Wikman H, Ruosaari S, Nymark P, Sarhadi VK, Saharinen J, et al. Gene expression and copy number profiling suggests the importance of allelic imbalance in 19p in asbestos-associated lung cancer. Oncogene. 2007;26:4730–4737. doi: 10.1038/sj.onc.1210270. [DOI] [PubMed] [Google Scholar]
- Williams SL, Lutz S, Charlie NK, Vettel C, Ailion M, et al. Trio’s Rho-specific GEF domain is the missing Galpha q effector in C. elegans. Genes Dev. 2007;21:2731–2746. doi: 10.1101/gad.1592007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan X, Zhou X, Han J. Mining Closed Relational Graphs with Connectivity Constraints. Proc. KDD; Chicago, Illinois, USA. 2005. pp. 324–333. [Google Scholar]
- Zheng Y, Brockie PJ, Mellem JE, Madsen DM, Maricq AV. Neuronal control of locomotion in C. elegans is modified by a dominant mutation in the GLR-1 ionotropic glutamate receptor. Neuron. 1999;24:347–361. doi: 10.1016/s0896-6273(00)80849-1. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.