Abstract
Post-translational modifications of proteins play key roles in eukaryotic growth, differentiation and environmental adaptation. In model systems the ubiquitination of specific proteins contributes to the control of cell cycle progression, stress adaptation and metabolic reprogramming. We have combined molecular, cellular and proteomic approaches to examine the roles of ubiquitination in Candida albicans, because little is known about ubiquitination in this major fungal pathogen of humans. Independent null (ubi4/ubi4) and conditional (MET3p-UBI4/ubi4) mutations were constructed at the C. albicans polyubiquitin-encoding locus. These mutants displayed morphological and cell cycle defects, as well as sensitivity to thermal, oxidative and cell wall stresses. Furthermore, ubi4/ubi4 cells rapidly lost viability under starvation conditions. Consistent with these phenotypes, proteins with roles in stress responses (Gnd1, Pst2, Ssb1), metabolism (Acs2, Eno1, Fba1, Gpd2, Pdx3, Pgk1, Tkl1) and ubiquitination (Ubi4, Ubi3, Pre1, Pre3, Rpt5) were among the ubiquitination targets we identified, further indicating that ubiquitination plays key roles in growth, stress responses and metabolic adaptation in C. albicans. Clearly ubiquitination plays key roles in the regulation of fundamental cellular processes that underpin the pathogenicity of this medically important fungus. This was confirmed by the observation that the virulence of C. albicans ubi4/ubi4 cells is significantly attenuated.
Introduction
Post-translational modifications of proteins play critical roles in the cellular adaptation of all organisms, as well as their growth, division, differentiation and development. These modifications provide essential mechanisms by which the functions, activities and stabilities of pre-existing proteins can be rapidly and specifically modulated, thereby controlling dynamic cellular processes. Regulatory protein modifications include phosphorylation, acetylation, sumoylation and ubiquitination.
Ubiquitin is a highly conserved, small polypeptide of 76 amino acids found in all eukaryotic cells. The amino acid sequence of ubiquitin is identical across the animal kingdom, and differs by only three residues between yeast and animals (Ozkaynak et al., 1987; Finley and Chau, 1991). Within the cell, ubiquitin may be free or covalently attached to a range of cytoplasmic and nuclear proteins (Finley and Chau, 1991). Ubiquitin becomes covalently attached to free amino groups on target proteins through its carboxy-terminal glycine, a reaction catalysed by ubiquitin ligases (Jentsch et al., 1990; Stadtman, 1990; Rechsteiner, 1991). This enzymatic reaction is driven by an E1 ubiquitin-activating enzyme, an E2 ubiquitin-conjugating enzyme and an E3 ubiquitin ligase (Weissman, 2001).
The formation and processing of protein–ubiquitin conjugates, termed ‘ubiquitination’, is important for a number of biological processes. For example, ubiquitination plays a key role in the targeting of specific proteins for cytoplasmic degradation. Abnormal or short-lived proteins are extensively polyubiquitinated, consequently targeting their degradation by the multicatalytic 26S proteasome complex (Ciechanover, 1998; Ciechanover et al., 2000a,b;). The ubiquitin system also plays an important role in the degradation of denatured proteins following environmental stress (Finley et al., 1987; Amerik et al., 2005). The ubiquitination of some proteins is controlled in response to specific stimuli, thereby regulating their activities via relocalization and/or degradation (Finley and Chau, 1991; Paiva et al., 2009). Thus, ubiquitination is essential for the accurate execution of many cellular processes involving protein degradation, such as the cell cycle, endocytosis and the stress response (Hershko, 1997; Wolf and Hilt, 2004; Staub and Rotin, 2006).
The major opportunistic pathogen of humans, Candida albicans, remains one of the most persistent yeast pathogens known to man. It is a relatively harmless commensal of the oral cavity, gastrointestinal tract and genitalia of humans, but frequently causes superficial mucosal infections such as oral thrush and vaginitis in otherwise healthy individuals (Odds, 1988; Calderone, 2002). In the immunocompromised host, C. albicans can cause deep-seated systemic infections of the bloodstream and internal organs. Hence, C. albicans occupies a variety of niches within the human body, encountering a range of stressful conditions as it interacts with the host and its immunological defences (Lorenz et al., 2004; Fradin et al., 2005). For example, C. albicans encounters potentially damaging reactive oxidative species in some niches through the action of host defences (Enjalbert et al., 2007). In an attempt to counteract these defences, C. albicans has evolved effective oxidative stress responses, and these promote the survival of this pathogen in the host (Alonso-Monge et al., 1999; Hwang et al., 2002; Chauhan et al., 2006; Brown et al., 2009). In addition, evolutionarily conserved thermal adaptation mechanisms help C. albicans tune the levels of essential chaperones to the ambient temperature of host niches (Nicholls et al., 2009). Furthermore, C. albicans modulates its metabolism, cell morphology, stress responses and other cellular processes to the diverse microenvironments it encounters in the host (Hube, 1996; Ernst, 2000; Mavor et al., 2005; Kim and Kim, 2010). Therefore, a powerful combination of virulence factors and fitness attributes promote the virulence of this major pathogen. Protein phosphorylation has been shown to play critical roles in the regulation of many of these processes (Liu et al., 1994; Leberer et al., 1996; Alonso-Monge et al., 1999; Smith et al., 2004; Nicholls et al., 2009). There is some evidence that ubiquitination might also contribute to the regulation of some virulence factors in C. albicans (Leng et al., 2000; Roig and Gozalbo, 2003).
In Saccharomyces cerevisiae ubiquitin is encoded by a multigene family of natural gene fusions (Ozkaynak et al., 1987). UBI1, UBI2 and UBI3 encode hybrid proteins in which ubiquitin is fused to unrelated amino acid sequences (Ozkaynak et al., 1987). The fourth ubiquitin gene, UBI4, encodes a polyubiquitin comprising five consecutive head-to-tail ubiquitin repeats (Ozkaynak et al., 1987). After translation of this polyubiquitin polypeptide it is rapidly cleaved to generate ubiquitin monomers. S. cerevisiae UBI4 is highly expressed under stress conditions such as elevated temperatures and starvation. Also, the inactivation of UBI4 results in reduced resistance to starvation, growth at higher temperatures and sensitivity to amino acid analogues (Finley et al., 1987; Tanaka et al., 1988; Fraser et al., 1991). The global roles of ubiquitination in C. albicans have not been examined.
In C. albicans, polyubiquitin is encoded by the UBI4 gene, which has three tandem repeats in a consecutive head-to-tail arrangement (Sepulveda et al., 1996; Roig et al., 2000). Downregulation of the UBI4 gene in C. albicans induces the growth of hyphae and pseudohyphae (Roig and Gozalbo, 2003). This was consistent with a previous report indicating that the ubiquitination via the E2 enzyme Rad6 inhibits hyphal development in C. albicans (Leng et al., 2000). Downregulation of UBI4 has also been reported to cause mild temperature sensitivity in stationary cells grown in glycerol, but not during growth on glucose (Roig and Gozalbo, 2003), suggesting that polyubiquitin might contribute to stress responses in C. albicans. It has been suggested that the UBI4 gene is essential for viability in C. albicans (Roig and Gozalbo, 2003). These authors have also reported the existence of a second ubiquitin-encoding gene in C. albicans (Roig et al., 2000). UBI3 encodes a protein fusion between a single unit of ubiquitin and ribosomal protein S34. blast searches of the C. albicans genome reveal no other ubiquitin-encoding sequences (Skrzypek et al., 2010).
Our aim in this study was to better understand the roles of the polyubiquitin gene in C. albicans. To achieve this we constructed independent null and conditional C. albicans ubi4 mutants, indicating that the UBI4 gene is not essential, as previously thought. C. albicans ubi4/ubi4 cells were sensitive to a number of different physiologically relevant stresses. They also displayed defective metabolic adaptation, significant morphological abnormalities and unusual nuclear segregation in filamentous cells. Using a proteomic approach we identified 19 ubiquitinated C. albicans proteins with roles in growth, stress responses and metabolic adaptation, a finding that was entirely consistent with the phenotypes of ubi4/ubi4 cells. Finally, we report that compromising Ubi4 function attenuates virulence in a murine model of systemic candidiasis. Clearly, ubiquitination plays important regulatory roles in key cellular processes that contribute to the pathogenicity of C. albicans.
Results
UBI4 is not essential for viability in C. albicans
The polyubiquitin gene, UBI4, is not essential in S. cerevisiae. However, previous attempts to create a homozygous ubi4/ubi4 null mutation in C. albicans had failed (Roig and Gozalbo, 2003). For this reason we first created a methionine conditional ubi4 mutant using the methionine/cysteine repressible MET3 promoter (Care et al., 1999; Roig and Gozalbo, 2003). C. albicans is constitutively diploid. Hence, the first UBI4 allele was placed under the control of the MET3 promoter, and the second UBI4 allele was deleted to create the independent conditional C. albicans MET3p-UBI4/ubi4 mutants, MLC03 and MLC12 (Fig. 1A, Table 1).
Fig. 1.
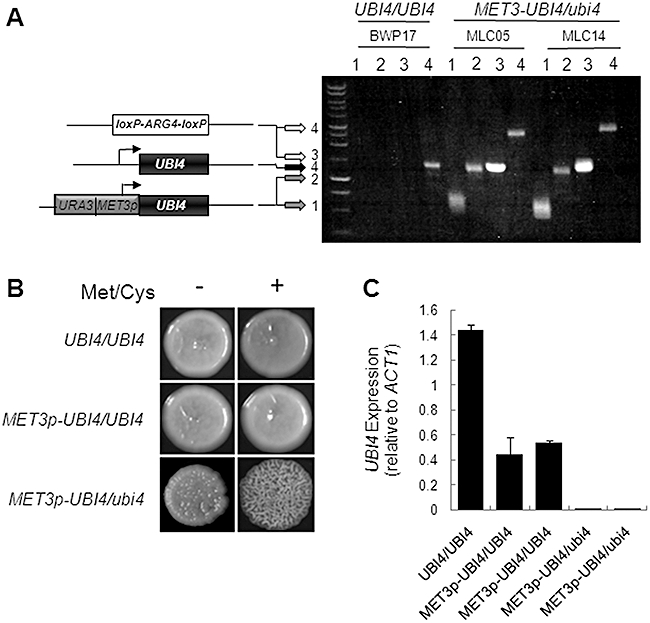
Downregulation of polyubiquitin expression in conditional C. albicans MET3p-UBI4/ubi4 mutants does not prevent growth.A. Construction of the methionine-conditional C. albicans mutants MLC05 and MLC14 from the parental strain BWP17 (Table 1). One UBI4 allele was disrupted by insertional inactivation using the loxP-ARG4-loxP cassette, and the other allele was placed under the control of the MET3 promoter. Cartoons represent these alleles, and the arrows indicate the lengths of the corresponding diagnostic PCR products on the agarose gel: PCR reactions 1: primers MET3p-F and UBI4d-R (Supporting information– Primers); PCR reactions 2: primers MET3p-F and UBI4d2-R; PCR reactions 3: primers UBI4d-F and LALd-R; PCR reactions 4: primers UBI4d-F and UBI4d2-R.B. Growth of conditional C. albicans MET3p-UBI4/ubi4 mutants in the presence of methionine and cysteine on plates. YPD plates contained (+) or lacked (−) 2.5 mM methionine (Met) and cysteine (Cys): UBI4/UBI4 (BWP17); MET3p-UBI4/UBI4 (MLC03); MET3p-UBI4/ubi4 (MLC05) (Table 1).C. qRT-PCR quantification of UBI4 mRNA levels, measured relative to the internal ACT1 mRNA control, in cells grown with 2.5 mM Met/Cys for 4 h. Independent MET3p-UBI4/UBI4 (MLC03, MLC12) and MET3p-UBI4/ubi4 strains (MLC05, MLC14) were analysed.
Table 1.
Candida albicans strains.
| Strain | Genotype | Source |
|---|---|---|
| BWP17 | ura3Δ::λimm434/ura3Δ::λimm434 his1::hisG/his1::hisG arg4Δ::hisG/arg4::hisG | Wilson et al. (1999) |
| THE1 | ade2::hisG/ade2::hisG, ura3::λ imm434/ura3::λ imm434, ENO1/eno1::ENO1-tetR-ScHAP4AD-3XHA-ADE2 | Nakayama et al. (2000) |
| MLC03 | ura3Δ::λimm434/ura3Δ::λimm434 his1::hisG/his1::hisG arg4Δ::hisG/arg4::hisG, URA3-MET3p-UBI4/UBI4 | This study |
| MLC05 | ura3Δ::λimm434/ura3Δ::λimm434 his1::hisG/his1::hisG arg4Δ::hisG/arg4::hisG, URA3-MET3p-UBI4/ubi4::loxP-ARG4-loxP | This study |
| MLC12 | ura3Δ::λimm434/ura3Δ::λimm434 his1::hisG/his1::hisG arg4Δ::hisG/arg4::hisG, URA3-MET3p-UBI4/UBI4 | This study |
| MLC14 | ura3Δ::λimm434/ura3Δ::λimm434 his1::hisG/his1::hisG arg4Δ::hisG/arg4::hisG, URA3-MET3p-UBI4/ubi4::loxP-ARG4-loxP | This study |
| MLC35 | ura3Δ::λimm434/ura3Δ::λimm434 his1::hisG/his1::hisG arg4Δ::hisG/arg4::hisG, UBI4/ubi4::loxP-URA3-loxP | This study |
| MLC36 | ura3Δ::λimm434/ura3Δ::λimm434 his1::hisG/his1::hisG arg4Δ::hisG/arg4::hisG, UBI4/ubi4::loxP-URA3-loxP | This study |
| MLC40 | ura3Δ::λimm434/ura3Δ::λimm434 his1::hisG/his1::hisG arg4Δ::hisG/arg4::hisG, ubi4::loxP-URA3-loxP/ubi4::loxP-ARG4-loxP | This study |
| MLC41 | ura3Δ::λimm434/ura3Δ::λimm434 his1::hisG/his1::hisG arg4Δ::hisG/arg4::hisG, ubi4::loxP-URA3-loxP/ubi4::loxP-ARG4-loxP | This study |
| MLC42 | ura3Δ::λimm434/ura3Δ::λimm434 his1::hisG/his1::hisG arg4Δ::hisG/arg4::hisG, ubi4::loxP-URA3-loxP/ubi4::loxP-ARG4-loxP | This study |
| MLC60 | ura3Δ::λimm434/ura3Δ::λimm434 his1::hisG/his1::hisG arg4Δ::hisG/arg4::hisG, ubi4::loxP-URA3-loxP/ubi4::loxP-ARG4-loxP, RPS1::CIp30 (URA3, HIS1, ARG4) | This study |
| MLC61 | ura3Δ::λimm434/ura3Δ::λimm434 his1::hisG/his1::hisG arg4Δ::hisG/arg4::hisG, ubi4::loxP-URA3-loxP/ubi4::loxP-ARG4-loxP, RPS1::CIp30-UBI4 (URA3, HIS1, ARG4) | This study |
| MLC125 | ura3Δ::λimm434/ura3Δ::λimm434 his1::hisG/his1::hisG arg4Δ::hisG/arg4::hisG, RPS1::CIp30 (URA3, HIS1, ARG4) | This study |
To test the essentiality of UBI4 in C. albicans, the wild-type parent (UBI4/UBI4), the heterozygous strain (MET3p-UBI4/UBI4) and the conditional mutants (MET3p-UBI4/ubi4) were plated on YPD medium containing or lacking 2.5 mM methionine and cysteine. As expected, all strains exhibited normal growth in the absence of methionine and cysteine (Fig. 1B). Also, the control strains grew normally in the presence of methionine and cysteine. Unexpectedly, MET3p-UBI4/ubi4 cells also grew in the presence of methionine and cysteine, albeit displaying a wrinkly colonial phenotype (Fig. 1B), suggesting that UBI4 is not essential for the growth of C. albicans. The strains were also examined in liquid culture, where they continued to grow in the presence of methionine and cysteine even after overnight incubation.
It was possible that the continued growth of MET3p-UBI4/ubi4 cells in the presence of methionine and cysteine was due to ineffective repression of the MET3p-UBI4 allele in our hands. To test this, we measured UBI4 mRNA levels by qRT-PCR relative to the internal ACT1 mRNA control (Fig. 1C). qRT-PCR was performed on RNA extracted from wild-type cells (BWP17: UBI4/UBI4) (Table 1), the conditional heterozygote (MLC03 and MLC12: MET3p-UBI4/UBI4) and the conditional mutants (MLC05 and MLC14: MET3p-UBI4/ubi4) grown for 4 h either in the absence or presence of 2.5 mM methionine and cysteine. As expected, following repression by methionine and cysteine, UBI4 mRNA levels remained high in control cells, but were reduced to negligible levels in the conditional MET3p-UBI4/ubi4 mutants (Fig. 1C).
Although the MET3p-UBI4 allele was effectively repressed by methionine and cysteine in the conditional mutant, some residual UBI4 expression remained (Fig. 1C). Hence formally it remained possible that this residual UBI4 expression was sufficient to support the growth of C. albicans (Fig. 1B). Therefore, we created independent C. albicans ubi4/ubi4 null mutants (ubi4::URA3/ubi4::ARG4: MLC40, MLC41, MLC42: Table 1), confirming their genotypes both by Southern blotting (Fig. 2A) and diagnostic PCR. These ubi4/ubi4 cells grew relatively normally on plates and in liquid media (Fig. 2B and C), thereby confirming that UBI4 is not an essential gene in C. albicans.
Fig. 2.
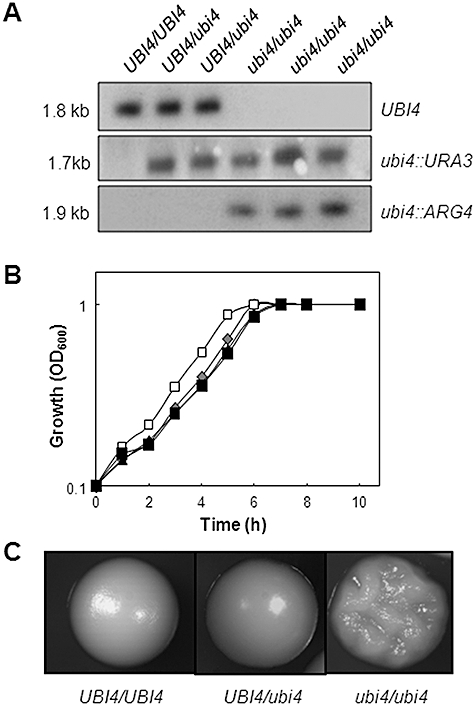
Polyubiquitin is not essential in C. albicans.A. The genotype of C. albicans ubi4/ubi4 mutants was confirmed by Southern blotting of HindIII digested C. albicans genomic DNA with PCR-amplified probes against the UBI4, ARG4 and URA3 open reading frames: UBI4/UBI4 (BWP17); UBI4/ubi4 (MLC35 and MLC36); ubi4/ubi4 (MLC40, MLC41, MLC42) (Table 1).B. Growth of C. albicans ubi4/ubi4 mutants in liquid YPD at 30°C: UBI4/UBI4, grey diamonds (BWP17); UBI4/ubi4, open squares (MLC35); ubi4/ubi4, closed triangles and closed squares (MLC40, MLC41).C. Colonial growth of C. albicans ubi4 mutants: UBI4/UBI4 (BWP17); UBI4/ubi4 (MLC35); ubi4/ubi4 (MLC40). Images were taken with the same magnification.
Polyubiquitin inactivation or depletion affects C. albicans cell morphology
Roig and Gozalbo (2003) demonstrated that downregulation of UBI4 using the methionine and cysteine-conditional MET3 promoter affects colony morphology in C. albicans. They also suggested that Ubi4 depletion promotes hyphal and pseudohyphal growth in liquid culture although, as they pointed out, they were unable to draw a clear conclusion because the morphology of their control strains was also affected by methionine and cysteine addition (Roig and Gozalbo, 2003). For this reason we re-examined the impact of polyubiquitin depletion upon C. albicans morphology. The C. albicans ubi4/ubi4 mutants formed wrinkly, rough, asymmetrical colonies compared with the smooth, waxy appearance of control colonies on YPD plates at 30°C (Fig. 2C). Furthermore, ubi4/ubi4 cells also showed abnormal morphological phenotypes in liquid culture. Exponentially growing wild-type cells showed the expected budding yeast morphology in YPD at 30°C, whereas ubi4/ubi4 cells formed a mixture of yeast, pseudohyphal and hyphal cells (Fig. 3A). Repression of the MET3p-UBI4 allele in the conditional MET3p-UBI4/ubi4 cells essentially phenocopied this behaviour of the ubi4/ubi4 null mutant (Fig. 3B). Furthermore, the addition of methionine and cysteine had no effect upon the morphology of wild-type cells under our experimental conditions. These observations reinforced the suggestion of Roig and Gozalbo (2003) that the downregulation of UBI4 promotes filamentous growth as well as our previous finding that ubiquitination is involved in morphogenetic regulation (Leng et al., 2000).
Fig. 3.
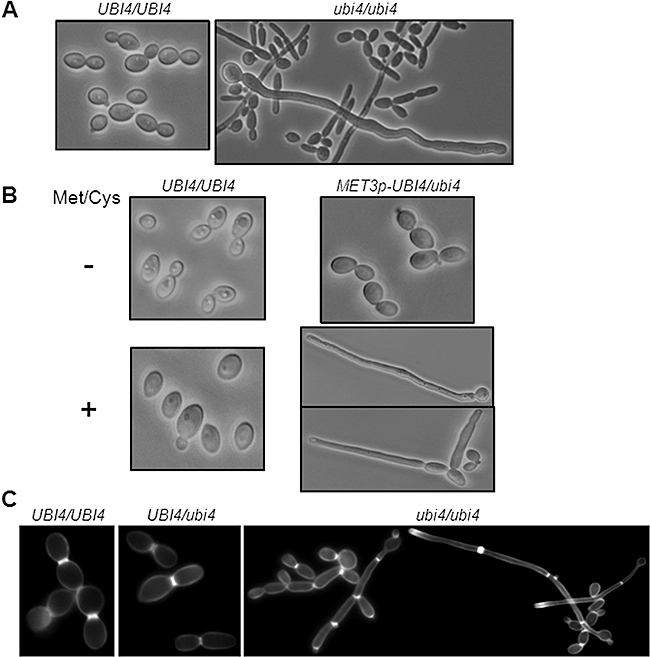
Inactivation or downregulation of polyubiquitin affects C. albicans cell morphology.A. Light microscopy of C. albicans ubi4/ubi4 cells grown in YPD at 30°C reveals morphological abnormalities: UBI4/UBI4 (BWP17); ubi4/ubi4 (MLC40) (Table 1).B. Downregulation of UBI4 in C. albicans MET3p-UBI4/ubi4 cells with 2.5 mM methionine and cysteine (Met/Cys) essentially phenocopies the ubi4/ubi4 null mutants: UBI4/UBI4 (BWP17); MET3p-UBI4/ubi4 (MLC05).C. Calcofluor White staining of C. albicans ubi4/ubi4 cells reveals true hypha formation in some cells: UBI4/UBI4 (BWP17); UBI4/ubi4 (MLC35); ubi4/ubi4 (MLC40).Images were taken at the same magnification and with the same exposures.
To examine septum formation in C. albicans ubi4/ubi4 cells, we stained cells with Calcofluor White (Fig. 3C). As expected, wild-type budding cells displayed normal chitin distributions at septal junctions. Some filamentous ubi4/ubi4 cells displayed septal plates within the germ tube, indicative of true hyphae (Sudbery, 2001). Other filamentous ubi4/ubi4 cells were confirmed as pseudohyphae based on the location of their septal junctions at mother–daughter junctions (Fig. 3C). Similar morphological phenotypes were observed in the conditional MET3p-UBI4/ubi4 mutant after growth in the presence of 2.5 mM methionine and cysteine. These data confirmed that the inactivation or depletion of polyubiquitin promotes mixed cell morphologies in C. albicans.
Polyubiquitin inactivation or depletion affects nuclear segregation in C. albicans
Ubiquitination has been implicated in cell cycle regulation, for example through the degradation of cyclins (Glotzer et al., 1991). For this reason we examined the distribution of nuclei in C. albicans ubi4/ubi4 cells by DAPI staining. Nuclear segregation appeared normal in ubi4/ubi4 cells that grew with a yeast-like, budding morphology (Fig. 4A). However, defective nuclear segregation was apparent in those ubi4/ubi4 cells displaying an aberrant filamentous morphology. In 91% of these filamentous cells, the ovoid mother cell lacked a nucleus, while the filamentous daughter cell contained an elongated nucleus. (Examples of this are illustrated in Fig. 4A.) Therefore, the altered morphology of some ubi4/ubi4 cells correlated closely with defects in nuclear segregation. Similar observations were made following polyubiquitin depletion in the conditional MET3p-UBI4/ubi4 mutant. We conclude that polyubiquitin contributes to the normal execution of the C. albicans cell cycle.
Fig. 4.
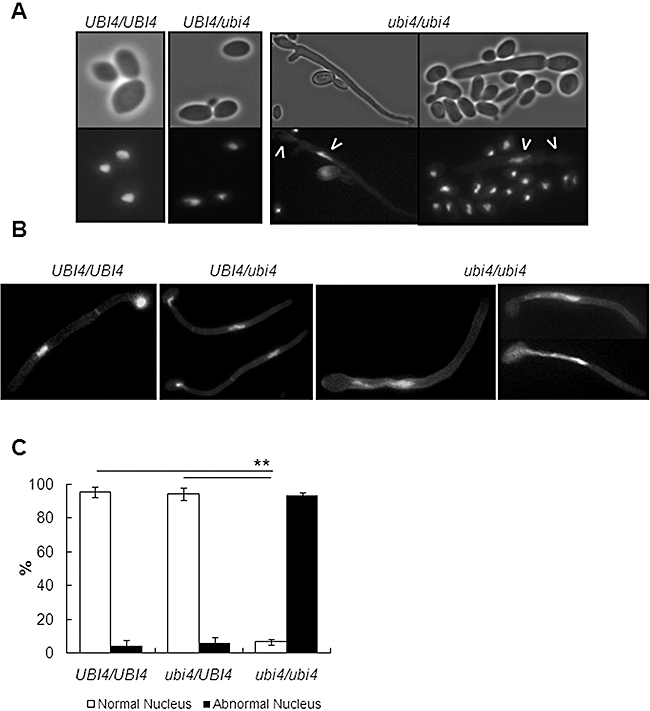
Inactivation of polyubiquitin affects C. albicans nuclear segregation.A. DAPI staining of C. albicans ubi4/ubi4 cells grown in YPD at 30°C reveals defects in mitosis and aberrant nuclear segregation in some cells: UBI4/UBI4 (BWP17); UBI4/ubi4 (MLC35); ubi4/ubi4 (MLC40).B. DAPI and Calcofluor White staining of ubi4/ubi4 cells under hypha-inducing conditions highlights the extent to which nuclear segregation is incomplete: UBI4/UBI4 (BWP17); UBI4/ubi4 (MLC35); ubi4/ubi4 (MLC40).C. Percentage of cells with aberrant nuclear segregation: UBI4/UBI4 (BWP17); UBI4/ubi4 (MLC35); ubi4/ubi4 (MLC40). **P < 0.01.
All of the above experiments were performed under conditions that normally promote growth of C. albicans in the yeast form. Do these observations hold under hypha-inducing conditions? To test this C. albicans ubi4/ubi4 cells were compared with wild-type (UBI4/UBI4) and heterozygous control cells (UBI4/ubi4) after 2 h of growth at 37°C in YPD containing 10% serum. As expected, wild-type (UBI4/UBI4) and heterozygous (ubi4/UBI4) cells contained a single nucleus in the mother cell, and a second nucleus in the germ tube (Fig. 4B). In contrast, we observed that the nucleus was significantly extended from the mother cell into the germ tube in most cells lacking Ubi4 (ubi4/ubi4) (Fig. 4B and C). While only a small proportion of wild-type and heterozygous cells displayed an extended nucleus lying between ovoid mother cell and the daughter hyphal compartment (5% and 7% respectively), most mutant ubi4/ubi4 cells (> 90%) displayed aberrant nuclear segregation (Fig. 4C). These data reinforce our conclusion that polyubiquitin contributes to the normal execution of the C. albicans cell cycle, particularly in filamentous growth.
Polyubiquitin is required for stress adaptation in C. albicans
In S. cerevisiae, ubiquitination is regulated by a number of different stresses, and UBI4 expression is increased in response to elevated temperatures and starvation. Furthermore, inactivating ScUBI4 causes sensitivity to starvation, high temperatures and amino acid analogues (Finley et al., 1987; Tanaka et al., 1988; Fraser et al., 1991). We reasoned therefore, that C. albicans ubi4/ubi4 cells might display a range of stress sensitivities. To test this, we compared the growth of wild-type (BWP17) and mutant cells (MLC40) under a variety of stress conditions.
Candida albicans ubi4/ubi4 cells reproducibly displayed temperature sensitivity at 37°C and 42°C (Fig. 5). This observation was consistent with the suggestion that increased ubiquitination and proteasome-dependent degradation can alleviate the functions of heat shock proteins during stress (Friant et al., 2003). Although there was no obvious effect upon osmotic stress resistance, UBI4 inactivation did render C. albicans cells more sensitive to peroxide (Fig. 5). This observation is significant in the context of infection as reactive oxygen species are important for the antimicrobial activity of host immune cells (Miller and Britigan, 1997). In addition, C. albicans ubi4/ubi4 cells consistently displayed cell wall stress phenotypes. They grew more slowly in the presence of Congo Red and the anti-fungal drug, caspofungin, both of which target glucan biosynthesis. The ubi4/ubi4 mutant was also sensitive to low concentrations of Calcofluor White, which affects chitin synthesis (Fig. 5). Furthermore, ubi4/ubi4 cells were also sensitive to tunicamycin, which triggers the unfolded protein response (Wimalasena et al., 2008).
Fig. 5.
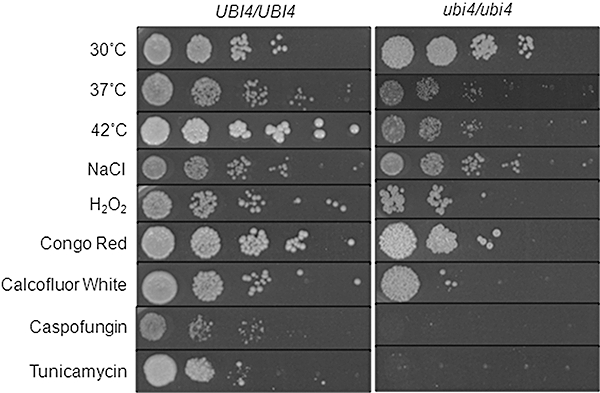
Candida albicans polyubiquitin mutants are sensitive to a range of stresses.A. Sensitivity of an ubi4/ubi4 null mutant to stresses: UBI4/UBI4 (BWP17); ubi4/ubi4 (MLC40) (Table 1). Serial dilutions of exponentially growing cells were spotted onto YPD plates containing the appropriate stress at the following concentrations and examined after 48 h: H2O2 (5 mM), NaCl (1 M), Calcofluor White (20 µg ml−1), Congo Red (100 µg ml−1), tunicamycin (4.73 mM), caspofungin (0.064 µg ml−1) and thermal stress (37°C and 42°C).
With the exception of peroxide and tunicamycin, these stress sensitivities were also examined in the conditional C. albicans MET3p-UBI4/ubi4 mutant (Supporting information–MET3p-UBI4 control). The impact of peroxide was not tested in the conditional MET3p-UBI4/ubi4 mutant because of the confounding effects of the reducing agents methionine and cysteine. C. albicans cells were especially sensitive to tunicamycin in the presence of methionine and cysteine because both tunicamycin and reducing agents trigger the unfolded protein response in C. albicans (Wimalasena et al., 2008). Under repressing conditions the C. albicans MET3p-UBI4/ubi4 mutant phenocopied ubi4/ubi4 cells with respect to the other stress conditions we examined, displaying temperature and cell wall stress sensitivities (Supporting information–MET3p-UBI4 control). In control experiments, the wild-type (BWP17) and MET3p-UBI4/ubi4 cells (MLC05) displayed no significant differences under non-repressing conditions, i.e. in the absence of methionine and cysteine (Supporting information–MET3p-UBI4 control). Collectively, our data indicate that polyubiquitin contributes to heat adaptation, the oxidative stress response and cell wall remodelling.
Polyubiquitin is required for metabolic adaptation in C. albicans
Metabolic adaptation is central to the interactions that occur between C. albicans and its mammalian host during the infection process (Lorenz and Fink, 2001; Lorenz et al., 2004; Brown, 2005; Fradin et al., 2005; Thewes et al., 2007; Zakikhany et al., 2007). In other organisms, ubiquitination is not only important for the degradation of short-lived or damaged proteins (Bachmair et al., 1986; Jentsch et al., 1991), but also for the turnover of proteins during metabolic adaptation (Finley et al., 1987; Werner-Washburne et al., 1993; Muratani et al., 2005; Santt et al., 2008). Therefore, we reasoned that ubiquitination might play an important role during metabolic adaptation in C. albicans.
To test this, wild-type (BWP17), heterozygous UBI4/ubi4 (MLC35) and homozygous ubi4/ubi4 null cells (MLC40 and MLC41) were grown to mid-exponential phase in rich medium (YPD), and then transferred to starvation conditions. The viabilities of the wild-type and mutant cells were then compared at various times thereafter (Fig. 6). Wild-type and UBI4/ubi4 cells retained their viability for a week after being starved in water, or after carbon deprivation (YNB-C: Experimental procedures). In contrast, the viabilities of two independent ubi4/ubi4 null strains declined after these different types of starvation were imposed (Fig. 6A and B). An even more dramatic phenotype was observed when cells were starved for nitrogen in the presence of excess carbon (YNB-N: Experimental procedures). The ubi4/ubi4 cells rapidly lost their viability under these conditions, whereas wild-type and UBI4/ubi4 control cells not only remained viable, but displayed limited growth (Fig. 6C). This phenotype was not due to marker effects. Strains in which all auxotrophic markers were reinstated displayed similar starvation sensitivities to those shown in Fig. 6 (MLC60, MLC61, MLC125: Table 1). We conclude that, like the situation in S. cerevisiae (Finley et al., 1987; Werner-Washburne et al., 1993), polyubiquitin is clearly required for the survival of C. albicans under starvation conditions, and particularly when nitrogen is limiting but carbon is in excess. This is entirely consistent with a major role for ubiquitin-mediated protein degradation during the metabolic adaptation of this pathogen to starvation conditions.
Fig. 6.
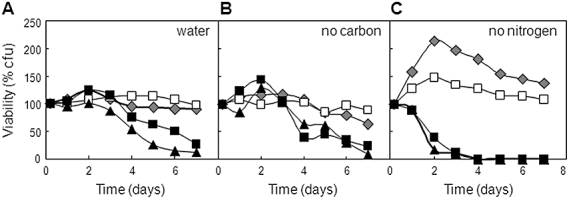
Inactivating polyubiquitin affects adaptation to nutrient starvation in C. albicans. Mid-exponential C. albicans cells were resuspended in minimal medium lacking a carbon and/or nitrogen source, and viability monitored thereafter (Experimental procedures): UBI4/UBI4, grey diamonds (BWP17); UBI4/ubi4, open squares (MLC35); ubi4/ubi4, closed triangles and closed squares (MLC40, MLC41) (Table 1).A. Carbon plus nitrogen starvation, where cells were incubated in water.B. Carbon starvation in YNB-C.C. Nitrogen starvation in YNB-N.
UBI4 is required for the ubiquitination of numerous proteins in C. albicans
In S. cerevisiae, ubiquitination influences numerous processes including metabolic adaptation, the cell cycle, protein transport, transcription and stress responses (Peng et al., 2003; Mayor et al., 2005; Tagwerker et al., 2006; Seyfried et al., 2008). However, little is known about ubiquitination targets in C. albicans, although ubiquitination probably affects many processes that underpin the virulence of this fungal pathogen. Based on our phenotypic analysis, we reasoned that a number of proteins must be targeted by UBI4, and therefore our next aim was to identify ubiquitinated proteins in C. albicans.
Epitope tagging of Ubi4 was not feasible because polyubiquitin is cleaved into single units for ubiquitination. Therefore, we used an anti-ubiquitin antibody to examine ubiquitinated protein targets in C. albicans. First, we examined ubiquitination in C. albicans cells under a range of experimental conditions: heat shock (30–42°C for 1 h), oxidative stress (50 mM H2O2 for 1 h), cell wall stress (SDS, Congo Red and Calcofluor White for 1 h), serum (10% fetal calf serum for 1 h) and control untreated cells. Protein extracts were then prepared from these cells and subjected to Western blotting with the anti-ubiquitin antibody.
A large number of ubiquitinated proteins were observed in untreated C. albicans cells, and these patterns of ubiquitination changed following heat shock, or after exposure to cell wall stress or peroxide (Fig. 7A). For example, peroxide stress led to the appearance of a number of ubiquitinated proteins of between 20 and 50 kDa. Also, a new band of about 23 kDa was apparent following heat stress, as well as additional ubiquitinated proteins of masses between 60 and 100 kDa. Following exposure to cell wall stresses, heavy ubiquitination was observed on proteins of between 50 and 100 kDa. These different ubiquitination patterns were consistent with our working hypothesis that ubiquitination plays significant roles in C. albicans during stress adaptation.
Fig. 7.
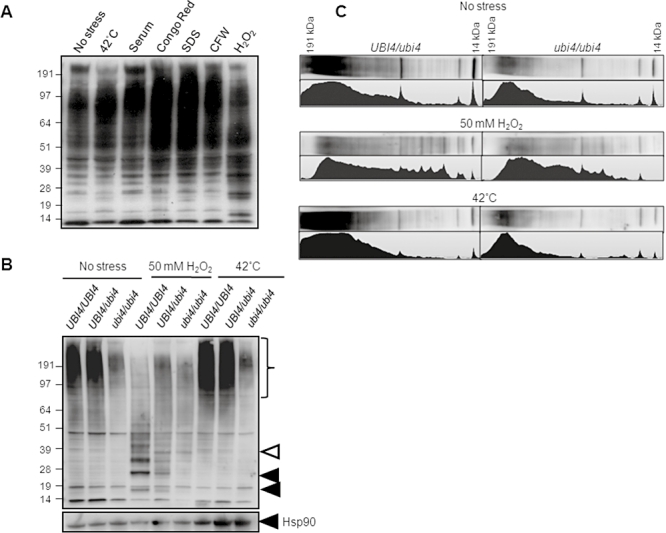
Many C. albicans proteins are ubiquitinated in a polyubiquitin-dependent fashion.A. Protein ubiquitination in wild-type C. albicans cells under different stress conditions. C. albicans THE1 was grown for 5 h and then exposed to the specified stress for 1 h (Experimental procedures). Protein extracts were prepared and analysed by Western blotting with an α-ubiquitin antibody.B. Impact of UBI4 inactivation upon protein ubiquitination in C. albicans. UBI4/UBI4 (THE1), UBI4/ubi4 (MLC35) and ubi4/ubi4 (MLC40) cells were grown for 5 h and then stressed for 1 h with heat or peroxide (Experimental procedures). Protein extracts were prepared and analysed by Western blotting. To control for loading, membranes were stripped and reprobed for Hsp90. Closed arrows highlight bands observed in wild-type cells that are absent following UBI4 inactivation, and the open arrow indicates a band observed in heat stressed cells that is absent in the ubi4/ubi4 mutant. The bracket indicates numerous ubiquitinated proteins, the levels of which are decreased in ubi4/ubi4 cells.C. Quantification of protein levels in the heterozygous UBI4/ubi4 (MLC35) and null ubi4/ubi4 (MLC40) strains under the three conditions tested.
We chose to examine ubiquitination following peroxide treatment and heat stress because C. albicans is exposed to oxidative stress during contact with host immune defences (Lorenz et al., 2004; Fradin et al., 2005; Enjalbert et al., 2007) and because ubiquitination has been shown to play a significant role in S. cerevisiae during the heat shock response (Friant et al., 2003). However, before doing so, we tested whether UBI4 contributes significantly to ubiquitination under these experimental conditions. This is because in C. albicans ubiquitin is derived from Ubi3 as well as Ubi4. To do this we examined the patterns of ubiquitination in wild-type (UBI4/UBI4), heterozygous (UBI4/ubi4) and homozygous null cells (ubi4/ubi4). Protein extracts from untreated, heat-shocked or peroxide-treated cells were examined by Western blotting and the signals quantified (Fig. 7B and C). UBI4 inactivation led to significant reductions in ubiquitination under all of the conditions examined, and in particular to the loss of some ubiquitinated proteins in the 20–50 kDa range following peroxide stress (Fig. 7C). This observation was strengthened by Ponceau S staining of the membranes, which confirmed that the protein loadings on the Western blots were reasonably even (Supporting information– Ponceau S Stain). We also reprobed the membranes for Hsp90 (Fig. 7B). Hsp90 levels were similar, except following heat shock, when Hsp90 levels increased as expected (Swoboda et al., 1995; Nicholls et al., 2009).
We conclude that a significant proportion of the ubiquitination observed during peroxide or heat stress is performed using ubiquitin derived from UBI4, and that residual ubiquitination under these conditions employs ubiquitin from UBI3. This conclusion is consistent with the fact that UBI4 expression is upregulated in response to stress (Brown et al., 2000; Roig and Gozalbo, 2002).
Identification of ubiquitinated proteins in C. albicans
Having established that UBI4 plays a significant role in ubiquitination during responses to heat shock and peroxide, we attempted to identify protein targets of ubiquitination under these conditions. Exponentially growing C. albicans cells were exposed to a 30–42°C heat shock or to 50 mM H2O2 for 1 h, while control cells remained untreated. Protein extracts from these cells were subjected to 2-D gel electrophoresis, and replicate 2-D gels were stained with Coomassie blue or subjected to Western blotting using the anti-ubiquitin antibody (Fig. 8). Ubiquitinated proteins were identified by aligning the Western blots with the Coomassie-stained gels, and only those spots that displayed reproducible regulation in two biologically independent experiments were examined further. These spots of interest were then cut from the Coomassie-stained gels and the corresponding proteins identified by tryptic digestion followed by tandem mass spectrometry (Experimental procedures).
Fig. 8.
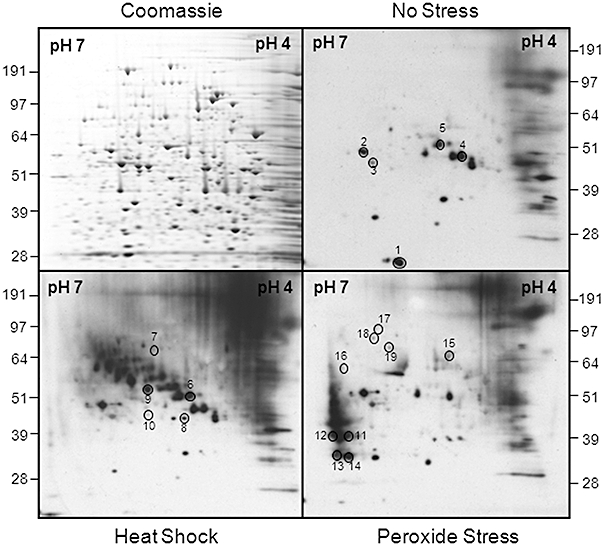
Identification of ubiquitinated proteins in C. albicans using a proteomic screen. C. albicans THE1 cells were grown for 5 h to mid-exponential phase and then exposed to stress for 1 h. Protein extracts were run on replicate 2-D gels, which were either stained with Coomassie blue or subjected to Western blotting with an α-ubiquitin antibody: Western blots of no stress control; peroxide-treated cells (50 mM H2O2); heat-shocked cells (30–42°C). Autoradiographs were aligned with the Coomassie-stained gels, spots chosen for analysis, and the corresponding proteins identified by tryptic digestion and LC-MS/MS (see text). The sample reference numbers for ubiquitination targets are shown (Table 2).
Table 2 lists those protein identifications that met several strict data quality criteria: a Mascot score of > 35 (P < 0.05), plus an Excess of Limit-Digested Peptides (EDLP) score of ≥ 1 (Stead et al., 2006). The quality metrics for these protein identifications are provided in the Supporting information (Ubiquitination Targets). For those C. albicans proteins that were identified using our proteomic approach, we looked for experimental evidence for ubiquitination of their S. cerevisiae orthologue. In addition, we screened for potential ubiquitination sites in the C. albicans protein sequence using the program UbPred (Radivojac et al., 2010) (Table 2).
Table 2.
Identification of ubiquitinated proteins in C. albicans.
| Sample ref | Accession number | Protein name | Molec. Wt. (Da) | pI | Function | Ubiquitinated in S. cerevisiae? | Ubiquitination detected in absence of stress? | Predicted ubiquitination site(s) |
|---|---|---|---|---|---|---|---|---|
| Constitutively ubiquitinated | ||||||||
| 1 | Ca2937 | RPS21B | 8 807 | 8.85 | Ribosomal protein S21 | ND | Yes | Medium confidence, position 5 |
| 2 | Ca1691 | PGK1 | 45 266 | 6.07 | Phosphoglycerate kinase | Yes | Yes | Medium confidence, position 6; low confidence, position 296 |
| 3 | Ca5180 | FBA1 | 39 362 | 5.69 | Fructose-bisphosphate aldolase | Yes | Yes | Medium confidence, position 72 and 192; low confidence, position 77 |
| 4 | Ca2939 | TIF1 | 44 742 | 5.22 | Translation initiation factor | Yes | Yes | Low confidence, position 24 |
| 4 | Ca0824 | GPD2 | 41 174 | 5.21 | Glycerol-3-phosphate dehydrogenase | Yes | Yes | Low confidence, position 274 and 318 |
| Increased ubiquitination in response to heat | ||||||||
| 5 | Ca3874 | ENO1 | 47 202 | 5.54 | Enolase | Yes | Yes | High confidence, position 273; medium confidence, position 265; low confidence, position 432 |
| 6 | Ca4389 | RPT5 | 47 814 | 5.25 | Regulatory particle triphosphatase | Yes | Yes | Medium confidence, position 55, 63, 163, 170, 284 and 290; low confidence, position 35 and 92 |
| 7 | Ca3924 | TKL1 | 73 841 | 5.48 | Transketolase | Yes | No | Low confidence, position 351 |
| 8 | Ca2895 | PRN4 | 37 072 | 5.31 | Putative pirin by homology | ND | No | High confidence, position 315; medium confidence, position 234; low confidence, position 240, 320 and 327 |
| 9 | Ca5239 | GND1a | 57 159 | 6.14 | 6-Phosphogluconate dehydrogenase | Yes | No | Medium confidence, position 6; low confidence, position 151 and 388 |
| 9 | Ca2011 | UBI3 | 17 485 | 9.86 | Functional homologue of S. cerevisiae RPS31 | ND | No | Medium confidence, position 105 |
| 10 | Ca0210 | orf19.5525 | 37 783 | 5.53 | Putative NADP(H) oxidoreductase | ND | No | Medium confidence, position 285; low confidence, position 268 and 344 |
| Increased ubiquitination in response to H2O2 | ||||||||
| 11 | Ca4261 | PDX3 | 28 773 | 6.02 | Pyridoxine (pyridoxamine) phosphate oxidase | Yes | No | Medium confidence, position 22 and 58 |
| 12 | Ca2675 | GSP1a | 24 470 | 6.53 | Small RAN G-protein | Yes | No | Medium confidence, position 193 |
| 12 | Ca5932 | UBI4 | 25 776 | 7.76 | Ubiquitin precursor (polyubiquitin) | Yes | No | Low confidence, position 33, 48, 63, 139, 179 and 215 |
| 13 | Ca1673 | PST2 | 21 714 | 6.51 | NADH : quinone oxidoreductase | Yes | No | None |
| 13 | Ca5037 | PRE3 | 23 629 | 6.42 | Beta-1 subunit of proteasome | ND | No | Low confidence, position 12 |
| 14 | Ca4791 | PRE1a | 22 119 | 6.43 | β4 subunit of the 20S proteasome | ND | No | Low confidence, position 19 |
| 14 | Ca4589 | ASC1a | 23 619 | 6.3 | 40S ribosomal subunit protein | Yes | No | None |
| 14 | Ca5932 | UBI4 | 25 776 | 7.76 | Ubiquitin precursor (polyubiquitin) | Yes | No | Low confidence, position 33, 48, 63, 139, 179 and 215 |
| Increased ubiquitination in response to heat and H2O2 | ||||||||
| 15 | CA3534 | SSB1 | 66 580 | 5.25 | HSP70 family chaperone | Yes | No | High confidence, position 528 and 545; medium confidence, position 521, 530, 543 and 597; low confidence, position 191, 314 and 393 |
| 16 | Ca1246 | IMH3 | 40 055 | 6.67 | Inosine monophosphate (IMP) dehydrogenase | ND | No | Medium confidence, position 7; low confidence, position 24, 175, 230 and 517 |
| 17 | Ca3546 | ACO1 | 84 625 | 5.96 | Aconitase | ND | No | Medium confidence, position 25. |
| 18 | Ca2858 | ACS2 | 74 215 | 5.73 | Acetyl-CoA synthetase | Yes | No | Medium confidence, position 32 and 596; low confidence, position 17 and 583 |
| 19 | Ca2470 | SDH12 | 70 930 | 6.03 | Flavoprotein subunit of succinate dehydrogenase | ND | No | Medium confidence, position 480; low confidence, position 417, 492 and 500 |
Protein detected with ubiquitin sequences.
ND, not detected.
This was not an exhaustive proteomic screen for ubiquitinated target proteins in C. albicans. For example, some ubiquitinated proteins in C. albicans might lie outside the pI range of our 2-D gels, or they might not be expressed under the conditions examined. Furthermore, some ubiquitin targets might be low abundance proteins that lie below the sensitivity of our analyses. Nevertheless, our aim in these experiments was to identify some proteins that are ubiquitinated in stressed or control cells, and some proteins whose ubiquitination increases either in response to heat shock or peroxide stress. Some of the gel spots we analysed contained more than one protein. In this case, we looked for evidence of ubiquitination in the S. cerevisiae orthologue, for potential ubiquitination sites in the C. albicans protein, and protein identifications with the most significant Mascot scores. In a small number of cases, we found that of two proteins identified, one is ubiquitinated in S. cerevisiae but contains no potential ubiquitination sites in the C. albicans sequence, whereas the other protein does contain a ubiquitination site but has not been shown to be ubiquitinated in S. cerevisiae. In these circumstances it was not possible to prioritize the likely ubiquitination target. Furthermore, it was conceivable that more than one protein in a particular 2-D spot was ubiquitinated. Therefore, under these circumstances both protein identifications were retained in Table 2. Nevertheless, 19 ubiquitination features were identified in our proteomic screen, the orthologues of many of which are ubiquitinated in S. cerevisiae. Furthermore, our identification of ubiquitin sequences with a number of these targets reinforced the validity of this screen (Table 2).
As mentioned, our proteomic approach revealed C. albicans proteins whose orthologues have been shown to be ubiquitinated in S. cerevisiae (Table 2). These included metabolic proteins (Eno1, Fba1, Gpd2, Pgk1, Pdx3, Tkl1, Acs2) a heat shock protein (Ssb1) and oxidative stress proteins (Gnd1, Pst2). Furthermore, proteins involved in the degradation of ubiquitinated substrates were also detected (Rpt5, Ubi4 and Ubi3), as well a translation factor (Tif1) and a cell cycle protein (Gsp1). Moreover, Pre3 and Pre1, proteins of the 20S proteosome involved in ubiquitin-dependent degradation were also detected. These proteins lend credence to our list of ubiquitination targets in C. albicans.
Some of the ubiquitination targets we identified appeared to be constitutively ubiquitinated in C. albicans based on the intensity of their signals on Western blots, whereas the ubiquitination of other targets appeared to increase in response to heat shock and/or oxidative stress (Fig. 8, Table 2). In many cases the functions of these ubiquitination targets appeared to correlate with their patterns of ubiquitination. For example, the ubiquitination of the heat shock protein (Ssb1) and oxidative stress proteins (Gnd1, Pst2) increased in response to stress, whereas other proteins, such as the metabolic enzymes Eno1, Pgk1 and Fba1, were constitutively ubiquitinated. In particular, a set of proteins of between 20 and 50 kDa were ubiquitinated in response to peroxide stress, and their ubiquitination was Ubi4-dependent (Fig. 7B). Our proteomic analyses identified four peroxide stress-induced features in this category (spots 8, 10–14: Fig. 8), including an oxidoreductase (Pst2) and two proteasomal subunits (Pre1, Pre3) (Table 2). Two of these spots contained ubiquitin sequences, thereby reinforcing their validity as ubiquitination targets. Obviously the aberrant ubiquitination of these targets in ubi4/ubi4 cells could contribute to their peroxide sensitivity (Fig. 5). Therefore, the proteomic data further supported the view that ubiquitination contributes significantly to environmental adaptation in C. albicans and provides some mechanistic explanations for the phenotypes of the ubi4/ubi4 mutants.
Polyubiquitin contributes significantly to virulence in C. albicans
The above observations suggested that ubiquitination might contribute significantly to the physiological fitness of C. albicans cells in vivo, and hence, that Ubi4 might contribute to the virulence of this pathogen. To test this mice were infected with C. albicans ubi4/ubi4 cells (MLC60), or with the control strains MLC125 (UBI4/UBI4) and MLC61 (ubi4/ubi4/UBI4). All of these strains were prototrophic, the markers having been restored by integration of the plasmid CIp30 (Dennison et al., 2005). Virulence was assayed by determining infection outcome scores, based upon weight change and kidney burdens at 3 days post infection (Experimental procedures; MacCallum et al., 2010). Statistical analyses revealed no significant difference between the wild-type (MLC125) and reintegrant control strains (MLC61) for any of the parameters tested (weight loss, kidney fungal burden and outcome score) (Fig. 9A). However, when mice were infected with the ubi4/ubi4 null mutant (MLC60), they exhibited significantly lower kidney fungal burdens and outcome scores compared with the control strains; P < 0.01 (Fig. 9A and B). Taken together these data clearly indicate that UBI4 inactivation attenuates the virulence of C. albicans.
Fig. 9.
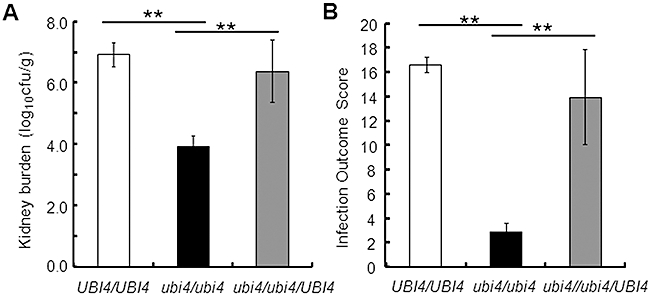
Inactivation of polyubiquitin significantly attenuates the virulence of C. albicans in a mouse model of systemic candidiasis.A. Kidney burdens measured at 72 h post infection (means and standard deviations for six animals): UBI4/UBI4, MLC125, white bar; ubi4/ubi4, MLC60, black bar; ubi4/ubi4/UBI4, MLC61, grey bar (Table 1).B. Infection outcome scores calculated after 3 days (means and standard deviations for six animals), where higher outcome scores reflect more severe infection.In both panels, **P < 0.01.
Discussion
In this study we combined molecular, cellular and proteomic approaches to examine the roles of ubiquitination, and polyubiquitin in particular, in the fungal pathogen C. albicans. A number of significant conclusions can be drawn from our datasets.
First we have shown that, despite the critical roles for ubiquitination in C. albicans, the polyubiquitin gene, UBI4, is not essential for viability (Fig. 2). The generation of a homozygous C. albicans ubi4/ubi4 mutant proved difficult in the past (Roig and Gozalbo, 2003). However, independent ubi4/ubi4 mutants were successfully generated in this study (Fig. 2), and their phenotypes were replicated by independent methionine-conditional C. albicans MET3p-UBI4/ubi4 mutants (Fig. 3 and Supporting information–MET3p-UBI4 control). The growth of ubi4/ubi4 cells is almost certainly supported by the expression of residual amounts of ubiquitin, even in the absence of the Ubi4 polyubiquitin protein (Fig. 7B and C). We presume that this residual ubiquitin is derived from the only other ubiquitin-encoding locus in the C. albicans genome, UBI3.
The second conclusion is that ubiquitination appears to contribute to cell cycle progression and morphogenesis in C. albicans. This conclusion is based on our observation that a subset of ubi4/ubi4 cells displays both aberrant nuclear segregation patterns and abnormal filamentous morphologies (Figs 3 and 4). However, if UBI4 plays a role in C. albicans cell cycle regulation, why did ubi4/ubi4 cells display population heterogeneity (Fig. 3)? Presumably the residual ubiquitin encoded by UBI3 is sufficient to drive cell cycle regulation in most ubi4/ubi4 cells. However, these residual ubiquitin levels might fall below critical levels in a subset of ubi4/ubi4 cells, thereby leading to the observed defects in morphology and nuclear segregation. Therefore, stochasticity in residual ubiquitin levels might account for the observed population heterogeneity of ubi4/ubi4 cells (Fig. 3).
Why might the defects in morphology and nuclear segregation correlate so strongly? Our data are consistent with the idea that ubiquitin depletion preferentially inhibits nuclear division in filamentous, rather than budding cells. According to this working model, once ubiquitin levels fall below a critical level, this probably triggers hyphal development in this C. albicans cell (Leng et al., 2000). The migration of the maternal nucleus into this new germ tube might proceed normally (Sudbery, 2001), but because ubiquitin levels are abnormally low in this cell, nuclear division is inhibited, thereby preventing the subsequent migration of the daughter nuclei into the mother and daughter compartments. This working model can account for our observations.
How might ubiquitination influence morphology and cell cycle-related events in C. albicans? Gsp1 is a ubiquitinated protein in C. albicans (Table 2). This Ran GTPase is also ubiquitinated in S. cerevisiae, where it is involved in the maintenance of nuclear and chromosome organization (Tagwerker et al., 2006). Overexpression of Gsp1 causes aberrant cell cycle progression in S. cerevisiae (Stevenson et al., 2001), and therefore aberrant ubiquitination of Gsp1 in C. albicans might account for the defective cell cycle progression in some ubi4/ubi4 cells (Fig. 4A). Genes encoding other ubiquitinated proteins in C. albicans have links to morphogenesis (Table 2). PDX3 is regulated during hyphal development (Nantel et al., 2002) and GPD2 is regulated by Ssn6, a negative regulator of morphogenesis (Hwang et al., 2003; Garcia-Sanchez et al., 2005). Interestingly, Ssn6 also regulates UBI4 expression (Garcia-Sanchez et al., 2005). While these links between ubiquitination and morphogenesis may appear relatively tenuous, they illustrate the important point that ubiquitination might influence C. albicans cell morphology indirectly via a number of cellular processes including cell cycle progression, defects in which are known to affect filamentation in C. albicans (Andaluz et al., 2001; Shi et al., 2007).
However, more direct links exist between ubiquitination and morphogenesis in C. albicans. Rad6, which is an E2 ubiquitin-conjugating enzyme that targets proteins for degradation via the proteasome (Madura and Varshavsky, 1994), is a negative regulator of hyphal development in C. albicans (Leng et al., 2000). The prevailing view is that Rad6 targets a component on the Ras-cAMP-Efg1 signalling pathway for ubiquitination-mediated protein degradation. Furthermore the inactivation of Doa1, which plays a role in the ubiquitin pathway (Hochstrasser et al., 1995; Johnson et al., 1995), promotes filamentous growth in C. albicans (Kunze et al., 2007). Therefore, ubiquitination probably influences C. albicans morphogenesis in several ways: directly through regulation of morphogenetic signalling events, and indirectly through modulation of cell cycle progression. Whatever the mechanisms, the observation that inactivation or downregulation of UBI4 affects colonial and cellular morphology in C. albicans is consistent with the work of Roig and Gozalbo (2003). It is also consistent with previous work from our laboratory implicating ubiquitination in the regulation of the yeast–hypha transition in C. albicans (Leng et al., 2000).
Our third main conclusion is that polyubiquitin is required for stress adaptation in C. albicans. C. albicans ubi4 mutants showed susceptibility to a wide range of stresses (Fig. 5). These defects were specific because, while C. albicans ubi4/ubi4 cells were sensitive to some conditions (such as elevated temperatures, cell wall and oxidative stresses), they were resistant to others (such as osmotic stress). The temperature-sensitive phenotype was consistent with the observation that UBI4 expression is upregulated in response to thermal upshifts (Roig and Gozalbo, 2002). The stress sensitivities of ubi4/ubi4 cells were also consistent with the findings that ubiquitination is stimulated by stresses in C. albicans (Fig. 7) and in S. cerevisiae (Cheng et al., 1994). Interestingly, although ubiquitination has been extensively studied in S. cerevisiae, there are no reports of ubiquitin-related osmotic stress phenotypes. This is entirely consistent with our observation that C. albicans ubi4/ubi4 cells are as resistant to osmotic stresses as wild-type cells (Fig. 5). Furthermore, proteomic screens in S. cerevisiae have shown that numerous heat shock proteins are ubiquitinated in this yeast. These include Hsc82, Hsp104 Ssa1, Ssa2, Ssa3, Ssa4, Ssb1, Ssb2 and Kar2 (Mayor et al., 2005; Tagwerker et al., 2006). Once again, this was consistent with our identification of Ssb1 as a ubiquitination target in C. albicans (Table 2), and other Hsp70 family members are likely to ubiquitinated in this pathogen as Hsp70, Ssa2 and Kar2, for example, all carry potential ubiquitination sites. In addition, we identified the oxidative stress response proteins Gnd1 and Pst2 as ubiquitination targets in C. albicans. The ubiquitination of Pst2 was induced by peroxide stress (Table 2). Furthermore, the peroxide-induced ubiquitination of several proteins was significantly attenuated in ubi4/ubi4 cells (Fig. 7B). Clearly, aberrant ubiquitination of Gnd1 and Pst2 could account, at least in part, for the stress sensitivities of C. albicans ubi4/ubi4 cells (Fig. 5).
The fourth conclusion is that polyubiquitin contributes significantly to metabolic adaptation in C. albicans. Our proteomic data and phenotypic analyses, combined with observations in other systems, strongly support this conclusion. Numerous metabolic enzymes were identified as ubiquitination targets in C. albicans, most of which are involved in carbon metabolism (Table 2). Enzymes on the following pathways were identified in our screen: glycolysis/gluconeogenesis (Eno1, Fba1, Pgk1); pentose phosphate pathway (Tkl1); fatty acid metabolism (Pdx3); acetate utilisation (Asc2); and glycerol synthesis (Gpd2). It should be noted that the loss of polyubiquitin affected the ability of C. albicans to survive under all of the starvation conditions examined (Fig. 6). However, the strongest metabolic phenotype was not observed when C. albicans cells were starved for carbon, but when cells were starved for nitrogen in the presence of excess carbon (Fig. 6C). Under these conditions ubi4/ubi4 cells rapidly lost their viability while control cells were able to support some growth.
The S. cerevisiae proteome undergoes considerable remodelling in response to nutrient limitation (Werner-Washburne et al., 1993), with proteins involved in protein turnover and nitrogen source scavenging being upregulated following nitrogen starvation (Kolkman et al., 2006). Furthermore, nitrogen recycling enzymes such as glutamine synthetase are ubiquitinated in S. cerevisiae (Peng et al., 2003; Tagwerker et al., 2006). We did not identify Gln1 in our screen. However, our bioinformatic analyses revealed that it contains four possible ubiquitination sites, indicating that it may still be a target in C. albicans. Nevertheless, we suggest that the ubiquitinated metabolic enzymes that we did identify reflect the recycling of available nitrogen in C. albicans, rather than the reprogramming of metabolism per se. Metabolic reprogramming undoubtedly takes place in C. albicans cells following starvation. This view is supported by the transcriptomic studies of Lorenz and co-workers (Lorenz et al., 2004), and by the observation that UBI4 is upregulated during starvation (Roig and Gozalbo, 2002). Nevertheless, the ubiquitination of glycolytic enzymes, for example, might reflect the fact that these are abundant proteins that are present in excess in C. albicans cells (Yin et al., 2004; Rodaki et al., 2006), thereby representing a ready source of organic nitrogen under conditions of nitrogen starvation.
The importance of ubiquitin-mediated protein turnover was further highlighted by our observation that the proteasomal components Pre1 and Pre3 are ubiquitinated in C. albicans (Table 2). Again this was consistent with findings in S. cerevisiae, where the proteasomal protein Pre5 is ubiquitinated (Kolkman et al., 2006).
Collectively, our data indicate that protein ubiquitination is highly dynamic in C. albicans, contributing to the regulation of key cellular processes. It is important to note that all of these processes underpin the virulence of this pathogen. Ubiquitination influences the growth and morphogenesis of C. albicans, which are fundamentally important for its virulence (Lo et al., 1997; Gow et al., 2002; Saville et al., 2003). Polyubiquitin is vital for robust stress adaptation in C. albicans, which is critical for pathogenicity (Alonso-Monge et al., 1999; Hwang et al., 2002; Smith et al., 2004; Hromatka et al., 2005; Enjalbert et al., 2006). Furthermore, effective metabolic adaptation in C. albicans is dependent upon polyubiquitin, and metabolic adaptation is central to host–fungus interactions during C. albicans infections (Lorenz and Fink, 2001; Lorenz et al., 2004; Brown, 2005; Fradin et al., 2005; Thewes et al., 2007; Zakikhany et al., 2007). It is not surprising therefore that our fifth conclusion is that polyubiquitin is important for the virulence of C. albicans in a mouse model of systemic candidiasis (Fig. 9). This study, and in particular our proteomic identification of ubiquitination targets, provides a platform for future studies to dissect the impact of specific target ubiquitination events upon the C. albicans cell and its virulence.
Experimental procedures
Strains, growth media and stress susceptibilities
Candida albicans strains (Table 1) were grown in YPD (Sherman, 1991) or synthetic complete medium lacking methionine and cysteine (Kaiser et al., 1994). The growth medium was supplemented with 2.5 mM methionine and cysteine for MET3 promoter shut-off assays.
Susceptibilities to H2O2 (5 mM), NaCl (1 M), Calcofluor White (20 µg ml−1), Congo Red (100 µg ml−1), tunicamycin (4.73 mM), caspofungin (0.064 µg ml−1) and thermal stress (37 and 42°C) were tested using cells grown in YPD medium at 30°C to exponential phase (OD600 = 0.5). Cells were serially diluted in YPD, spotted onto YPD plates containing the appropriate supplements and examined after incubation for 48 h at 30°C (or at the temperatures specified). Experiments were done in duplicate and with independent mutants.
Metabolic adaptation phenotypes were examined by growing cells in YPD medium at 30°C to exponential phase (OD600 = 0.5). These cells were harvested, washed in 20 ml water and then resuspended at an OD600 of 0.5 in water (starvation), YNB-C (carbon starvation: containing ammonium sulphate but no carbon source or amino acids) or YNB-N (nitrogen starvation: containing 2% glucose but no amino acids or ammonium sulphate). These cell suspensions were then incubated with shaking at 30°C, and cell viability measured at various times thereafter by plating samples onto YPD to determine colony-forming units (cfu).
To test the impact of stresses upon ubiquitination levels, cells were examined after exposing them to the following stresses for 1 h in YPD: heat shock (30–42°C) by addition of an equal volume of pre-warmed YPD at 54°C; hyphal inducing conditions (37°C containing 10% fetal calf serum); Congo Red (100 µg ml−1); SDS (10%); Calcofluor White (100 µg ml−1); and H2O2 (50 mM).
Strain construction
To generate conditional C. albicans UBI4 mutants, one UBI4 allele was placed under the control of the methionine/cysteine (Met/Cys) repressible MET3 promoter (Care et al., 1999), and the other allele was deleted using the lox-ARG4-lox marker (Dennison et al., 2005). To achieve this, the URA3-MET3p cassette from plasmid pURA3-MET3 (Rodaki et al., 2006) was PCR amplified with Pfu Turbo (Promega; Southampton, UK) using the primers MetUS-F and MetUS-R (Supporting information– Primers) to generate a URA3-MET3p-UBI4 cassette with 80 bp of homology to the UBI4 5′ upstream region and 80 bp of homology to the beginning of the UBI4 open reading frame. Similarly, the loxP-ARG4-loxP sequence from plasmid pLAL (Dennison et al., 2005) was PCR amplified using primers LAL-F and LAL-R (Supporting information– Primers) to generate an ubi4::ARG4 disruption cassette with 90 bp of flanking homology to the 5′ upstream and 3′ downstream regions of the UBI4 gene. C. albicans strain BWP17 (Table 1) was then transformed with these cassettes as described previously (Gietz et al., 1995; Walther and Wendland, 2003), to generate the independent strains MLC03 and MLC12 (MET3p-UBI4/UBI4). These strains were then transformed with the ubi4::ARG4 cassette to generate the independent conditional mutants MLC05 and MLC14 (MET3p-UBI4/ubi4::ARG4).
To generate C. albicans ubi4 null mutants, the two UBI4 alleles in the C. albicans strain BWP17 (UBI4/UBI4: Table 1) were inactivated sequentially with the loxP-URA3-loxP (LUL) and loxP-ARG4-loxP (LAL) markers (Dennison et al., 2005). This led to the construction of the independent ubi4/ubi4 mutants, MLC40, MLC41 and MLC42 (Table 1). The ubi4::LUL and ubi4::LAL disruption cassettes, which were designed to delete the complete UBI4 open reading frame, were generated by PCR amplification with the primers LAL-F, LAL-R and LUL-F, LUL-R (Supporting information– Primers). The ubi4::LUL cassette was transformed into C. albicans, transformants selected on the basis of their uridine protrophy, and correct integration confirmed by diagnostic PCR with primers UBI4d2-F and LULd-R (Supporting information– Primers). The resultant heterozygotes (ubi4::URA3/UBI4) were then transformed with the ubi4::LAL cassette, and accurate disruption of the remaining UBI4 allele confirmed by diagnostic PCR using primers UBI4d2-F and LALd-R (Supporting information– Primers). This yielded the homozygous null mutants, MLC40, MLC41 and MLC42 (ubi4::URA3/ubi4::ARG4) (Table 1). The genotypes of all of mutants made in this study were confirmed both by PCR and Southern analysis using the ECL Direct Nucleic Acid Labelling and Detection System (GE Healthcare; Chalfont St Giles, Buckinghamshire, UK).
To generate a reintegrant control strain (ubi4/ubi4/UBI4), the UBI4 locus was PCR amplified using the primers UBI4C-F and UBI4C-R (Supporting information– Primers), and ligated between the MluI and SalI sites of CIp30, a derivative of CIp10 (Murad et al., 2000; Dennison et al., 2005). The resulting CIp30-UBI4 plasmid was digested with StuI and integrated at the RPS10 locus in the ubi4/ubi4 mutant (MLC41) to generate the control strain MLC61 (Table 1). To generate prototrophic control strains for the virulence studies, the empty CIp30 vector was integrated at the RPS10 locus in MLC41 to generating MLC60, and CIp30 was transformed into BWP17 to generate MLC125 (Table 1).
mRNA analyses
UBI4 mRNA levels were measured by qRT-PCR. C. albicans cells were grown overnight at 30°C in YPD containing 2.5 mM Met/Cys. Cells were then reinoculated at OD600 = 0.1 into 50 ml of fresh YPD either containing or lacking Met/Cys, and regrown at 30°C to mid-log phase (OD600 = 0.5). Cells were harvested and frozen rapidly in liquid N2. RNA was extracted with Trizol (GibcoBRL, Grand Island, NY, USA) as described previously (Hauser et al., 1998), and RNA integrity assayed on an Agilent Bioanalyser 2100 (Stockport, UK). For qRT-PCR, samples were incubated at room temperature for 15 min in a 20 µl reaction mix containing 2 µg RNA, 2 µl DNase I buffer (Invitrogen; Paisley, UK), 1.5 µl DNase I and 1.5 µl RNase OUT (Invitrogen). cDNA was prepared using Superscript II (Invitrogen) as per the manufacturer's protocol. Real-time PCR was performed in triplicate in optical multiwall plate 384 using the LightCycler 480 Probes Master (Roche Applied Science; Burgess Hill, UK) as per the manufacturer's guidelines. Briefly, for the target transcripts UBI4 and ACT1, probes were chosen using the ProbeFinder Software Version 2.45 (Roche, http://www.universalprobelibrary.com). PCR was performed in a 20 µl reaction containing 10 µl LightCycler 480 Probes Master Mix, 3 µl of 1:5 diluted cDNA, 0.2 µl of forward and reverse primers, 0.2 µl selective probe (Roche) and 6.4 µl water, PCR grade. Negative controls were performed using water instead of cDNA. Reactions were performed in a LightCycler 480 system (Roche Applied Science) using the following parameters: preincubation at 95°C for 5 min, 50 cycles of amplification at 95°C for 10 s and 60°C for 30 s, and a final cooling at 40°C for 1 min. Standard curves were prepared using four dilutions of the control, wild-type samples. UBI4 mRNA levels were normalized against the internal ACT1 mRNA control (in arbitrary units).
Microscopy
Samples of exponentially growing C. albicans cells (OD600 = 0.5) were collected, fixed in 3.7% paraformaldehyde and examined by phase differential interference contrast microscopy. Cells were stained with 2 µg ml−1 CFW to visualize chitin. Nuclei were stained by overlaying samples with mounting media containing 1.5 µg ml−1 DAPI (Vector Laboratories, Peterborough, UK). All samples were examined by differential interference contrast and fluorescence microscopy using a Zeiss Axioplan 2 microscope. Images were recorded digitally using the Openlab system (Openlab v 4.04, Improvision, Coventry, UK) using a Hamamatsu C4742-95 digital camera (Hamamatsu Photonics, Hamamatsu, Japan).
1-D western analysis
Total soluble protein was extracted and subjected to Western blotting using published protocols (Smith et al., 2004). Briefly, cells were resuspended in 250 µl lysis buffer (0.1 M Tris-HCl, pH 8, 10% glycerol, 1 mM DTT, pepstatin A, protease inhibitor cocktail) and sheared with glass beads in a mini-bead beater (6 × 30 s with 1 min intervals on ice). Lysates were centrifuged at 13 000 r.p.m. for 10 min at 4°C. Proteins (15 µg) were separated by SDS-PAGE using the XCell SureLock™ Mini-Cell system (Invitrogen) with NuPAGE®Novex Bis-Tris 4–12% precast gels (Invitrogen) in NuPAGE® MOPS-SDS Running Buffer (Invitrogen) containing NuPAGE® Antioxidant (Invitrogen) as per the manufacturer's instructions. Proteins were transferred to Invitrolon™ PVDF Membranes (Invitrogen) in NuPAGE® Transfer Buffer containing methanol using the XCell II™ Blot Module (Invitrogen) as per the manufacturer's instructions. Following transfer, the membranes were rinsed in PBS and blocked in PBS-T + 5% BSA [PBS 0.1% Tween-20, 5% (w/v) BSA] for 1 h at room temperature. The membranes were then incubated overnight at 4°C in PBS-T + 5% BSA containing antibody. To detect ubiquitin, a 1:3000 dilution of anti-ubiquitin antibody was used (Stressgen, SPA-200, Exeter, UK). To detect Hsp90, a 1:10 000 dilution of anti-Hsp90 antibody was used (courtesy of Peter Piper) in PBS-T + 5% Milk [PBS 0.1% Tween-20, 5% (w/v) milk]. Membranes were incubated for 1 h at room temperature. Then an anti-rabbit HRP conjugated antibody was used at a 1:2000 dilution in PBS-T + 5% BSA for 1 h at room temperature. Membranes were washed in PBS-T and signals detected using an ECL Western blotting kit (Amersham, UK) as per the manufacturer's instructions. Signals were quantified using a Fusion FX7/SL Chemiluminescence and Fluorescence Combination Imaging System.
Proteomics
Candida albicans protein extracts were prepared as described previously (Yin et al., 2004). Briefly, cells were disrupted with glass beads in 160 µl lysis solution [7.5 M urea, 2.5 M thio-urea, 1.25 mM EDTA, 1.75 µg ml−1 pepstatin A, 62.5 mM DTT, 25 mM Tris-Cl, pH 10.8 and 3.7 × protease inhibitor cocktail tablets (Roche, Lewes, UK)] using a bead beater. After cell disruption, proteins were further solubilized by adding 40 µl of Lysis 2 [20% w/v CHAPS, 50% v/v glycerol, 10% v/v carrier ampholyte (pH 4–7)], and incubated on ice for 1 h. After repeated bead beating, cell debris were removed by centrifugation (20 min, 13 000 r.p.m., 4°C) and protein extracts were stored at −20°C.
Two dimensional gel electrophoresis was performed as described previously (Cash and Argo, 2009). After rehydration, IPG strips (pH 4–7 linear) were used to separate proteins on a Multiphor II (75–100 µg) in the first dimension according to the following protocol: 200 V for 1 min, 3500 V for 90 min, 3500 V for 65 min. After isoelectric focusing, IPG strips were equilibrated in two steps: (i) 50 mM Tris-Cl (pH 6.8), 6 M urea, 30% v/w glycerol, 2% w/w SDS, 62.5 mM DTT for 25 min; and (ii) 50 mM Tris-Cl (pH 6.8), 6 M urea, 30% v/w glycerol, 2% w/w SDS, 2.5% w/v iodoacetamide for 25 min. Proteins were then separated in the second dimension on precast 4–12% SDS polyacrylamide gels (Invitrogen) using MOPS buffer at first with 100 V per gel for 30 min, followed by 200 V per gel for a further 90 min. Gels were fixed (2% H3PO4, 50% ethanol) overnight, rinsed three times with water and equilibrated [1.3 M (NH4)2SO4, 2% w/w H3PO4, 34% w/v methanol] for 60 min, before staining with colloidal Coomassie blue G250 (0.67 g l−1). Equivalent 2-D gels were subjected to Western blotting to detect ubiquitin-conjugated proteins, as described above for 1-D gels. Independent biological replicates were performed for all experiments and only reproducible observations are reported.
Ubiquitinated proteins were selected by aligning the Western blots with the corresponding Coomassie-strained gels, and these proteins identified by liquid chromatography – tandem mass spectrometry (LC-MS/MS). The corresponding spots (1.2 mm diameter) were cut from gels and transferred to 96-well microtitre plates using an Investigator ProPic robotic workstation (Genomic Solutions; Huntingdon, UK). Proteins in gel plugs were digested with trypsin (Promega, UK) using an Investigator ProGest robotic workstation (Genomic Solutions). Peptides were extracted, dried in a SpeedVac SC110A (Savant Instruments) and dissolved in 0.1% formic acid for LC-MS/MS. The LC-MS system comprised an UltiMate 3000 LC system (Dionex) coupled to an HCTultra ion trap mass spectrometer with ESI source fitted with a low-flow nebulizer (Bruker Daltonics). Peptides were separated on a PepSwift monolithic capillary column (Dionex) at 2 µl min−1 using a linear gradient of acetonitrile. Eluent A was 3% acetonitrile in 0.05% formic acid and Eluent B was 80% acetonitrile in 0.04% formic acid. The gradient was 3–45% Eluent B over 12 min at a flow rate of 2 µl min−1.
Tandem mass spectra were acquired in data-dependent AutoMS(2) mode using the following parameters: MS scan range = 300–1500 m/z; averages = 3, max. no. of precursors = 3; MS(2) scan range = 100–2200 m/z, averages = 2; active exclusion on (max. spectra = 2, release after 1 min). Peptide peaks were detected and deconvoluted automatically using DataAnalysis software (Bruker). Mass lists were used as input for Mascot MS/MS Ions searches using Mascot Server version 2.2 (Matrix Science, London, UK). Our in-house protein sequence database (containing 6166 sequences) was built from the flat file <CALBI_prot.txt> downloaded from the CandidaDB web server ftp://ftp.pasteur.fr/pub/GenomeDB/CandidaDB/FlatFiles. Protein annotations were taken from the Candida Genome Database (http://www.candidagenome.org/) and CandidaDB (http://genolist.pasteur.fr/CandidaDB/). Potential ubiquitination sites were predicted using the program UbPred (Radivojac et al., 2010).
Our proteomics dataset can be accessed via the PRoteomics IDEntifications database (PRIDE) data repository at the European Bioinformatics Institute (http://www.ebi.ac.uk/pride) (Vizcaino et al., 2010) using Accession Number 13 697.
C. albicans virulence assays
All animal experimentation conformed to the requirements of the UK Home Office legislation and of the Ethical Review Committee of the University of Aberdeen.
The virulence of C. albicans strains MLC125, MLC60 and MLC61 (Table 1) was assessed in the mouse intravenous challenge model (MacCallum et al., 2010). Female BALB/c mice (6–8 weeks old; Charles River, UK) were infected intravenously with a saline suspension of C. albicans cells (4 × 104 cfu g−1 mouse body weight in a 100 µl volume; six mice per C. albicans strain), prepared from cells grown for 18–24 h in NGY medium at 30°C (MacCallum and Odds, 2005). At 72 h post infection, mice were weighed, humanely terminated, the kidneys removed aseptically and renal fungal burdens determined. Infection outcome scores were calculated on the basis of weight change and renal fungal burdens, as described previously (MacCallum et al., 2010). For virulence assays, kidney burdens and infection outcome scores were compared by the Kruskall–Wallis and Mann–Whitney U-tests. Statistical analyses were carried out using PASW statistics 18.0.
Acknowledgments
We are grateful to Brian Morgan, Jan Quinn, Neil Gow and Phil Cash for stimulating discussions and helpful advice. We would also like to thank Peter Piper for providing us with the anti-Hsp90 antibody. M.L. was supported by a Carnegie/Caledonian Scholarship from The Carnegie Trust. This work was also supported by grants from the UK Biotechnology and Biological Research Council (BB/D009308/1; BB/F00513X/1), the Wellcome Trust (080088) and the European Commission (PITN-GA-2008-214004; ERC-2009-AdG-249793). We thank Susan Budge and Jan Walker for excellent technical support.
Supporting information
Additional supporting information may be found in the online version of this article.
Please note: Wiley-Blackwell are not responsible for the content or functionality of any supporting materials supplied by the authors. Any queries (other than missing material) should be directed to the corresponding author for the article.
References
- Alonso-Monge R, Navarro-Garcia F, Molero G, Diez-Orejas R, Gustin M, Pla J, et al. Role of the mitogen-activated protein kinase Hog1p in morphogenesis and virulence of Candida albicans. J Bacteriol. 1999;181:3058–3068. doi: 10.1128/jb.181.10.3058-3068.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amerik AY, Li S-J, Hochstrasser M. Analysis of the deubiquitinating enzymes of the yeast Saccharomyces cerevisiae. Biol Chem. 2005;381:981–992. doi: 10.1515/BC.2000.121. [DOI] [PubMed] [Google Scholar]
- Andaluz E, Calderone R, Reyes G, Larriba G. Phenotypic analysis and virulence of Candida albicans LIG4 mutants. Infect Immun. 2001;69:137–147. doi: 10.1128/IAI.69.01.137-147.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachmair A, Finley D, Varshavsky A. In vivo half-life of a protein is a function of its amino-terminal residue. Science. 1986;234:179–186. doi: 10.1126/science.3018930. [DOI] [PubMed] [Google Scholar]
- Brown AJP, Haynes K, Quinn J. Nitrosative and oxidative stress responses in fungal pathogenicity. Curr Opin Microbiol. 2009;12:384–391. doi: 10.1016/j.mib.2009.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown AJP. Integration of metabolism with virulence in Candida albicans. In: Brown AJP, editor. Fungal Genomics. In Mycota XIII. Heidelberg: Springer-Verlag; 2005. pp. 185–203. [Google Scholar]
- Brown AJP, Barelle CJ, Budge S, Duncan J, Harris S, Lee PR, et al. Gene regulation during morphogenesis in Candida albicans. In: Ernst JF, Schmidt A, editors. Contributions to Microbiology: Dimorphism in Human Pathogenic and Apathogenic Yeasts. Basel: Karger AG; 2000. pp. 112–125. [DOI] [PubMed] [Google Scholar]
- Calderone RA. Candida and Candidiasis. Washington, DC: ASM Press; 2002. [Google Scholar]
- Care RS, Trevethick J, Binley KM, Sudbery PE. The MET3 promoter: a new tool for Candida albicans molecular genetics. Mol Microbiol. 1999;34:792–798. doi: 10.1046/j.1365-2958.1999.01641.x. [DOI] [PubMed] [Google Scholar]
- Cash P, Argo E. Analysis of bacterial proteins by 2DE. Methods Mol Biol. 2009;519:131–144. doi: 10.1007/978-1-59745-281-6_9. [DOI] [PubMed] [Google Scholar]
- Chauhan N, Latge JP, Calderone R. Signalling and oxidant adaptation in Candida albicans and Aspergillus fumigatus. Nat Rev Microbiol. 2006;4:435–444. doi: 10.1038/nrmicro1426. [DOI] [PubMed] [Google Scholar]
- Cheng L, Watt R, Piper PW. Polyubiquitin gene expression contributes to oxidative stress resistance in respiratory yeast (Saccharomyces cerevisiae) Mol Gen Genet. 1994;243:358–362. doi: 10.1007/BF00301072. [DOI] [PubMed] [Google Scholar]
- Ciechanover A. The ubiquitin-proteasome pathway: on protein death and cell life. EMBO J. 1998;17:7151–7160. doi: 10.1093/emboj/17.24.7151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciechanover A, Orian A, Schwartz AL. Ubiquitin-mediated proteolysis: biological regulation via destruction. Bioessays. 2000a;22:442–451. doi: 10.1002/(SICI)1521-1878(200005)22:5<442::AID-BIES6>3.0.CO;2-Q. [DOI] [PubMed] [Google Scholar]
- Ciechanover A, Orian A, Schwartz AL. The ubiquitin-mediated proteolytic pathway: mode of action and clinical implications. J Cell Biochem Suppl. 2000b;34:40–51. doi: 10.1002/(sici)1097-4644(2000)77:34+<40::aid-jcb9>3.0.co;2-6. [DOI] [PubMed] [Google Scholar]
- Dennison PM, Ramsdale M, Manson CL, Brown AJ. Gene disruption in Candida albicans using a synthetic, codon-optimised Cre-loxP system. Fungal Genet Biol. 2005;42:737–748. doi: 10.1016/j.fgb.2005.05.006. [DOI] [PubMed] [Google Scholar]
- Enjalbert B, Smith DA, Cornell MJ, Alam I, Nicholls S, Brown AJ, Quinn J. Role of the Hog1 stress-activated protein kinase in the global transcriptional response to stress in the fungal pathogen Candida albicans. Mol Biol Cell. 2006;17:1018–1032. doi: 10.1091/mbc.E05-06-0501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enjalbert B, MacCallum DM, Odds FC, Brown AJ. Niche-specific activation of the oxidative stress response by the pathogenic fungus Candida albicans. Infect Immun. 2007;75:2143–2151. doi: 10.1128/IAI.01680-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ernst JF. Transcription factors in Candida albicans– environmental control of morphogenesis. Microbiology. 2000;146:1763–1774. doi: 10.1099/00221287-146-8-1763. [DOI] [PubMed] [Google Scholar]
- Finley D, Chau V. Ubiquitination. Ann Rev Cell Biol. 1991;7:25–69. doi: 10.1146/annurev.cb.07.110191.000325. [DOI] [PubMed] [Google Scholar]
- Finley D, Özkaynak E, Varshavsky A. The yeast polyubiquitin gene is essential for resistance to high temperatures, starvation, and other stresses. Cell. 1987;48:1035–1046. doi: 10.1016/0092-8674(87)90711-2. [DOI] [PubMed] [Google Scholar]
- Fradin C, Groot PD, MacCallum D, Schaller M, Klis F, Odds FC, Hube B. Granulocytes govern the transcriptional response, morphology and proliferation of Candida albicans in human blood. Mol Microbiol. 2005;56:397–415. doi: 10.1111/j.1365-2958.2005.04557.x. [DOI] [PubMed] [Google Scholar]
- Fraser J, Luu HA, Neculcea J, Thomas DY, Storms RK. Ubiquitin gene expression: response to environmental changes. Curr Genet. 1991;20:17–23. doi: 10.1007/BF00312760. [DOI] [PubMed] [Google Scholar]
- Friant S, Meier KD, Riezman H. Increased ubiquitin-dependent degradation can replace the essential requirement for heat shock protein induction. EMBO J. 2003;22:3783–3791. doi: 10.1093/emboj/cdg375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Sanchez S, Mavor AL, Russell CL, Argimon S, Dennison P, Enjalbert B, Brown AJP. Global roles of Ssn6 in Tup1- and Nrg1-dependent gene regulation in the fungal pathogen, Candida albicans. Mol Biol Cell. 2005;16:2913–2925. doi: 10.1091/mbc.E05-01-0071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gietz RD, Schiestl RH, Willems AR, Woods RA. Studies on the transformation of intact yeast cells by the LiAc/SS-DNA/PEG procedure. Yeast. 1995;11:355–360. doi: 10.1002/yea.320110408. [DOI] [PubMed] [Google Scholar]
- Glotzer M, Murray AW, Kirschner MW. Cyclin is degraded by the ubiquitin pathway. Nature. 1991;349:132–138. doi: 10.1038/349132a0. [DOI] [PubMed] [Google Scholar]
- Gow NA, Brown AJ, Odds FC. Fungal morphogenesis and host invasion. Curr Opin Microbiol. 2002;5:366–371. doi: 10.1016/s1369-5274(02)00338-7. [DOI] [PubMed] [Google Scholar]
- Hauser NC, Vingron M, Scheideler M, Krems B, Hellmuth K, Entian KD, Hoheisel JD. Transcriptional profiling on all open reading frames of Saccharomyces cerevisiae. Yeast. 1998;14:1209–1221. doi: 10.1002/(SICI)1097-0061(19980930)14:13<1209::AID-YEA311>3.0.CO;2-N. [DOI] [PubMed] [Google Scholar]
- Hershko A. Roles of ubiquitin-mediated proteolysis in cell cycle control. Curr Opin Cell Biol. 1997;9:788–799. doi: 10.1016/s0955-0674(97)80079-8. [DOI] [PubMed] [Google Scholar]
- Hochstrasser M, Papa FR, Chen P, Swaminathan S, Johnson P, Stillman L, et al. The DOA pathway: studies on the functions and mechanisms of ubiquitin-dependent protein degradation in the yeast Saccharomyces cerevisiae. Cold Spring Harb Symp Quant Biol. 1995;60:503–513. doi: 10.1101/sqb.1995.060.01.054. [DOI] [PubMed] [Google Scholar]
- Hromatka BS, Noble SM, Johnson AD. Transcriptional response of Candida albicans to nitric oxide and the role of the YHB1 gene in nitrosative stress and virulence. Mol Biol Cell. 2005;16:4814–4826. doi: 10.1091/mbc.E05-05-0435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hube B. Candida albicans secreted aspartyl proteinases. Curr Top Med Mycol. 1996;7:55–69. [PubMed] [Google Scholar]
- Hwang CS, Rhie GE, Oh JH, Huh WK, Yim HS, Kang SO. Copper- and zinc-containing superoxide dismutase (Cu/ZnSOD) is required for the protection of Candida albicans against oxidative stresses and the expression of its full virulence. Microbiology. 2002;148:3705–3713. doi: 10.1099/00221287-148-11-3705. [DOI] [PubMed] [Google Scholar]
- Hwang C-S, Oh J-H, Huh W-K, Yim H-S, Kang S-O. Ssn6, an important factor of morphological conversion and virulence in Candida albicans. Mol Microbiol. 2003;47:1029–1043. doi: 10.1046/j.1365-2958.2003.03353.x. [DOI] [PubMed] [Google Scholar]
- Jentsch S, Seufert W, Sommer T, Reins HA. Ubiquitin-conjugating enzymes: novel regulators of eukaryotic cells. Trends Biochem Sci. 1990;15:195–198. doi: 10.1016/0968-0004(90)90161-4. [DOI] [PubMed] [Google Scholar]
- Jentsch S, Seufert W, Hauser H-P. Genetic analysis of the ubiquitin system. Biochim Biophys Acta. 1991;1089:127–139. doi: 10.1016/0167-4781(91)90001-3. [DOI] [PubMed] [Google Scholar]
- Johnson ES, Ma PC, Ota IM, Varshavsky A. A proteolytic pathway that recognizes ubiquitin as a degradation signal. J Biol Chem. 1995;270:17442–17456. doi: 10.1074/jbc.270.29.17442. [DOI] [PubMed] [Google Scholar]
- Kaiser C, Michaelis S, Mitchell A. Methods in Yeast Genetics. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1994. [Google Scholar]
- Kim SY, Kim J. Roles of dihydrolipoamide dehydrogenase Lpd1 in Candida albicans filamentation. Fungal Genet Biol. 2010;47:782–788. doi: 10.1016/j.fgb.2010.06.005. [DOI] [PubMed] [Google Scholar]
- Kolkman A, Daran-Lapujade P, Fullaondo A, Olsthoorn MMA, Pronk JT, Slijper M, Heck AJR. Proteome analysis of yeast response to various nutrient limitations. Mol Syst Biol. 2006;2:2006.0026. doi: 10.1038/msb4100069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunze D, MacCallum D, Odds FC, Hube B. Multiple functions of DOA1 in Candida albicans. Microbiology. 2007;153:1026–1041. doi: 10.1099/mic.0.2006/002741-0. [DOI] [PubMed] [Google Scholar]
- Leberer E, Harcus D, Broadbent ID, Clark KL, Dignard D, Ziegelbauer K, et al. Signal transduction through homologs of the Ste20p and Ste7p protein kinases can trigger hyphal formation in the pathogenic fungus Candida albicans. Proc Natl Acad Sci USA. 1996;93:13217–13222. doi: 10.1073/pnas.93.23.13217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leng P, Sudbery PE, Brown AJP. Rad6p represses yeast-hypha morphogenesis in the human fungal pathogen Candida albicans. Mol Microbiol. 2000;35:1264–1275. doi: 10.1046/j.1365-2958.2000.01801.x. [DOI] [PubMed] [Google Scholar]
- Liu H, Kohler J, Fink GR. Suppression of hyphal formation in Candida albicans by mutation of a STE12 homolog. Science. 1994;266:1723–1726. doi: 10.1126/science.7992058. [DOI] [PubMed] [Google Scholar]
- Lo HJ, Kohler JR, DiDomenico B, Loebenberg D, Cacciapuoti A, Fink G. Nonfilamentous C. albicans mutants are avirulent. Cell. 1997;90:939–949. doi: 10.1016/s0092-8674(00)80358-x. [DOI] [PubMed] [Google Scholar]
- Lorenz MC, Fink GR. The glyoxylate cycle is required for fungal virulence. Nature. 2001;412:83–86. doi: 10.1038/35083594. [DOI] [PubMed] [Google Scholar]
- Lorenz MC, Bender JA, Fink GR. Transcriptional response of Candida albicans upon internalization by macrophages. Eukaryot Cell. 2004;3:1076–1087. doi: 10.1128/EC.3.5.1076-1087.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacCallum DM, Odds FC. Temporal events in the intravenous challenge model for experimental Candida albicans infections in female mice. Mycoses. 2005;48:151–161. doi: 10.1111/j.1439-0507.2005.01121.x. [DOI] [PubMed] [Google Scholar]
- MacCallum DM, Coste A, Ischer F, Jacobsen MD, Odds FC, Sanglard D. Genetic dissection of azole resistance mechanisms in Candida albicans and their validation in a mouse model of disseminated infection. Antimicrob Agents Chemother. 2010;54:1476–1483. doi: 10.1128/AAC.01645-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madura K, Varshavsky A. Degradation of G alpha by the N-end rule pathway. Science. 1994;265:1454–1458. doi: 10.1126/science.8073290. [DOI] [PubMed] [Google Scholar]
- Mavor AL, Thewes S, Hube B. Systemic fungal infections caused by Candida Species: epidemiology, infection process and virulence attributes. Curr Drug Targets. 2005;6:863–874. doi: 10.2174/138945005774912735. [DOI] [PubMed] [Google Scholar]
- Mayor T, Lipford JR, Graumann J, Smith GT, Deshaies RJ. Analysis of polyubiquitin conjugates reveals that the Rpn10 substrate receptor contributes to the turnover of multiple proteasome targets. Mol Cell Proteomics. 2005;4:741–751. doi: 10.1074/mcp.M400220-MCP200. [DOI] [PubMed] [Google Scholar]
- Miller RA, Britigan BE. Role of oxidants in microbial pathophysiology. Clin Microbiol Rev. 1997;10:1–18. doi: 10.1128/cmr.10.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murad AM, Lee PR, Broadbent ID, Barelle CJ, Brown AJ. CIp10, an efficient and convenient integrating vector for Candida albicans. Yeast. 2000;16:325–327. doi: 10.1002/1097-0061(20000315)16:4<325::AID-YEA538>3.0.CO;2-#. [DOI] [PubMed] [Google Scholar]
- Muratani M, Kung C, Shokat KM, Tansey WP. The F box protein Dsg1/Mdm30 is a transcriptional coactivator that stimulates Gal4 turnover and cotranscriptional mRNA processing. Cell. 2005;120:887–899. doi: 10.1016/j.cell.2004.12.025. [DOI] [PubMed] [Google Scholar]
- Nakayama H, Mio T, Nagahashi S, Kokado M, Arisawa M, Aoki Y. Tetracycline-regulatable system to tightly control gene expression in the pathogenic fungus Candida albicans. Infect Immun. 2000;68:6712–6719. doi: 10.1128/iai.68.12.6712-6719.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nantel A, Dignard D, Bachewich C, Harcus D, Marcil A, Bouin A-P, et al. Transcription profiling of Candida albicans cells undergoing the yeast-to-hyphal transition. Mol Biol Cell. 2002;13:3452–3465. doi: 10.1091/mbc.E02-05-0272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicholls S, Leach MD, Priest CL, Brown AJ. Role of the heat shock transcription factor, Hsf1, in a major fungal pathogen that is obligately associated with warm-blooded animals. Mol Microbiol. 2009;74:844–861. doi: 10.1111/j.1365-2958.2009.06883.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Odds FC. Candida and Candidosis. London: Bailliere Tindall; 1988. [Google Scholar]
- Ozkaynak E, Finley D, Solomon MJ, Varshavsky A. The yeast ubiquitin genes: a family of natural gene fusions. EMBO J. 1987;6:1429–1439. doi: 10.1002/j.1460-2075.1987.tb02384.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paiva S, Vieira N, Nondier I, Haguenauer-Tsapis R, Casal M, Urban-Grimal D. Glucose-induced ubiquitylation and endocytosis of the yeast Jen1 transporter: role of lysine 63-linked ubiquitin chains. J Biol Chem. 2009;284:19228–19236. doi: 10.1074/jbc.M109.008318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng J, Schwartz D, Elias JE, Thoreen CC, Cheng D, Marsischky G, et al. A proteomics approach to understanding protein ubiquitination. Nat Biotechnol. 2003;21:921–926. doi: 10.1038/nbt849. [DOI] [PubMed] [Google Scholar]
- Radivojac P, Vacic V, Haynes C, Cocklin RR, Mohan A, Heyen JW, et al. Identification, analysis, and prediction of protein ubiquitination sites. Proteins. 2010;78:365–380. doi: 10.1002/prot.22555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rechsteiner M. Natural substrates of the ubiquitin proteolytic pathway. Cell. 1991;66:615–618. doi: 10.1016/0092-8674(91)90104-7. [DOI] [PubMed] [Google Scholar]
- Rodaki A, Young T, Brown AJ. Effects of depleting the essential central metabolic enzyme fructose-1,6-bisphosphate aldolase on the growth and viability of Candida albicans: implications for antifungal drug target discovery. Eukaryot Cell. 2006;5:1371–1377. doi: 10.1128/EC.00115-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roig P, Gozalbo D. Candida albicans UBI3 and UBI4 promoter regions confer differential regulation of invertase production to saccharomyces cerevisiae cells in response to stress. Int Microbiol. 2002;5:33–36. doi: 10.1007/s10123-002-0056-1. [DOI] [PubMed] [Google Scholar]
- Roig P, Gozalbo D. Depletion of polyubiquitin encoded by the UBI4 gene confers pleiotropic phenotype to Candida albicans cells. Fungal Gen Biol. 2003;39:70–81. doi: 10.1016/s1087-1845(03)00004-5. [DOI] [PubMed] [Google Scholar]
- Roig P, Martínez JP, Gil ML, Gozalbo D. Molecular cloning and characterization of the Candida albicans UBI3 gene coding for a ubiquitin-hybrid protein. Yeast. 2000;16:1413–1419. doi: 10.1002/1097-0061(200011)16:15<1413::AID-YEA632>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- Santt O, Pfirrmann T, Braun B, Juretschke J, Kimmig P, Scheel H, et al. The yeast GID complex, a novel ubiquitin ligase (E3) involved in the regulation of carbohydrate metabolism. Mol Biol Cell. 2008;19:3323–3333. doi: 10.1091/mbc.E08-03-0328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saville SP, Lazzell AL, Monteagudo C, Lopez-Ribot JL. Engineered control of cell morphology in vivo reveals distinct roles for yeast and filamentous forms of Candida albicans during infection. Eukaryot Cell. 2003;2:1053–1060. doi: 10.1128/EC.2.5.1053-1060.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sepulveda P, Cervera AM, Lopez-Ribot JL, Chaffin WL, Martinez JP, Gozalbo D. Cloning and characterization of a cDNA coding for Candida albicans polyubiquitin. Med Mycol. 1996;34:315–322. [PubMed] [Google Scholar]
- Seyfried NT, Xu P, Duong DM, Cheng D, Hanfelt J, Peng J. Systematic approach for validating the ubiquitinated proteome. Anal Chem. 2008;80:4161–4169. doi: 10.1021/ac702516a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherman F. Getting started with yeast. Methods Enzymol. 1991;194:3–21. doi: 10.1016/0076-6879(91)94004-v. [DOI] [PubMed] [Google Scholar]
- Shi QM, Wang YM, Zheng XD, Lee RT, Wang Y. Critical role of DNA checkpoints in mediating genotoxic-stress-induced filamentous growth in Candida albicans. Mol Biol Cell. 2007;18:815–826. doi: 10.1091/mbc.E06-05-0442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skrzypek MS, Arnaud MB, Costanzo MC, Inglis DO, Shah P, Binkley G, et al. New tools at the Candida Genome Database: biochemical pathways and full-text literature search. Nucleic Acids Res. 2010;38:D428–D432. doi: 10.1093/nar/gkp836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith DA, Nicholls S, Morgan BA, Brown AJ, Quinn J. A conserved stress-activated protein kinase regulates a core stress response in the human pathogen Candida albicans. Mol Biol Cell. 2004;15:4179–4190. doi: 10.1091/mbc.E04-03-0181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stadtman ER. Covalent modification reactions are marking steps in protein turnover. Biochemistry. 1990;29:6323–6331. doi: 10.1021/bi00479a001. [DOI] [PubMed] [Google Scholar]
- Staub O, Rotin D. Role of ubiquitylation in cellular membrane transport. Physiol Rev. 2006;86:669–707. doi: 10.1152/physrev.00020.2005. [DOI] [PubMed] [Google Scholar]
- Stead DA, Preece A, Brown AJ. Universal metrics for quality assessment of protein identifications by mass spectrometry. Mol Cell Proteomics. 2006;5:1205–1211. doi: 10.1074/mcp.M500426-MCP200. [DOI] [PubMed] [Google Scholar]
- Stevenson LF, Kennedy BK, Harlow E. A large-scale overexpression screen in Saccharomyces cerevisiae identifies previously uncharacterized cell cycle genes. Proc Natl Acad Sci USA. 2001;98:3946–3951. doi: 10.1073/pnas.051013498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sudbery PE. The germ tubes of Candida albicans hyphae and pseudohyphae show different patterns of septin ring localization. Mol Microbiol. 2001;41:19–31. doi: 10.1046/j.1365-2958.2001.02459.x. [DOI] [PubMed] [Google Scholar]
- Swoboda RK, Bertram G, Budge S, Gooday GW, Gow NA, Brown AJ. Structure and regulation of the HSP90 gene from the pathogenic fungus Candida albicans. Infect Immun. 1995;63:4506–4514. doi: 10.1128/iai.63.11.4506-4514.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tagwerker C, Flick K, Cui M, Guerrero C, Dou Y, Auer B, et al. A tandem affinity tag for two-step purification under fully denaturing conditions. Mol Cell Proteom. 2006;5:737–748. doi: 10.1074/mcp.M500368-MCP200. [DOI] [PubMed] [Google Scholar]
- Tanaka K, Matsumoto K, Toh-e A. Dual regulation of the expression of the polyubiquitin gene by cyclic AMP and heat shock in yeast. EMBO J. 1988;7:495–502. doi: 10.1002/j.1460-2075.1988.tb02837.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thewes S, Kretschmar M, Park H, Schaller M, Filler SG, Hube B. In vivo and ex vivo comparative transcriptional profiling of invasive and non-invasive Candida albicans isolates identifies genes associated with tissue invasion. Mol Microbiol. 2007;63:1606–1628. doi: 10.1111/j.1365-2958.2007.05614.x. [DOI] [PubMed] [Google Scholar]
- Vizcaino JA, Cote R, Reisinger F, Barsnes H, Foster JM, Rameseder J, et al. The proteomics identifications database: 2010 update. Nucleic Acids Res. 2010;38:D736–D742. doi: 10.1093/nar/gkp964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walther A, Wendland J. An improved transformation protocol for the human fungal pathogen Candida albicans. Curr Genet. 2003;42:339–343. doi: 10.1007/s00294-002-0349-0. [DOI] [PubMed] [Google Scholar]
- Weissman AM. Themes and variations on ubiquitylation. Nat Rev Mol Cell Biol. 2001;2:169–178. doi: 10.1038/35056563. [DOI] [PubMed] [Google Scholar]
- Werner-Washburne M, Braun E, Johnston GC, Singer RA. Stationary phase in the yeast Saccharomyces cerevisiae. Microbiol Rev. 1993;57:383–401. doi: 10.1128/mr.57.2.383-401.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson RB, Davis D, Mitchell AP. Rapid hypothesis testing with Candida albicans through gene disruption with short homology regions. J Bacteriol. 1999;181:1868–1874. doi: 10.1128/jb.181.6.1868-1874.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wimalasena TT, Enjalbert B, Guillemette T, Plumridge A, Budge S, Yin Z, et al. Impact of the unfolded protein response upon genome-wide expression patterns, and the role of Hac1 in the polarized growth, of Candida albicans. Fungal Genet Biol. 2008;45:1235–1247. doi: 10.1016/j.fgb.2008.06.001. [DOI] [PubMed] [Google Scholar]
- Wolf DH, Hilt W. The proteasome: a proteolytic nanomachine of cell regulation and waste disposal. Biochem Biophys Acta. 2004;1695:19–31. doi: 10.1016/j.bbamcr.2004.10.007. [DOI] [PubMed] [Google Scholar]
- Yin Z, Stead D, Selway L, Walker J, Riba-Garcia I, McLnerney T, et al. Proteomic response to amino acid starvation in Candida albicans and Saccharomyces cerevisiae. Proteomics. 2004;4:2425–2436. doi: 10.1002/pmic.200300760. [DOI] [PubMed] [Google Scholar]
- Zakikhany K, Naglik JR, Schmidt-Westhausen A, Holland G, Schaller M, Hube B. In vivo transcript profiling of Candida albicans identifies a gene essential for interepithelial dissemination. Cell Microbiol. 2007;9:2938–2954. doi: 10.1111/j.1462-5822.2007.01009.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


