Abstract
A subset of cells called leukemia stem cells (LSCs) possess limitless self-renewal and are responsible for maintenance of leukemia. Selective eradication of LSCs could offer significant therapeutic benefit and there is great interest in identifying the signaling pathways that control their development. Here, LSCs were studied in mouse models of acute myelogenous leukemia (AML) induced either by co-expression of the Hoxa9 and Meis1a oncogenes or the fusion oncoprotein MLL-AF9. We show that the Wnt/β-catenin signaling pathway is required for self-renewal of LSCs derived from either hematopoietic stem cells (HSC) or more differentiated granulocyte macrophage progenitors (GMP). Since the Wnt/β-catenin pathway is normally active in HSCs, but not in GMP, reactivation of β-catenin signaling is required for transformation of progenitor cells by certain oncogenes. β-catenin is not absolutely required for self-renewal of adult HSCs, thus targeting the Wnt/β-catenin pathway may represent a new therapeutic opportunity in AML.
Acute myelogenous leukemia (AML) is the most common acute leukemia in adults and most people with AML are not cured with current therapies. Only a small subset of AML cells are capable of extensive proliferation and self-renewal (1). Such cells are referred to as leukemia stem cells (LSC) because they share properties, including extensive self-renewal potential, with normal stem cells. LSC are being studied as potential therapeutic targets (2, 3), and the signaling pathways that control their development and survival are of particular interest. The Wnt/β-catenin is active in some human leukemias (4–7) and in hematopoietic stem cells (HSC) (8–10). While β-catenin is not required for self-renewal of adult HSC (11, 12), it is unclear if β-catenin is required for LSC development and maintenance in AML. We have studied well characterized mouse models to address whether β-catenin is necessary for LSC development from either HSC or more differentiated committed myeloid progenitor cells.
Hox genes have been implicated in the regulation of normal stem cell self-renewal (16, 17), and enforced co-expression of Hoxa9 and Meis1a in murine bone marrow leads to rapid AML development (18). Previous studies demonstrated that leukemogenic fusion proteins, such as those involving the mixed lineage leukemia (MLL) protein e.g. MLL-AF9, can transform non-self renewing granulocyte/macrophage restricted progenitors (GMP) and activate genes encoding homeobox (Hox) proteins such as HoxA9 and Meis1 (13–15). To determine whether a combination of Hoxa9 and Meis1a (HoxA9/M) can transform GMP, we co-transduced sorted Lin− c-Kit+ Sca-1+ (KLS) cells, which are enriched for HSC, or Lin− c-Kit+ Sca-1− FcRγ+ CD34+ (GMP) cells with viruses encoding HoxA9 (MSCV-HoxA9-GFP) and Meis1a (MSCV-Meis1a-puro), and used either a bulk approach (15, 18, 26) or a single cell approach (Fig. S1). When single cells were expanded in vitro up to 6 weeks, no significant differences were observed between KLS cells transduced with HoxA9/M (KLS-HoxA9/M), GMP transduced with HoxA9/M (GMP-HoxA9/M), or GMP transduced with MLL-AF9 (GMP-MLLAF9) in proliferation, cell cycle profile, apoptosis levels, gross morphology or immunophenotype (Fig. S1). However there was a proliferative defect upon extended culture of GMP-HoxA9/M cells. When cells were injected into recipient mice, 13 of 14 (93%) mice that received GMP-MLLAF9 cells developed AML, 14 of 20 (70%) of mice that received KLS-HoxA9/M cells developed AML, whereas only 1 of 23 mice that received GMP-HoxA9/M cells developed AML (Fig. S2). These differences in leukemogenic potential could not be ascribed to differential expression of HoxA9 and Meis1 (Fig. S3)
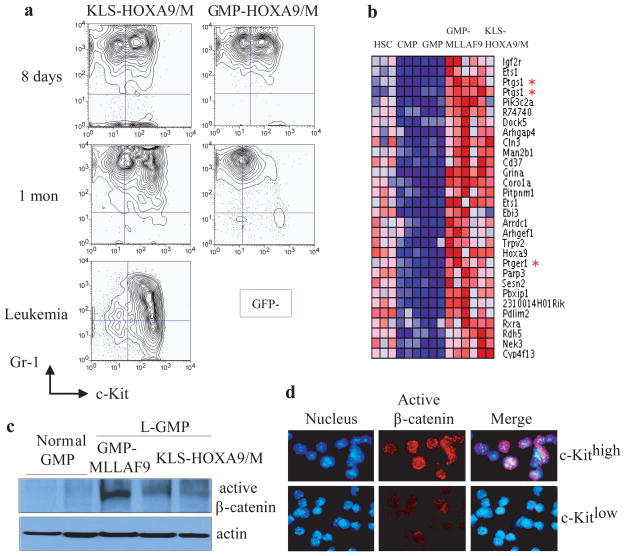
To characterize the in vivo defect in HoxA9/M transduced GMP we assessed for GFP+ cells and their immunophenotypes in recipient mice at multiple timepoints after transplantation. By 18 hours KLS-HoxA9/M and GMP-HoxA9/M cells homed to the bone marrow with similar efficiency (Fig. S2). By eight days both cell types displayed heterogeneity of expression of the stem cell marker, c-Kit (c-Kithigh and c-Kitlow) (Fig. 1a). KLS-HoxA9/M cells sustained the two c-Kit populations through one month until ultimately a lethal leukemia developed (Fig 1a). Conversely, at 1 month post-transplantation, GMP-HoxA9/M only retained the c-Kitlow population that was eventually lost from bone marrow (Fig. 1a).
Figure 1. β-catenin is activated in developing leukemia cells and L-GMP derived from KLS-HoxA9/M and GMP-MLLAF9.
a, Immunophenotypic analyses of GFP+ cells from bone marrow of mice transplanted with pre-leukemia KLS-HoxA9/M or GMP-HoxA9/M cells at 8 days, 1 month or 4 months (leukemia) post-transplantation.
b, The heat map displays the top 30 probe sets for genes that show increased expression in the HSC population (KLS cells) and L-GMP using Comparative Marker Selection and permutation testing (200 probe sets passed a cutoff of P < 0.002). Note expression of Ptgs-1 (Cox-1) and Ptger1 (prostaglandin E receptor 1).
c, Immunoblot analysis for active (dephosphorylated) β-catenin in normal GMP or L-GMP derived from GMP-MLLAF9 or KLS-HoxA9/M mediated leukemias.
d, Immunofluorescence assessment for active β-catenin in c-Kithigh or c-Kitlow populations isolated from mice 1 month after injection of pre-leukemia KLS-HoxA9/M cells. (active β-catenin: red, Hoechst: blue; active β-catenin/Hoechst: merge).
To determine whether the c-Kithigh and c-Kitlow populations identified 1 month post injection of KLS-HoxA9/M cells are functionally distinct, we utilized in vivo limiting dilution analysis to measure the frequency of leukemia initiating cells (LIC) in these two populations. LIC are defined as cells isolated prior to the development of clinically evident leukemia that can initiate leukemia in subsequent recipient mice. A high frequency of LIC was observed in the c-Kithigh population. In contrast, c-Kitlow cells were very inefficient at inducing leukemia (Fig. S4). We next assessed for LSC activity in the fully developed HoxA9/M induced leukemias. Limiting dilution analysis demonstrated the Gr1−/(low)c-Kithigh population was over 100-fold enriched for LSC as compared to the Gr1+c-Kithigh cells (Fig. S4, S5), and had an immunophenotype similar to MLL-AF9 LSC (L-GMP) (15,19) (Fig. S6). Thus, maintenance of the c-Kithigh population is associated with leukemia development, and cellular heterogeneity is found throughout the process of leukemia development. The inability of GMP-HoxA9/M cells to induce leukemia prompted us to search for a self-renewal pathway active in LSC that might be deficient in the GMP-HoxA9/M cells.
We previously demonstrated that Lin− Sca-1− c-Kithigh FcRγ+ CD34+ cells (termed L-GMP) are enriched for LSC in the MLL-AF9 AML model, and that L-GMP are globally similar to normal cells at a mid myeloid stage of development but express a subset of genes normally highly expressed in HSC (15). Given the LSC population in HoxA9/M driven leukemia described here has a similar immunophenotype, we reasoned that genes highly expressed in MLL-AF9 L-GMP, HoxA9/M L-GMP, and HSC but not in GMP might pinpoint genes important for leukemic self-renewal. Supervised analysis of gene expression data demonstrated a group of genes that are highly expressed in normal and leukemia stem cells, and expressed at lower levels in normal myeloid progenitors (Fig. 1b). Prostaglandin-endoperoxide synthase 1 (Ptgs1) (also known as Cycloxygenase-1 or Cox-1) and one of the prostaglandin receptors (Ptger1) were upregulated in both GMP-MLLAF9-derived and KLS-HoxA9/M-derived L-GMP, which was confirmed by qPCR (Fig. 1b; Fig. S6). As studies have highlighted a critical connection between prostaglandin synthesis and the Wnt/β-catenin pathway (20–23), we assessed for β-catenin activation. An antibody specific for activated β-catenin demonstrated activity in GMP-MLLAF9-derived and KLS-HoxA9/M-derived L-GMPs but not normal GMP as assessed by immunoblot or immunofluorescence (Fig. 1c; Fig. S7). Also, β-catenin activity was found in GMP-MLLAF9-derived and KLS-HoxA9/M-derived cells grown in vitro but not in GMP-HoxA9/M -derived cells (Fig. S7). Remarkably, activated/nuclear β-catenin is found in c-Kithigh, but not c-Kitlow cells isolated from mice 1 month after injection with KLS-HoxA9/M cells (Fig. 1d).
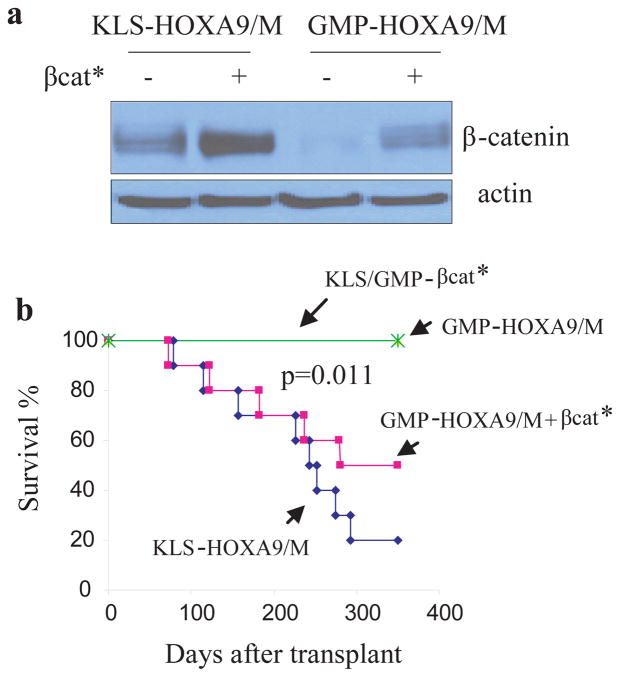
Given the activation pattern of β-catenin in GMP-MLLAF9-derived and KLS-HoxA9/M-derived leukemias, and its minimal activation in GMP-HoxA9/M-derived cells, we hypothesized that the inability of HoxA9/M to transform GMP might be due to the inability to sufficiently activate β-catenin. To test this we transduced GMP-HoxA9/M cells with a retrovirus encoding a constitutively active form of β-catenin (βcat*) (Fig. 2a). Transplantation of GMP-HoxA9/M cells transduced with βcat* produced AML similar to KLS-HoxA9/M cells (Fig. 2b). Expression of βcat* in GMP cells did not induce leukemia (Fig. 2b).
Figure 2. Constitutively active β-catenin cooperates with HoxA9/M to induce AML from GMP cells.
a, Constitutively active βcat* was transduced into pre-leukemia KLS-HoxA9/M or GMP-HoxA9/M cells and protein levels were assessed by immunoblot analysis.
b, Survival curves of mice receiving KLS or GMP cells transduced with active βcat* (controls), GMP-HoxA9/M, GMP-HoxA9/M cells transduced with active βcat*, or KLS-HoxA9/M. 5 × 105 cells were transplanted into sublethally irradiated recipients (n=10 in each group). P determined using the log-rank test
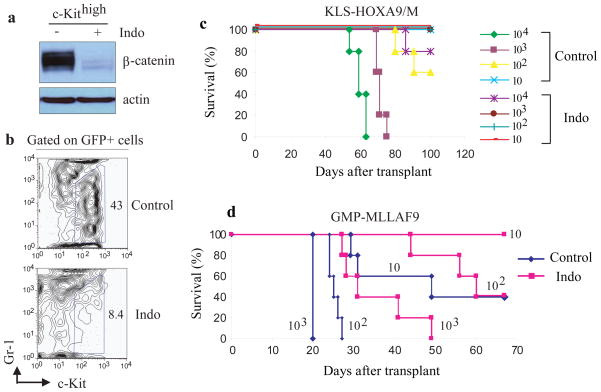
We next tested the effect of indomethacin (Indo), a reversible COX inhibitor which blocks activation of the Wnt pathway by suppressing β-catenin expression (22). Exposure of LIC-enriched c-Kithigh cells to indomethacin reduced β-catenin levels in vitro (Fig. 3a), and mice treated with indo showed a dramatic reduction in the c-Kithigh population after 7 days of treatment (Fig. 3b). We performed a limiting dilution assay using flow-sorted GFP+ cells from BM of control and Indo-treated mice which demonstrated a 100-fold decrease in LIC after indo treatment (Fig. 3c). Furthermore, Indo treatment of a fully developed MLL-AF9 leukemia partially suppressed β-catenin (Fig. S8) and reduced LSC frequency (Fig 3d). Thus, indomethacin treatment can suppress the LIC compartment critical for development of AML and also influences established LSC suggesting that AML is dependent on β-catenin signaling. However, the therapeutic effect appears greatest in situations of minimal disease (Fig. 3c) as compared to treatment of more extensive disease (Fig. 3d).
Figure 3. Pharmacological inhibition of β-catenin impairs LSC function.
a, Immunoblot analysis of β-catenin levels in c-Kithigh cells sorted from bone marrow at 1 month post-injection of mice with KLS-HoxA9/M cells which were subsequently grown in methylcellulose with and without exposure to indomethacin for 3 weeks (25,26).
b, Immunophenotypic analysis of bone marrow GFP+ cells after injection of KLS-HoxA9/M cells and treatment with either vehicle or indomethacin (repeated with 2 independent clones) for 7 days. Treatment began at day 3 post-injection and mice were sacrificed at day 10 post-transplantation (22,26).
c,d Survival curves of mice (5 mice for each group) injected with the indicated number (10 – 104) of GFP+ marrow cells sorted from control or Indomethacin (Indo)-treated mice that had received pre-leukemia KLS-HoxA9/M cells (c) or leukemic GMP-MLLAF9 cells (d) (repeated with 2 independent clones).
Next, we examined the ability of HoxA9/M to induce leukemia from β-catenin-deficient KLS cells using BM from βcatloxP/loxP mice (24). We transduced βcatloxP/loxP KLS cells with HoxA9/M, sorted single cells (KLS-HoxA9/M βcatloxP/loxP), and subsequently transduced the cells with a retroviral vector encoding either Cre recombinase, or an empty vector. Cells transduced with Cre recombinase (KLS-HoxA9/M βcat −/−) were devoid of wild-type β-catenin (Fig. S9). These cells were injected into recipient mice and we monitored expansion of KLS-HoxA9/M βcatloxP/loxP, KLS-HoxA9/M βcat −/− and control cells in bone marrow. KLS-HoxA9/M βcatloxP/loxP cells were able to expand efficiently in murine BM (Fig. 4a) and ultimately produced leukemia. However, loss of β-catenin led to impaired expansion, and KLS-HoxA9/M βcat −/− cells ultimately lost their ability to expand in bone marrow (Fig. 4a). Importantly βcat* rescued the proliferative defect of KLS-HoxA9/M βcat −/− cells (Fig. S9). Additionally, βcatWT KLS-HoxA9/M cells, βcatWT KLS-HoxA9/M cells expressing cre recombinase, and KLS-HoxA9/M βcatloxP/loxP cells induced AML in all recipients with similar latencies, whereas KLS-HoxA9/M βcat −/− cells were dramatically impaired in their ability to initiate AML (Fig. 4b). Three mice injected with KLS-HoxA9/M βcat −/− cells did ultimately develop AML, however in the 2 cases that were captured prior to the mouse succumbing to the disease, there was incomplete excision of β-catenin (Fig. S9). Finally, a similar experiment demonstrated a dramatic reduction in the ability of MLL-AF9 to induce leukemia from GMP (Fig. 4c).
Figure 4. Deletion of β-catenin impairs the development of KLS-HoxA9/M and GMP-MLLAF9 induced AML.
a, GFP + cells in bone marrow of mice injected with floxed β-catenin (βcatloxp/loxp) or β-catenin-deficient (βcat−/−) KLS-HoxA9/M cells at 18 hr, 1 month, 4 months post-transplantation. HoxA9/M-βcatloxP/loxP KLS cells were transduced with a retroviral vector encoding Cre recombinase to generate βcat −/− KLS-HoxA9/M cells or an empty vector to generate control βcatloxp/loxp KLS-HoxA9/M cells. After selection, 1 × 106 infected cells were transplanted into sublethally irradiated recipients (n=5 in each group). Mice were sacrificed at the indicated time points to assess for GFP+ cells.
b, Survival curves of mice transplanted with βcat−/− KLS-HoxA9/M cells, βcatloxp/loxp KLS-HoxA9/M cells, and empty vector or Cre-infected WT KLS-HoxA9/M (KLS cells were sorted from wild-type C57BL/6 mice and transformed with HoxA9/M to assess the effect of Cre on KLS-HoxA9/M leukemia development; n=10 for each group).
c, Survival curves of mice transplanted with βcat−/− GMP-MLLAF9 cells and βcatloxp/loxp GMP-MLLAF9 cells (n=10 for each group). P determined using the log-rank test
These data demonstrate that β-catenin is required for either Hox-gene mediated transformation of HSC or MLL-AF9 mediated transformation of committed progenitor cells. Furthermore, lack of β-catenin activation in normal GMP limits the ability of Hox genes to efficiently transform committed progenitor cells. This is in contrast to MLL-AF9 that activates sufficient β-catenin to transform GMP. This supports the concept that activation or maintenance of HSC-associated developmental pathways in downstream myeloid cells is an important component of AML development. Given that β-catenin activity is important for HSC during fetal development, but not for adult HSC survival/proliferation (9, 11, 12), targeting the β-catenin pathway may be an attractive therapeutic avenue in this disease.
Supplementary Material
Acknowledgments
We thank D. Fisher, X. He and H. Widlund for kindly providing the plasmid pcDNA3.1-βcat-*-Flag. We also thank D. Williams for helpful discussion and advice, and M. Smith for administrative assistance. This work was supported by the NIH grants 5P01CA66996 to S.A.A. and 5R01HL048801 to L.I.Z., the American Cancer Society, and the Harvard Stem Cell Institute. SAA is a Leukemia and Lymphoma Society Scholar. Microarray data was submitted to the NCBI Gene Expression Omnibus (GEO) # GSE20377.
References and Notes
- 1.Bonnet D, Dick JE. Nat Med. 1997 Jul;3:730. doi: 10.1038/nm0797-730. [DOI] [PubMed] [Google Scholar]
- 2.Dick JE. Blood. 2008 Dec 15;112:4793. doi: 10.1182/blood-2008-08-077941. [DOI] [PubMed] [Google Scholar]
- 3.Park CY, Tseng D, Weissman IL. Mol Ther. 2009 Feb;17:219. doi: 10.1038/mt.2008.254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jamieson CH, et al. N Engl J Med. 2004 Aug 12;351:657. doi: 10.1056/NEJMoa040258. [DOI] [PubMed] [Google Scholar]
- 5.Majeti R, et al. Proc Natl Acad Sci U S A. 2009 Mar 3;106:3396. doi: 10.1073/pnas.0900089106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Muller-Tidow C, et al. Mol Cell Biol. 2004 Apr;24:2890. doi: 10.1128/MCB.24.7.2890-2904.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ysebaert L, et al. Leukemia. 2006 Jul;20:1211. doi: 10.1038/sj.leu.2404239. [DOI] [PubMed] [Google Scholar]
- 8.Reya T, et al. Nature. 2003 May 22;423:409. doi: 10.1038/nature01593. [DOI] [PubMed] [Google Scholar]
- 9.Zhao C, et al. Cancer Cell. 2007 Dec;12:528. doi: 10.1016/j.ccr.2007.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Malhotra S, Kincade PW. Cell Stem Cell. 2009 Jan 9;4:27. doi: 10.1016/j.stem.2008.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jeannet G, et al. Blood. 2008 Jan 1;111:142. doi: 10.1182/blood-2007-07-102558. [DOI] [PubMed] [Google Scholar]
- 12.Cobas M, et al. J Exp Med. 2004 Jan 19;199:221. doi: 10.1084/jem.20031615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cozzio A, et al. Genes Dev. 2003 Dec 15;17:3029. doi: 10.1101/gad.1143403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Huntly BJ, et al. Cancer Cell. 2004 Dec;6:587. [Google Scholar]
- 15.Krivtsov AV, et al. Nature. 2006 Aug 17;442:818. doi: 10.1038/nature04980. [DOI] [PubMed] [Google Scholar]
- 16.Sauvageau G, et al. Genes Dev. 1995 Jul 15;9:1753. doi: 10.1101/gad.9.14.1753. [DOI] [PubMed] [Google Scholar]
- 17.Abramovich C, Pineault N, Ohta H, Humphries RK. Ann N Y Acad Sci. 2005 Jun;1044:109. doi: 10.1196/annals.1349.014. [DOI] [PubMed] [Google Scholar]
- 18.Kroon E, et al. Embo J. 1998 Jul 1;17:3714. doi: 10.1093/emboj/17.13.3714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Somervaille TC, Cleary ML. Cancer Cell. 2006 Oct;10:257. doi: 10.1016/j.ccr.2006.08.020. [DOI] [PubMed] [Google Scholar]
- 20.Castellone MD, Teramoto H, Williams BO, Druey KM, Gutkind JS. Science. 2005 Dec 2;310:1504. doi: 10.1126/science.1116221. [DOI] [PubMed] [Google Scholar]
- 21.North TE, et al. Nature. 2007 Jun 21;447:1007. doi: 10.1038/nature05883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Goessling W, et al. Cell. 2009 Mar 20;136:1136. doi: 10.1016/j.cell.2009.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Eisinger AL, Prescott SM, Jones DA, Stafforini DM. Prostaglandins Other Lipid Mediat. 2007 Jan;82:147. doi: 10.1016/j.prostaglandins.2006.05.026. [DOI] [PubMed] [Google Scholar]
- 24.Brault V, et al. Development. 2001 Apr;128:1253. doi: 10.1242/dev.128.8.1253. [DOI] [PubMed] [Google Scholar]
- 25.Orido T, et al. J Pharmacol Sci. 2008 Nov;108:389. doi: 10.1254/jphs.08167sc. [DOI] [PubMed] [Google Scholar]
- 26.Please refer to Supplemental Data and Methods for details.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.