Abstract
Acetyl group turnover on specific lysine ε-amino groups of the core chromosomal histones regulates DNA accessibility function, and the acetylating and deacetylating enzymes that govern the turnover provide important targets for the development of anti-cancer drugs. Histone deacetylase (HDAC) inhibitors have been developed and evaluated extensively in clinical trials, while the development of inhibitors of histone acetyltransferase (HAT) has proceeded more slowly. Here we have examined the cellular effects of an S-substituted coenzyme A (CoA) inhibitor of histone acetylation, consisting of spermidine (Spd) linked to the S-terminus of CoA through a thioglycolic acid linkage (adduct abbreviated as Spd-CoA), as well as the effects of a truncated Spd-CoA derivative lacking the negatively charged portion of the CoA moiety. While exposure of cancer cells to Spd-CoA has little effect on cell viability, it causes a rapid inhibition of histone acetylation that correlates with a transient arrest of DNA synthesis, a transient delay in S-phase progression, and an inhibition of nucleotide excision repair and DNA double strand break repair. These effects correlate with increased cellular sensitivity to the DNA-targeted chemotherapeutic drugs, cisplatin (Platinol™) and 5-fluorouracil, to the DNA damaging drug, camptothecin, and to UV-C irradiation. The sensitization effects of Spd-CoA are not observed in normal cells due to a barrier to uptake. The truncated Spd-CoA derivative displays similar but enhanced chemosensitization effects, suggesting that further modifications of the Spd-CoA structure could further improve potency. The results demonstrate that Spd-CoA and its truncated version are efficiently and selectively internalized into cancer cells, and suggest that the resulting inhibition of acetylation-dependent DNA repair enhances cellular sensitivity to DNA damage. These and related inhibitors of histone acetylation could therefore constitute a novel class of potent therapy sensitizers applicable to a broad range of conventional cancer treatments.
Keywords: histone acetylation, HAT inhibitor, DNA repair, chemosensitization, radiosensitization
Introduction
Post-translational acetylation of histones, primarily involving the ε-amino groups on specific lysine side chains in the N-terminal domains (N tails) of the core chromosomal histones H2A, H2B, H3 and H4 regulates chromatin structure and function.1 The N tails are conformationally variable and protrude from the nucleosome, the fundamental chromosomal unit, forming regulatable contacts with DNA and proteins2-5 (reviewed in refs. 6 and 7). The turnover of histone acetyl groups is required for transcription,8,9 DNA repair,10 histone deposition after DNA synthesis,11 and replication fork initiation,12 and therefore has broad relevance to chromatin function.
Acetyl group turnover is controlled by the opposing actions of histone acetyltransferases (HATs), which transfer an acetyl group from acetyl-CoA to the lysine side chain, and histone deacetylases (HDACs), which catalyze amide hydrolysis and release the acetyl group.2 Together, these two classes of enzymes provide for the coordinated changes in chromatin structure that are needed to carry out its functions.13 Based on such a mechanistic view it would be expected that the inhibition of the HATs as well as of the HDACs would interfere with essential chromatin activities and be beneficial for cancer therapy. HDAC inhibitors have been shown to fulfill such expectations and have undergone extensive clinical evaluation (reviewed in refs. 14 and 15). In contrast, progress in developing HAT inhibitors has been slower.
HAT inhibition in vitro was initially reported with a bisubstrate adduct, spermidine-CO-CH2-CoA (abbreviated Spd-CoA), formed by joining spermidine (Spd) covalently to the S atom of coenzyme A (CoA) through a thioglycolic acid linkage.16 Each of two isomeric forms of Spd-CoA, linking the N1 or N8 atom of spermidine to CoA, respectively,17 have subsequently been shown to be HAT inhibitors in vitro.18 Similar inhibitors in which a peptide appendage replaces the Spd moiety have been described, although they do not penetrate the cell.19,20 Several natural products have been found to inhibit histone acetyltransferase activity when added to whole cells, including garcinol,21 curcumin22 and anacardic acid,23 and synthetic analogs of anacardic acid have been developed.24
Spd-CoA has been shown to be active against histone acetytransferase activity in isolated nuclei, in permeabilized cells, and isolated polynucleosomes.16,18 However, because CoA itself carries negative charges that impede its transport across the cellular membrane, the effects of Spd-CoA-type conjugates on whole cells have not been extensively explored. Nevertheless, because polyamines such as spermidine are efficiently transported across cellular membranes,25 we hypothesized that Spd-CoA could also be internalized into whole cells despite its negatively-charged CoA moiety, and that internalization would lead to inhibition of histone acetyltransferase activity and acetylation-dependent chromatin function. Consistent with this hypothesis, we find in this study that the bisubstrate histone acetyltransferase inhibitor, Spd(N1)-CoA, acts on whole cells to inhibit histone acetylation, DNA synthesis and DNA repair. Furthermore, Spd(N1)-CoA synergizes with a variety of DNA damaging treatments, including several commonly used chemotherapeutic agents, to induce cancer cell killing. We find that the effects are specific for cancer cells and extend to a truncated derivative of Spd-CoA that lacks the negatively charged portion of the CoA moiety, suggesting that this class of inhibitors may be amenable to further refinement and have considerable clinical potential, particularly to reduce therapy toxicity and reverse therapy resistance.
Results
Spd-CoA (1a) structure
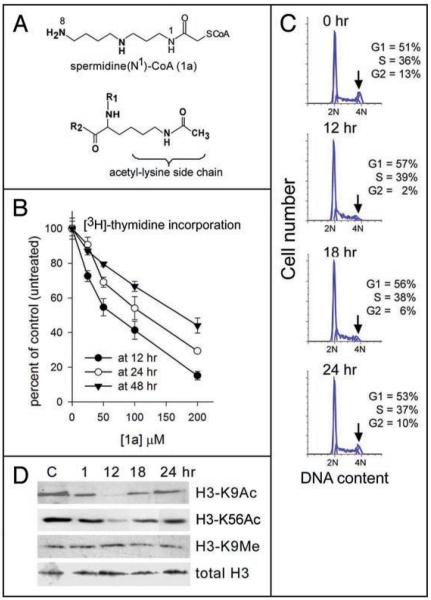
Following Roblot et al.17 we synthesized Spd(N1)-CoA (named 1a throughout this work) a bisubstrate spermidine-type histone acetyltransferase inhibitor based on the R-CoA design initially described by Cullis et al.16 Treatment of solubilized chromatin with 1a has been shown to inhibit endogenous HAT activity that co-isolates with the chromatin, resulting in complete suppression of acetyl group incorporation into histones.18 The structure of 1a and its resemblance to the reaction product of the HAT-mediated acetyl transfer to the lysine side chain is shown in Figure 1A.
Figure 1.
1a structure, inhibition of DNA synthesis, S-phase progression and histone acetylation in vivo by 1a. (A) Structures of 1a (Spd(N1)-CoA), and the acetylated side chain of a lysine residue within the polypeptide chain of a protein targeted by the acetylating enzyme (R1 and R2 are the main chain elements on both sides of the lysyl residue). (B) [3H]-thymidine incorporation into DNA of H358 cells measured after a 6 hour pulse carried out at intervals 6–12 hrs (12 hr curve), at 18–24 hrs (24 hr curve), or at 42–48 hrs (48 hr curve) after the addition of variable concentrations of 1a, and represented as % of control (untreated) cells. (C) Cell cycle analysis of propidium iodide-stained cells at the indicated times after addition of 50 μM 1a. Percent of cells in G1, S and G2 represents the average of two independent profiles. Arrows indicate major effects observed in the G2 (4N) population. (D) SDS-PAGE/western analysis of histones H3-K9 acetylated, H3-K56 acetylated, H3-K9 methylated and total histone H3, before treatment (“C”, control), or at the indicated times post-addition of 50 μM 1a. Each lane = 60 μg protein.
Spd-CoA (1a) inhibits DNA synthesis, S-phase progression and histone acetylation in whole cells
We examined DNA synthesis by [3H]-thymidine incorporation into H358 cells during 6 hour pulses at intervals 6–12 hours, 18–24 hours and 42–48 hours after the addition of increasing amounts of 1a. [3H]-thymidine incorporation decreased with increasing concentrations of 1a at all three pulse intervals, but the inhibition achieved at a given 1a concentration was maximal at 6–12 hours (Fig. 1B), where we observed 50% inhibition of [3H]-thymidine incorporation at about 50–60 μM 1a. To avoid possible non-specific effects that could be introduced at higher concentrations, we carried out additional studies using 50 μM 1a.
Treatment with 1a caused a transient slowing of S-phase progression as shown by the cell cycle analyses of propidium iodide-stained cells at various times after adding 50 μM 1a (Fig. 1C). H358 cells underwent a depletion of G2-phase cells (arrows) that was maximal at 12 hours post-start of treatment. By 24 hours, the cell cycle distribution had returned to normal.
To determine how 1a-mediated suppression of DNA synthesis and cell cycle correlate with suppression of histone acetylation, we followed the acetylation status over time of a representative N-terminal histone acetylation site, histone H3 lysine 9 (H3-K9Ac), and an acetylation site in the globular domain of histone H3 (H3-K56Ac) that has been linked to histone deposition after DNA synthesis in yeast (reviewed in ref. 32), and recently to genome instability and DNA repair in mammalian cells, including human cells.33,34 Acetylation levels were examined at 1, 12, 18 and 24 hours post-start of treatment of H358 cells with 50 μM 1a. As shown by the immunoblot analysis in Figure 1D, acetylation levels of both histone H3-K9 and H3-K56 fell at 1 and 12 hours post-start of treatment. A digital analysis of band intensities showed that over 50% of H3-K9 and H3-K56 acetyl groups were lost within the first hour and some 98% at both sites were lost within 12 hours post-start of treatment. Acetylation levels were restored by 24 hours. Over the same time period, levels of total histone H3 remained unchanged, as expected, as did levels of histone H3 methylated on lysine 9 (H3-K9Me), confirming that inhibition was specific for acetylation. The time frame observed for inhibition of DNA synthesis, cell cycle and histone acetylation following treatment of cells with 1a, is consistent with rapid internalization of 1a, leading to inhibition of acetylation-dependent DNA synthesis.
Spd-CoA (1a) synergizes with DNA damage to kill cancer cells
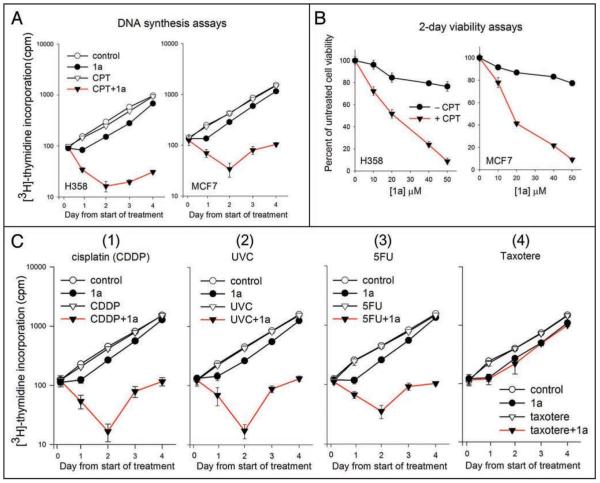
We examined DNA synthesis in H358 cells at various times after the addition of 50 μM 1a, either alone or together with an 18 hour exposure to 5 nM camptothecin (CPT), a DNA damaging plant alkaloid from which an important class of topoisomerase I-targeted chemotherapeutic agents [Hycamtin (topotecan) and Camptosar (irinotecan)] is derived.35 Treatment with 5 nM CPT alone for 18 hours has been previously shown to have minimal effects on growth of H358 cells.36 For the combined CPT/1a treatments, the medium containing both 1a and CPT was removed from cells after 18 hours and replaced with fresh medium containing 1a only, and incubation was continued. Triplicate wells of cells were then pulsed 6 hours with [3H]-thymidine at intervals 18–24 hr (Day 1), 42–48 hr (Day 2), 66–72 hr (Day 3) and 90–96 hr (Day 4) post-start of drug treatment. For the first time point, untreated wells at the start of the experiment were pulsed 6 hours with [3H]-thymidine to determine an initial rate of [3H]-thymidine incorporation. Total [3H]-thymidine incorporation (average of three wells) at the end of each pulse is plotted in Figure 2A. As shown in Figure 2A (left, open circles), [3H]-thymidine incorporation nearly doubled on each successive day in untreated cells. Treatment with 50 μM 1a alone (Fig. 2A, left, closed circles) caused a delay relative to untreated cells in the rate of increase in [3H]-thymidine incorporation until the day 1 point, consistent with the transient inhibition of DNA synthesis observed in Figure 1B. From day 1 to day 4, the [3H]-thymidine incorporation curve paralleled the curve for untreated cells.
Figure 2.
Effect of 1a and 1a-combination treatments on DNA synthesis. (A) [3H]-thymidine incorporation (counts per minute) into DNA of H358 cells (left) or MCF7 cells (right) during a 6 hour pulse ending at the indicated time points after initiation of treatment. (Time 0 measured in untreated cells). Treatments were 50 μM 1a alone (present throughout), 5 nM camptothecin (CPT) alone (18 hours), 50 μM 1a + 5 nM CPT (18 hours). Control cells were left untreated. (B) 2-day 96-well viability assays of H358 cells (left) or MCF7 cells (right) grown in presence of increasing concentrations of 1a, with or without 5 nM CPT (18 hrs). Cell viability 2 days post-start of treatment is represented as a percent of untreated cell viability (i.e., no 1a, no CPT). Each data point represents average of triplicate wells. Conditions were such that untreated cells remained in exponential growth. (C) [3H]-thymidine incorporation into DNA of H358 cells carried out as in part A. Treatment consisted of 1a alone (50 μM, present throughout), or cisplatin (2 μM, 1 hr 15 min) alone or together with 1a (Panel 1); or UV-C (40/J/m2), alone or together with 1a (Panel 2); or 5-fluorouracil (5FU, 2 μM, 18 hours) alone or together with 1a (Panel 3); or Taxotere™ (5 nM, 18 hours), alone or together with 1a (Panel 4). Control cells were left untreated. Symbols are as follows: (-○-) untreated control; (-●-) 50 μM 1a, (-▽-) drug or UV-C alone; (-▼-) combination 1a + drug or UV-C.
We observed no detectable effect of an 18-hour treatment with 5 nM CPT alone on [3H]-thymidine incorporation compared to untreated cells (Fig. 2A, left, open triangles), over the 4-day period of the assay, consistent with previous observations.36 In contrast, when cells were treated with 50 μM 1a and 5 nM CPT for 18 hours, followed by removal of the medium and replacement with medium containing 1a alone, [3H]-thymidine incorporation decreased progressively on days 1 and 2. By day 2 post-start of treatment [3H]-thymidine incorporation was less than 20% of untreated cells (Fig. 2A, left, closed triangles). Cell viabilities 18 hours post-start treatment (measured by the Trypan blue exclusion assay, a measure of cellular membrane integrity) were ≥96% viability for untreated cells or single agent-treated cells, and 36% for cells receiving the combined treatment. Similar results were observed in the breast cancer cell line, MCF7 (Fig. 2A, right), establishing that the results were not unique to H358.
We carried out 2-day 96-well assays to further evaluate how various treatments affected cell viability. As shown in Figure 2B, closed circles, treatment of H358 or MCF7 cells with increasing concentrations of 1a alone had little effect on 2-day cell viability up to 50 μM 1a, further showing that the cells quickly recovered and continued growing following the transient inhibition of [3H]-thymidine incorporation, cell cycle progression and histone acetylation seen in Figure 1B–D. Similarly, treatment of either cell line with 5 nM CPT (18 hours) without 1a (closed triangles at 0 μM 1a) had no effect on 2-day cell viability relative to untreated cells. However, treatment of both cells lines with increasing concentrations of 1a together with an 18 hr exposure to 5 nM CPT, followed by removal of the medium and replacement with medium containing 1a alone, led to a progressive loss of 2-day cell viability with increasing concentrations of 1a to less than 10% of untreated cell viability at 50 μM 1a (Fig. 2B, closed triangles).
We then extended the [3H]-thymidine incorporation assays to include the combined effects of 1a with other DNA-targeted chemotherapeutic drugs, namely cisplatin [CDDP, cis-diamminedichloroplatinum (II), Platinol™], UV-C radiation and 5-fluorouracil (5FU, Adrucil™). To determine if the synergistic effects applied to agents that do not target DNA, we examined the combined effect of 1a with the chemotherapeutic agent, Taxotere™, which targets microtubules and does not damage DNA. As with CPT, the drug and UV-C treatment conditions were chosen so as to have minimal effects on cell viability when used singly. As shown in Figure 2C, treatment with 1a sensitized H358 cells to CDDP, UV-C irradiation and 5FU (panels 1–3, respectively), but not to Taxotere™ (panel 4), suggesting that a common mechanism, relevant to DNA damage, underlies the ability of histone acetylation inhibition to synergize with drugs and radiation.
Spd-CoA (1a) inhibits DNA repair
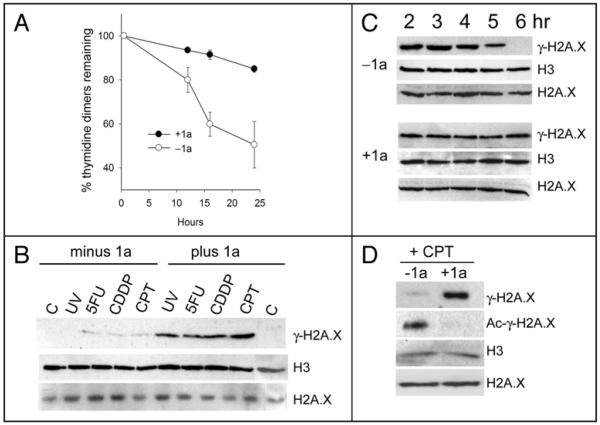
DNA repair can promote cell survival following DNA damage. Nucleotide excision repair (NER) requires histone acetylation37 (reviewed in ref. 38), and removes bulky DNA adducts such as cisplatin (CDDP)-induced intra-strand guanine-guanine or adenine-guanine crosslinks, or UV-C-induced intra-strand thymidine dimers. Furthermore, expression of the essential NER repair protein, ERCC1, is associated with cisplatin resistance in patients with non small cell lung cancer.39 An immunoblot assay of thymidine dimers in cellular DNA of H358 cells prepared at fixed times after exposure to 40 J/m2 UV-C radiation is shown in Figure 3A. About 50% of thymidine dimers were removed from the DNA of untreated H358 cells within 24 hours post-treatment, consistent with rates observed in other cancer cell lines.31 In contrast, thymidine dimers persisted in cells treated with 50 μM 1a, decreasing by only about 10% over the same time frame. The results support a model in which inhibition of histone acetylation by 1a prevents recruitment of repair factors, or prevents the localized relaxation of chromatin needed to increase DNA accessibility to the NER repair complex.
Figure 3.
Effect of 1a on DNA repair. (A) Nucleotide Excision Repair assay: Time course of thymidine dimer loss from DNA of H358 cells following treatment with 40 J/m2 UV-C followed by incubation in the presence or absence of 50 μM 1a for the indicated times. (B) Induction of γ-H2A.X by the combination treatments used for DNA synthesis assays in Figure 2: western analysis shows γ-H2A.X or total H3 levels (control) in acid extracted histones from H358 cells (plus or minus 1a) 18 hours after treatment with UV-C (40 J/m2), 5-fluorouracil (5FU, 2 μM, 18 hours), cisplatin (CDDP, 2 μM, 1 hour 15 min), or camptothecin (CPT, 5 nM, 18 hours). Also shown are controls for no treatment (minus 1a, lane “C”) or treatment with 1a only (plus 1a, lane “C”). Each lane = 60 μg protein. (C) DSB repair assay: SDS-PAGE/western of γ-H2A.X, H3 and H2A.X levels in H358 cells at various times post-start of treatment with 10 μM CPT (1 hour), followed by incubation in the absence (upper 2 rows) or presence (lower 2 rows) of 50 μM 1a. Each lane = 60 μg protein. (D) Inhibition of γ-H2A.X acetylation by 1a: Top row—SDS-PAGE/western of γ-H2A.X in H358 cells treated with 10 μM CPT (1 hour) followed by incubation for an additional hour in the presence (right) or absence (left) of 50 μM 1a (60 μg/lane). Second row—Immunoprecipitation of 60 μg aliquots of the same extracts with anti-acetyl-lysine followed by SDS-PAGE/western of γ-H2A.X in the immunoprecipitated material. Third and fourth rows—SDS-PAGE/western of total histone H3 and total H2A.X in 60 μg aliquots of the same extracts (control).
Double strand breaks (DSBs) can occur as a result of a variety of DNA damaging treatments, including CPT, cisplatin, 5FU and UV-C radiation,40-42 and DSB repair may play a role in reducing their cellular toxicity. The histone variant, H2A.X becomes phosphorylated at DSB sites (denoted γ-H2A.X) and serves as an indicator of DSBs following immunoblot analysis of acid extracted cells.43 Cells treated with UV-C, 5FU, cisplatin or CPT at the low doses used for the growth assays in Figure 2, either as single agents, or in combination with 50 μM 1a, were analyzed 18 hours post-start of treatment. γ-H2A.X was undetectable in untreated cells or cells treated with 1a or UV-C alone and was weakly detectable in cells treated with 5FU, cisplatin or CPT alone (Fig. 3B). γ-H2A.X levels increased dramatically when these treatments were combined with 1a, indicating increased levels of DSBs, while total H3 and H2A.X.levels (control) were essentially unaffected (Fig. 3B). γ-H2A.X levels remained undetectable following treatment with Taxotere™ (5 nM, 18 hours) either in the presence or absence of 50 μM 1a (data not shown), in agreement with the results in Figure 2C (panel 4). To follow DSB repair, H358 cells were treated with 10 μM CPT for 1 hour to induce a high level of DSBs, followed by incubation in the presence or absence of 50 μM 1a for various lengths of time. Acid extracts of cells were prepared at regular time intervals following CPT treatment and subjected to immunoblot analysis for γ-H2A.X. As shown in Figure 3C, γ-H2A.X levels decreased over time in the absence of 1a, and were undetectable after 6 hours post-start of treatment, indicating that double strand breaks had been repaired. In contrast, γ-H2A.X levels persisted in cells treated with 1a, indicating persistence of DSBs. Levels of total histone H3 and H2A.X (controls) remained unchanged in both cases over the same period (Fig. 3C).
Acetylation of γ-H2A.X is required for its removal and replacement with non-phosphorylated H2A.X during the chromatin remodeling that accompanies DNA repair,44 and failure of histone acetylation adjacent to DSBs has been reported to impair the efficiency of DNA repair.45 Thus, treatment with 1a may block DSB repair by interfering with acetylation of histone γ-H2A.X. To test this hypothesis we treated H358 cells for 18 hours with 5 nM CPT in the presence or absence of 50 μM 1a as in Figure 3B. We then prepared acid extracted histones from nuclei of treated cells and carried out an immunoprecipitation with an immobilized anti-histone acetyl antibody, followed by an immunoblot analysis of histone γ-H2A.X. As shown in Figure 3D, cells treated with CPT plus 1a accumulate DSBs, as manifested by the elevated level of γ-H2A.X, but fail to acetylate γ-H2A.X. This result supports a model in which inhibition of γ-H2A.X acetylation by 1a prevents chromatin remodeling during DSB repair, resulting in a failure to complete DSB repair and leading to persistent elevation of γ-H2A.X. In contrast, cells treated with CPT alone accumulate only a weakly detectable level of γ-H2A.X that appears to be highly acetylated, consistent with the efficient completion of DNA repair. Levels of total H3 and H2A.X remain unchanged (controls).
Normal human cells are not sensitized to DNA damage by Spd-CoA (1a)
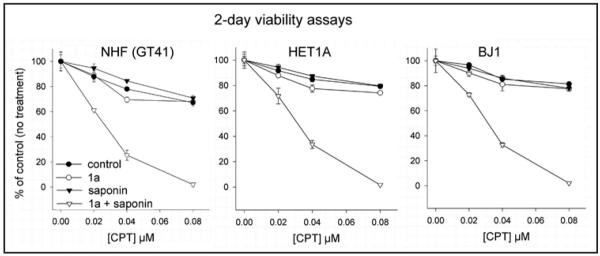
To determine if sensitization to DNA damage by 1a also occurred with normal cells we carried out 2-day viability assays of normal human fibroblasts (GT41), immortalized normal human epithelial cells (HET1A) and immortalized normal human fibroblasts (BJ1) treated with increasing concentrations of CPT for 18 hours in the absence of 1a, or increasing concentrations of CPT for 18 hours in the presence of 50 μM 1a (followed by replacement of the medium with medium containing 1a only). All three cell lines displayed a high level of resistance to increasing doses of CPT in the absence of 1a (Fig. 4, control curves, closed circles), that was virtually unaffected by the addition of 50 μM 1a (Fig. 4, open circles). However, treatment of cells with the detergent, saponin, to permeabilize the membrane, resulted in marked synergistic effect of the 1a/CPT combination (Fig. 4, open triangles). Treatment with saponin in the absence of 1a did not enhance the effects of CPT (Fig. 4, closed triangles). The results indicate that normal cells present a barrier to uptake of 1a, and suggest that this differential uptake can be exploited to achieve cancer cell-specific sensitization.
Figure 4.
Effects of 1a on normal human cells. 2-day 96-well viability of non-permeabilized (circles) or saponin-permeabilized (triangles) GT41 normal human fibroblasts (NHF, left), immortalized human epithelial cells (HET1A, middle), and immortalized human fibroblasts (BJ1, right) following an 18 hour exposure to increasing concentrations of CPT without 1a (-●-, -▼-, for non-permeabilized and permeabilized cells, respectively), or CPT together with 50 μM 1a (-○-, -▽-, for non-permeabilized and permeabilized, respectively). In the latter case, fresh medium with fresh 50 μM 1a was replaced after removal of CPT. Cell viability 2 days post-start of treatment is represented as a percent of untreated cell viability (i.e., no 1a, no CPT, no saponin). Each data point represents average of triplicate wells. Conditions were such that untreated cells remained in exponential growth.
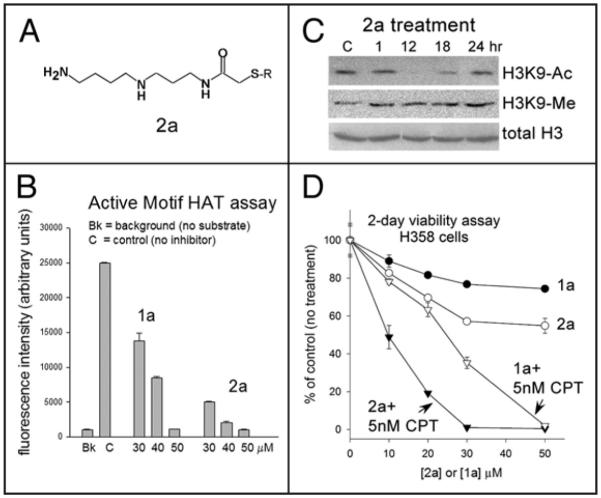
A truncated Spd-CoA analog (2a) displays enhanced cellular effects
Because the negative charges on the CoA moiety could reduce cellular uptake, we investigated the effects of a modified inhibitor, designated 2a (see Materials and Methods). The inhibitor was synthesized as described in reference,17 and has previously been shown to inhibit endogenous HAT activity in isolated rat liver polynucleosomes.18 2a is based on the Spd-CoA structure of the 1a compound, but the CoA moiety has been truncated so that only the cysteamine-beta-alanine portion of the CoA moiety (=S-R in Fig. 5A) is retained. The rationale is to retain many of the critical HAT/CoA contacts, while providing improved cellular uptake. Interestingly, 2a appears to be slightly more inhibitory of p300/CBP HAT activity in vitro than 1a using the commercially available 96-well p300/CBP HAT activity assay (Fig. 5B) and suppresses histone acetylation in vivo in H358 cells (Fig. 5C) in a time frame similar to that of 1a (see Fig. 1D), with greatest inhibition occurring at 12 hours post-start of treatment. Furthermore, 2a is more active than 1a against whole cells and synergizes more efficiently with CPT to kill H358 lung cancer cells (Fig. 5D). The results indicate that 2a itself or related versions of 2a could provide a basis for further drug discovery.
Figure 5.
Effects of compound 2a. (A) Structure of compound 2a. R = CH2-CH2-NH-CO-CH2-CH2-NH-CO-CH3. (B) In vitro assay of p300 activity using a 96-well assay kit (Active Motif) carried out with 2a at 30-50 μM, in comparison to 1a. Averages of duplicate samples with standard deviations are shown. (C) Western analysis of histone H3K9 acetylation and methylation, and total histone H3, before (“C”, control), or at the indicated times after addition of 50 μM 2a (60 μg lysate/lane). (D) 2-day 96-well viability assay of H358 cells incubated with increasing concentrations of 1a or 2a, +/− 5 nM CPT (18 hrs). Fresh medium with 1a and 2a was replaced after removal of CPT.
Discussion
This study is the first to show that Spd(N1)-CoA, or 1a,17 initially described by Cullis et al. to be a well-defined inhibitor of histone acetylation in vitro16 sensitizes whole tumor cells to UV-C treatment and to DNA targeted drugs. Treatment of cells with 1a inhibits histone acetylation, DNA synthesis and DNA repair, suggesting that sensitization results from an inhibition of one or more acetylation-dependent steps in DNA repair. Consistent with this possibility is our observation that 1a treatment inhibits acetylation of the H2A variant, γ-H2A.X, following DSB induction, resulting in a prolonged elevation in γ-H2A.X levels, a histone that accumulates at sites of DSBs. Furthermore, treatment with 1a also inhibits NER, suggesting that 1a treatment has global effects on multiple repair mechanisms relevant to the therapy response of cancer cells. Such global effects would be an advantage for an eventual broad clinical application of this and related polyamine-based compounds. As expected, based on our model, treatment with 1a did not sensitize cells to Taxotere™, a drug that stabilizes microtubules. However, given that Taxotere™ acts primarily on cells in M phase, it is possible that the lack of chemosensitization is at least partially due to the reduction of cells in G2/M as shown in Figure 1C.
The ability of 1a to penetrate the cell membrane despite the presence of a negatively charged CoA moiety is most likely due to the presence of the positively charged spermidinyl moiety in the Spd-CoA conjugate. A variety of polyamine conjugates are readily internalized into mammalian cells via the polyamine transporter,46 and the polyamine transport system has recently been exploited for drug delivery to cancer cells.47 Our finding that the truncated compound, 2a, displays enhanced sensitization to camptothecin, suggests that removal of the negatively-charged region of the CoA moiety improves cellular uptake, a finding that provides impetus for further improvements in this region of the compound to increase potency. Importantly, normal cell lines of fibroblast origin (GT41 and BJ1) and epithelial cell origin (HET1A) are not sensitized to camptothecin by treatment with 1a unless their cellular membranes are permeabilized by brief detergent treatment. The cancer cell specificity of the sensitization effect may therefore derive from a reduced ability of normal cells to take up the compound compared to cancer cells, and further studies are underway to establish the generality of these effects for other cancer cell types. Tumors often display elevated polyamine content as well as upregulated polyamine uptake through the polyamine transporter.48 Our own studies show that spermidine uptake is greater in a cultured lung cancer cell line compared to the normal GT41, BJ1 and HET1A cell lines, under conditions where growth rates for the cell lines are comparable (Bandyopadhyay K and Gjerset RA, unpublished). The structures of 1a and 2a suggest that they may be preferentially internalized into cancer cells through the polyamine transporter as adducts of spermidine (the molecules are still diamines). In addition, the bioavailability of 1a, 2a and related compounds following intravenous administration would be expected to be high, based on the high solubility of these compounds, and their rapid uptake by cancer cells (see Fig. 1D).
The cellular effects of 1a and 2a are observed within the first 24 hours and are maximal at 12 hours post-start of treatment, where some 98% of histone acetylation is suppressed at 50 μM. Nevertheless, the marked synergy we observe when these compounds are combined with DNA damaging treatments indicates that even short lived effects can dramatically impact the cellular response to therapeutic agents. The transient nature of the inhibitory effects on histone acetylation and DNA synthesis suggests that these compounds are likely to be metabolically unstable, and this may account for the failure of the inhibitors alone to suppress 2-day cell viability. Separate studies showed that suppression of viability by 50% could only be achieved at 500 μM 1a, although such a dose could introduce significant non-specific effects (Bandyopadhyay K, unpublished observations). Our finding that daily replenishment of cell cultures with fresh 1a inhibitor leads to reduced cell viability is consistent with metabolic instability (Bandyopadhyay K, unpublished observations). While the IC50 for suppression of [3H]-thymidine incorporation by 2a was in the range of 50 μM, similar to 1a (Bandyopadhyay K et al., unpublished observations), the slightly longer suppression of histone H3-K9 acetylation seen with 2a (Fig. 5C) compared with 1a (Fig. 1D), with 2a continuing to suppress at the 18 hour time point, suggests that 2a may be more metabolically stable than 1a, and further experiments will be needed to confirm this.
Most conventional cancer treatments target DNA. However, enzyme-mediated DNA repair can promote chemo- and radio-resistance and is a promising target for the development of approaches to reverse therapy resistance. Nucleotide excision repair has been linked to cisplatin resistance,49 and both the non-homologous endjoining (NHEJ) and homologous recombination (HR) mechanisms of DSB repair have been linked to chemo- and radio-resistance.50,51 Radiosensitization has been achieved with inhibitors of DNA Protein Kinase, an essential enzyme in NHEJ, and with inhibitors of poly(ADP-ribose) polymerase-1 (PARP-1), a component of the base excision repair (BER) complex, and a possible player in DSB repair as well (reviewed in ref. 52). Recently, anacardic acid, a plant-derived inhibitor of the histone acetyltransferase, Tip60, has been found to block DNA damage signaling through ATM, (Ataxia-Telangiectasia-Mutated) kinase, and to sensitize to ionizing radiation.53 Of particular interest is the fact that cancer stem cells, or cancer initiating cells, display a high degree of therapy resistance54 and appear to possess elevated DNA repair capacity compared to other cells within the tumor.55,56 An approach that targets DNA repair may therefore be particularly efficient in eliminating residual disease due to the stem cell population. Furthermore, sensitization occurs in both p53-null H358 cells and in MCF7 cells expressing wild-type p53, and therefore does not require p53-mediated apoptosis.
The tight temporal correlation of 1a-mediated inhibition of histone acetylation, DNA synthesis and S-phase progression in whole cells suggests that these events are interlinked mechanistically. Together with the large body of evidence that implicates histone acetylation as an essential step in DNA synthesis and repair,10,12 our data are consistent with a model in which inhibition of histone acetylation by 1a is causal in the chain of events that disrupts DNA metabolism. However, the molecular mechanism underlying the cellular effects of 1a is likely to be complex, and further investigation will be required to identify the critical enzymes and targets. These could include multiple potential acetylation sites within the N-terminal core histone regions mediated by a variety of HAT enzymes, including p300/CBP (reviewed in ref. 57, see Fig. 5B and Suppl. Data showing in vitro inhibition of HAT activity in solubilized chromatin by 1a and its isomeric form 1b), as well as DNA repair-linked acetylation of γ-H2A.X by Tip60.44 Our preliminary studies have confirmed Tip60 as one potential target of inhibition by 1a (Bandyopadhyay K and Gjerset RA, unpublished). In addition to acetylation of N-terminal sites such as H3-K9, acetylation has been observed in the globular domain of histone H3 on lysine 56 in yeast, where it has been linked to histone deposition during chromatin assembly and DNA repair,58,59 and recently in mammalian cells, including human cells.33,34 H3-K56 acetylation in human cells is mediated by p300/CBP.33 It is important to note that the two types of sites are topologically very distinct in the nucleosome assembly. In particular, in vitro studies with selectively proteolyzed nucleosome core particles suggest that while N-terminal sites such as H3K9 can be acetylated within the intact nucleosome, the acetylation of sites such as H3-K56 within the globular domain requires the absence of the immediate DNA environment.60 Thus DNA repair may involve a concerted action by p300/CBP acting first on peripheral (N-terminal) sites of DNA-bound histones to expose damaged DNA to repair complexes and then on internal sites of DNA-free histones to target them for deposition onto DNA and to signal the completion of the process. The ability of the compounds studied here to suppress acetylation at both sites (Figs. 1D and 5C) may contribute to their biological effects.
HAT enzymes act on non-histone substrates such as the p53 tumor suppressor as well (reviewed in ref. 57), and we find that 1a treatment of wild-type p53-expressing MCF7 cells reduces p53 acetylation (Bandyopadhyay and Gjerset, unpublished). However, this effect is unlikely to contribute to the therapy sensitization effects reported here, as p53-null H358 cells are as efficiently sensitized to DNA damage by 1a treatment as are MCF7 cells, which express wild-type p53. Finally, Spd-CoA is a potent inhibitor of spermine/spermidine acetyltransferase (SSAT),16 an important regulator of intracellular polyamine content and subcellular distribution. Treatment with 1a could therefore interfere with polyamine-mediated regulation of histone acetylation, a process observed with isolated chromatin,61,62 and in vivo.63
Inhibition of DNA repair, accompanied by radiosensitization, has been previously observed in cells lacking the histone deacetylase, Hdac3,64 a likely target of therapeutic HDAC inhibitors,64 and in cells treated with HDAC inhibitors.65 The fact that sensitization to DNA damage occurs when either HDAC or HAT activity is inhibited, suggests that the successful completion of DNA repair requires dynamic changes in chromatin involving both acetylation and deacetylation events. An advantage of the approach discussed here is that the synthesis of Spd-CoA and analogs from orthogonally protected polyamines is well documented,17 a situation highly advantageous for the development of modified compounds with improved therapeutic activity. One of the challenges of future work will be to identify the enzymatic activities and specific substrates responsible for the sensitization effects, and to design more targeted and more biologically stable versions of this promising class of therapy sensitizers.
Materials and Methods
Spermidine-CoA inhibitors
Spermidine(N1)-CoA or 1a, was synthesized as in ref.17 at the laboratory of one of us (C.B.). The truncated derivative spermidine(N1)-CO-CH2-S-(CH2)2-NH-CO-(CH2)2-NH-COCH3 or 2a was synthesized by A.M. and J.P. as in ref.17
Chemicals
Camptothecin (CPT) was purchased from Sigma, St. Louis, MO. Platinol™ [cisplatin, cis-diamminedichloroplatinum (II), CDDP] and Adrucil™ (5-fluorouracil, 5FU) were obtained from local pharmacies. Taxotere™ was provided by the late Dr. Pierre Potier.
In vitro assay of inhibition of p300HAT activity
Assays were performed in duplicate using a HAT assays kit (Active Motif, Carlsbad, CA), which provides an H3 peptide substrate (Ac-H3(5-23)-NH2, as communicated by the vendor) and the catalytic domain of p300(965-1810).
Cell lines and growth conditions
H358 human lung cancer cells, MCF7 human breast cancer cells, HET1A immortalized human epithelial cells, and BJ1 immortalized human fibroblasts were obtained from the American Type Culture Collection (ATCC). GT41 human skin fibroblasts (NHF) were obtained from a skin punch biopsy and cultured at the Sidney Kimmel Cancer Center, San Diego. All cell lines were maintained at 37°C in 10% CO2 in DMEM supplemented with non-essential amino acids, pyruvate, L-glutamine, gentamicin and 10% fetal bovine serum.
DNA synthesis assays
Cells (2,000/well) in 96-well plates were treated as follows: 1a (50 μM); camptothecin (CPT, 5 nM, 18 hours); cisplatin (Platinol™, 2 μM, 1 hour 15 minutes), 5-fluorouracil (5FU, Adrucil™, 2 μM, 1 hour 15 minutes), Taxotere™ (5 nM, 18 hours). For combined 1a/drug treatments, medium was removed after drug treatment and replaced with fresh medium containing 1a only. UV-C treatment (40 J/m2 using a Stratalinker™, Stratagene, La Jolla, CA) was carried using medium lacking phenol red. Triplicate wells of cells were pulsed 6 hours with 0.5 μCi [3H]-thymidine (NEN, Boston MA) at intervals 18–24 hr (Day 1), 42–48 hr (Day 2), 66–72 hr (Day 3) and 90–96 hr (Day 4) post-start of drug treatment, and harvested onto filter paper using a Brandel Harvester and subjected to scintillation counting. Untreated cells were pulsed from 0–6 hrs to establish the initial rate of incorporation. Trypan blue exclusion assays for cell membrane integrity have been described.26
Two-day cell growth and viability assays
Cells (3,000/well) in 96-well plates (exponential growth conditions for untreated cells) were treated in triplicate under conditions described in the text. Controls received no treatment. 2-day viability (expressed as percent control) was determined by the MTS assay MTS = [3-(4,5-dimethylthiazol-2-yl)-5-(3-carboxymethoxylphenyl)-2-(4-sulfophenyl)-2H-tetrazolium, inner salt], as described.26 For cell permeabilization, cells were exposed to saponin (Sigma, St. Louis, MO) as previously described27 prior to subsequent treatments.
Cell cycle analysis
Propidium iodide-stained cells were analyzed by a FACScan flow cytometer (Becton Dickenson, Franklin Lakes, NJ) as described.28 DNA histograms were analyzed with the FlowJo program using doublet discrimination. Each analysis was performed on two independent samples.
Western immunoblot analyses
Histones were isolated by extraction of cells in 0.2 M H2SO4 as described,29 followed by SDS-PAGE/western analysis.26 Protein concentrations were determined by a fluorescence-based quantitation assay (Quant-iT™ assay kit and Qubit™ fluorimeter, Invitrogen, Carlsbad, CA). Antibodies used were rabbit monoclonal anti-histone H3 antibody, rabbit polyclonal anti-trimethyl histone H3 (lysine 9), rabbit polyclonal anti-acetyl histone H3 (lysine 56), or rabbit polyclonal anti-acetyl histone H3 (lysine 9) (all purchased from Upstate Biotechnology Inc., Lake Placid, NY), or rabbit polyclonal anti-histone H2A.X (Bethyl Laboratories, Montgomery, TX).
DNA repair assays
To assay nucleotide excision repair (NER), a slot blot immunoassay was used to detect thymidine dimers in DNA of UV-C treated cells as described,30 using an anti-thymidine dimer antibody (1:40 dilution) (clone KTM53, Kamiya Biomedical Co., Seattle, WA). To eliminate possible cross-reactivity with a secondary pyrimidine-pyrimidone (6-4) photoproducts, DNA was treated with hot alkali as described.31
To assay double strand break (DSB) repair, cells were incubated for 1 hour with 10 μM CPT to induce DSBs, followed by incubation with or without 50 μM 1a. At fixed times, histones were isolated by extraction in 0.2 M H2SO4.29 60 μg extract was analyzed by SDS-PAGE/western,26 using rabbit polyclonal anti-γ-H2A.X [Ser 139] (Novus Biologicals, Littleton, CO) or anti-H3 (as described for western analyses). Band intensities were quantitated digitally. To study acetylation of γ-H2A.X, 60 μg protein extract in 10 mM sodium phosphate pH 7, 0.15 M NaCl, 0.1% SDS, 1% NP40, 1% sodium deoxycholate, aprotinin, 1 mM PMSF, complete protease inhibitors (Roche) was immunoprecipitated with immobilized anti-acetyl-lysine (Millipore, Temecula, CA), and analyzed by SDS-PAGE/western with anti γ-H2A.X.
Supplementary Material
Acknowledgements
California Tobacco-Related Disease Research Program 11RT-0074 and 18XT-0164 (to R.G.), National Cancer Institute R01CA111868 and R01CA135369 (to R.G.). Department of Pharmacology, Vanderbilt University (to J.P.) and Centre National de la Recherche Scientifique (CNRS) (to C.B. and J.-L.B.). We thank Marie Maturano for technical assistance in the preparation of the 1a compound.
We dedicate this work to the memory of Professor Pierre Potier.
Abbreviations
- Spd
spermidine
- CoA
coenzyme A
- HAT
histone acetyltransferase
- HDAC
histone deacetylase
- CDDP
cisplatin
- CPT
camptothecin
- 5-FU
5-fluorouracil
- DSB
double strand break
- NER
nucleotide excision repair
- NHF
normal human fibroblast
Footnotes
Note Supplementary materials can be found at: www.landesbioscience.com/supplement/BandyopadhyayCC8-17-Sup.pdf
References
- 1.Shahbazian MD, Grunstein M. Functions of site-specific histone acetylation and deacetylation. Annu Rev Biochem. 2007;76:75–100. doi: 10.1146/annurev.biochem.76.052705.162114. [DOI] [PubMed] [Google Scholar]
- 2.Berndsen CE, Tsubota T, Lindner SE, Lee S, Holton JM, Kaufman PD, et al. Molecular functions of the histone acetyltransferase chaperone complex Rtt109-Vps75. Nat Struct Mol Biol. 2008;15:948–56. doi: 10.1038/nsmb.1459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Davey CA, Sargent DF, Luger K, Maeder AW, Richmond TJ. Solvent mediated interactions in the structure of the nucleosome core particle at 1.9 Å resolution. J Mol Biol. 2002;319:1097–113. doi: 10.1016/S0022-2836(02)00386-8. [DOI] [PubMed] [Google Scholar]
- 4.Ko ER, Ko D, Chen C, Lipsick JS. A conserved acidic patch in the Myb domain is required for activation of an endogenous target gene and for chromatin binding. Mol Cancer. 2008;7:77. doi: 10.1186/1476-4598-7-77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Baneres JL, Martin A, Parello J. The N tails of histones H3 and H4 adopt a highly structured conformation in the nucleosome. J Mol Biol. 1997;273:503–8. doi: 10.1006/jmbi.1997.1297. [DOI] [PubMed] [Google Scholar]
- 6.Kornberg RD, Lorch Y. Twenty-five years of the nucleosome, fundamental particle of the eukaryote chromosome. Cell. 1999;98:285–94. doi: 10.1016/s0092-8674(00)81958-3. [DOI] [PubMed] [Google Scholar]
- 7.Shogren-Knaak M, Peterson CL. Switching on chromatin: mechanistic role of histone H4-K16 acetylation. Cell Cycle. 2006;5:1361–5. doi: 10.4161/cc.5.13.2891. [DOI] [PubMed] [Google Scholar]
- 8.Allfrey VG, Mirsky AE. Structural Modifications of Histones and their Possible Role in the Regulation of RNA Synthesis. Science. 1964;144:559. doi: 10.1126/science.144.3618.559. [DOI] [PubMed] [Google Scholar]
- 9.Jacobson S, Pillus L. Modifying chromatin and concepts of cancer. Curr Opin Genet Dev. 1999;9:175–84. doi: 10.1016/S0959-437X(99)80027-6. [DOI] [PubMed] [Google Scholar]
- 10.Wurtele H, Verreault A. Histone post-translational modifications and the response to DNA double-strand breaks. Curr Opin Cell Biol. 2006;18:137–44. doi: 10.1016/j.ceb.2006.02.008. [DOI] [PubMed] [Google Scholar]
- 11.Allis CD, Chicoine LG, Richman R, Schulman IG. Deposition-related histone acetylation in micronuclei of conjugating Tetrahymena. Proc Natl Acad Sci USA. 1985;82:8048–52. doi: 10.1073/pnas.82.23.8048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Suter B, Pogoutse O, Guo X, Krogan N, Lewis P, Greenblatt JF, et al. Association with the origin recognition complex suggests a novel role for histone acetyltransferase Hat1p/Hat2p. BMC Biol. 2007;5:38. doi: 10.1186/1741-7007-5-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Luo RX, Dean DC. Chromatin remodeling and transcriptional regulation. J Natl Cancer Inst. 1999;91:1288–94. doi: 10.1093/jnci/91.15.1288. [DOI] [PubMed] [Google Scholar]
- 14.Lindemann RK, Gabrielli B, Johnstone RW. Histone-deacetylase inhibitors for the treatment of cancer. Cell Cycle. 2004;3:779–88. [PubMed] [Google Scholar]
- 15.Marks PA, Richon VM, Miller T, Kelly WK. Histone deacetylase inhibitors. Adv Cancer Res. 2004;91:137–68. doi: 10.1016/S0065-230X(04)91004-4. [DOI] [PubMed] [Google Scholar]
- 16.Cullis PM, Wolfenden R, Cousens LS, Alberts BM. Inhibition of histone acetylation by N-[2-(S-coenzyme A)acetyl] spermidine amide, a multisubstrate analog. J Biol Chem. 1982;257:12165–9. [PubMed] [Google Scholar]
- 17.Roblot G, Wylde R, Martin A, Parello J. Regioselective synthesis of inhibitors of histone acetyl transferase covalently linking spermidine to the S-terminus of coenzyme A and fragments. Tetrahedron. 1993;49:6381–98. [Google Scholar]
- 18.Parello J, Roblot G, Wylde R, Martin A. Chemical Synthesis of Multisubstrate Inhibitors of Histone Acetyltransferase Covalently Linking Spermidine to an S-Terminal Fragment of Coenzyme-A. Comptes Rendus de l’Academie des Sciences Serie II. 1990;310:1441–6. [Google Scholar]
- 19.Zheng Y, Balasubramanyam K, Cebrat M, Buck D, Guidez F, Zelent A, et al. Synthesis and evaluation of a potent and selective cell-permeable p300 histone acetyltransferase inhibitor. J Am Chem Soc. 2005;127:17182–3. doi: 10.1021/ja0558544. [DOI] [PubMed] [Google Scholar]
- 20.Lau OD, Kundu TK, Soccio RE, Ait-Si-Ali S, Khalil EM, Vassilev A, et al. HATs off: selective synthetic inhibitors of the histone acetyltransferases p300 and PCAF. Mol Cell. 2000;5:589–95. doi: 10.1016/s1097-2765(00)80452-9. [DOI] [PubMed] [Google Scholar]
- 21.Balasubramanyam K, Altaf M, Varier RA, Swaminathan V, Ravindran A, Sadhale PP, et al. Polyisoprenylated benzophenone, garcinol, a natural histone acetyltransferase inhibitor, represses chromatin transcription and alters global gene expression. J Biol Chem. 2004;279:33716–26. doi: 10.1074/jbc.M402839200. [DOI] [PubMed] [Google Scholar]
- 22.Balasubramanyam K, Varier RA, Altaf M, Swaminathan V, Siddappa NB, Ranga U, et al. Curcumin, a novel p300/CREB-binding protein-specific inhibitor of acetyltransferase, represses the acetylation of histone/nonhistone proteins and histone acetyltransferase-dependent chromatin transcription. J Biol Chem. 2004;279:51163–71. doi: 10.1074/jbc.M409024200. [DOI] [PubMed] [Google Scholar]
- 23.Balasubramanyam K, Swaminathan V, Ranganathan A, Kundu TK. Small molecule modulators of histone acetyltransferase p300. J Biol Chem. 2003;278:19134–40. doi: 10.1074/jbc.M301580200. [DOI] [PubMed] [Google Scholar]
- 24.Eliseeva ED, Valkov V, Jung M, Jung MO. Characterization of novel inhibitors of histone acetyltransferases. Mol Cancer Ther. 2007;6:2391–8. doi: 10.1158/1535-7163.MCT-07-0159. [DOI] [PubMed] [Google Scholar]
- 25.Cohen SS. A Guide to Polyamines. Oxford University Press; New York, Oxford: 1998. pp. 467–9. [Google Scholar]
- 26.Saadatmandi N, Tyler T, Huang Y, Haghighi A, Frost G, Borgstrom P, et al. Growth suppression by a p14(ARF) exon 1beta adenovirus in human tumor cell lines of varying p53 and Rb status. Cancer Gene Ther. 2002;9:830–9. doi: 10.1038/sj.cgt.7700505. [DOI] [PubMed] [Google Scholar]
- 27.Johnson JA, Gray MO, Karliner JS, Chen CH, Mochly-Rosen D. An improved permeabilization protocol for the introduction of peptides into cardiac myocytes. Application to protein kinase C research. Circ Res. 1996;79:1086–99. doi: 10.1161/01.res.79.6.1086. [DOI] [PubMed] [Google Scholar]
- 28.Gjerset RA, Turla ST, Sobol RE, Scalise JJ, Mercola D, Collins H, et al. Use of wild-type p53 to achieve complete treatment sensitization of tumor cells expressing endogenous mutant p53. Mol Carcinog. 1995;14:275–85. doi: 10.1002/mc.2940140408. [DOI] [PubMed] [Google Scholar]
- 29.Furuta T, Takemura H, Liao ZY, Aune GJ, Redon C, Sedelnikova OA, et al. Phosphorylation of histone H2AX and activation of Mre11, Rad50 and Nbs1 in response to replication-dependent DNA double-strand breaks induced by mammalian DNA topoisomerase I cleavage complexes. J Biol Chem. 2003;278:20303–12. doi: 10.1074/jbc.M300198200. [DOI] [PubMed] [Google Scholar]
- 30.Lee C, Smith BA, Bandyopadhyay K, Gjerset RA. DNA damage disrupts the p14ARF-B23(nucleophosmin) interaction and triggers a transient subnuclear redistribution of p14ARF. Cancer Res. 2005;65:9834–42. doi: 10.1158/0008-5472.CAN-05-1759. [DOI] [PubMed] [Google Scholar]
- 31.McCready S. A dot-blot immunoassay for measuring repair of ultraviolet photoproducts. Methods Mol Biol. 2006;314:229–38. doi: 10.1385/1-59259-973-7:229. [DOI] [PubMed] [Google Scholar]
- 32.Ozdemir A, Masumoto H, Fitzjohn P, Verreault A, Logie C. Histone H3 lysine 56 acetylation: a new twist in the chromosome cycle. Cell Cycle. 2006;5:2602–8. doi: 10.4161/cc.5.22.3473. [DOI] [PubMed] [Google Scholar]
- 33.Das C, Lucia MS, Hansen KC, Tyler JK. CBP/p300-mediated acetylation of histone H3 on lysine 56. Nature. 2009;459:113–7. doi: 10.1038/nature07861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yuan J, Pu M, Zhang Z, Lou Z. Histone H3-K56 acetylation is important for genomic stability in mammals. Cell Cycle. 2009;8:1747–53. doi: 10.4161/cc.8.11.8620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Garcia-Carbonero R, Supko JG. Current perspectives on the clinical experience, pharmacology and continued development of the camptothecins. Clin Cancer Res. 2002;8:641–61. [PubMed] [Google Scholar]
- 36.Bandyopadhyay K, Lee C, Haghighi A, Baneres JL, Parello J, Gjerset RA. Serine phosphorylation-dependent coregulation of topoisomerase I by the p14ARF tumor suppressor. Biochemistry. 2007;46:14325–34. doi: 10.1021/bi7013618. [DOI] [PubMed] [Google Scholar]
- 37.Cazzalini O, Perucca P, Savio M, Necchi D, Bianchi L, Stivala LA, et al. Interaction of p21(CDKN1A) with PCNA regulates the histone acetyltransferase activity of p300 in nucleotide excision repair. Nucleic Acids Res. 2008;36:1713–22. doi: 10.1093/nar/gkn014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yu Y, Waters R. Histone acetylation, chromatin remodelling and nucleotide excision repair: hint from the study on MFA2 in Saccharomyces cerevisiae. Cell Cycle. 2005;4:1043–5. doi: 10.4161/cc.4.8.1928. [DOI] [PubMed] [Google Scholar]
- 39.Olaussen KA, Dunant A, Fouret P, Brambilla E, Andre F, Haddad V, et al. DNA repair by ERCC1 in non-small-cell lung cancer and cisplatin-based adjuvant chemotherapy. N Engl J Med. 2006;355:983–91. doi: 10.1056/NEJMoa060570. [DOI] [PubMed] [Google Scholar]
- 40.Avemann K, Knippers R, Koller T, Sogo JM. Camptothecin, a specific inhibitor of type I DNA topoisomerase, induces DNA breakage at replication forks. Mol Cell Biol. 1988;8:3026–34. doi: 10.1128/mcb.8.8.3026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bogdanov KV, Chukhlovin AB, Zaritskey AY, Frolova OI, Afanasiev BV. Ultraviolet irradiation induces multiple DNA double-strand breaks and apoptosis in normal granulocytes and chronic myeloid leukaemia blasts. Br J Haematol. 1997;98:869–72. doi: 10.1046/j.1365-2141.1997.2913109.x. [DOI] [PubMed] [Google Scholar]
- 42.Pavon MA, Parreno M, Leon X, Sancho FJ, Cespedes MV, Casanova I, et al. Ku70 predicts response and primary tumor recurrence after therapy in locally advanced head and neck cancer. Int J Cancer. 2008;123:1068–79. doi: 10.1002/ijc.23635. [DOI] [PubMed] [Google Scholar]
- 43.Rogakou EP, Boon C, Redon C, Bonner WM. Megabase chromatin domains involved in DNA double-strand breaks in vivo. J Cell Biol. 1999;146:905–16. doi: 10.1083/jcb.146.5.905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kusch T, Florens L, Macdonald WH, Swanson SK, Glaser RL, Yates JR, 3rd, et al. Acetylation by Tip60 is required for selective histone variant exchange at DNA lesions. Science. 2004;306:2084–7. doi: 10.1126/science.1103455. [DOI] [PubMed] [Google Scholar]
- 45.Herceg Z, Wang ZQ. Rendez-vous at mitosis: TRRAPed in the chromatin. Cell Cycle. 2005;4:383–7. doi: 10.4161/cc.4.3.1546. [DOI] [PubMed] [Google Scholar]
- 46.Bolognesi ML, Calonghi N, Mangano C, Masotti L, Melchiorre C. Parallel Synthesis and Cytotoxicity Evaluation of a Polyamine-Quinone Conjugates Library. J Med Chem. 2008;51:5463–7. doi: 10.1021/jm800637b. [DOI] [PubMed] [Google Scholar]
- 47.Barret JM, Kruczynski A, Vispe S, Annereau JP, Brel V, Guminski Y, et al. F14512, a potent antitumor agent targeting topoisomerase II vectored into cancer cells via the polyamine transport system. Cancer Res. 2008;68:9845–53. doi: 10.1158/0008-5472.CAN-08-2748. [DOI] [PubMed] [Google Scholar]
- 48.Wolf M, Bauder-Wust U, Pipkorn R, Eskerski H, Eisenhut M. Fluorophor-labeled spermidine derivatives as fluorescent markers in optical tumor imaging. Bioorg Med Chem Lett. 2006;16:3193–6. doi: 10.1016/j.bmcl.2006.03.046. [DOI] [PubMed] [Google Scholar]
- 49.Martin LP, Hamilton TC, Schilder RJ. Platinum resistance: the role of DNA repair pathways. Clin Cancer Res. 2008;14:1291–5. doi: 10.1158/1078-0432.CCR-07-2238. [DOI] [PubMed] [Google Scholar]
- 50.Biedermann KA, Sun JR, Giaccia AJ, Tosto LM, Brown JM. scid mutation in mice confers hypersensitivity to ionizing radiation and a deficiency in DNA double-strand break repair. Proc Natl Acad Sci USA. 1991;88:1394–7. doi: 10.1073/pnas.88.4.1394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Deriano L, Guipaud O, Merle-Beral H, Binet JL, Ricoul M, Potocki-Veronese G, et al. Human chronic lymphocytic leukemia B cells can escape DNA damage-induced apoptosis through the nonhomologous end-joining DNA repair pathway. Blood. 2005;105:4776–83. doi: 10.1182/blood-2004-07-2888. [DOI] [PubMed] [Google Scholar]
- 52.Boulton S, Kyle S, Durkacz BW. Interactive effects of inhibitors of poly(ADP-ribose) polymerase and DNA-dependent protein kinase on cellular responses to DNA damage. Carcinogenesis. 1999;20:199–203. doi: 10.1093/carcin/20.2.199. [DOI] [PubMed] [Google Scholar]
- 53.Sun Y, Jiang X, Chen S, Price BD. Inhibition of histone acetyltransferase activity by anacardic acid sensitizes tumor cells to ionizing radiation. FEBS Lett. 2006;580:4353–6. doi: 10.1016/j.febslet.2006.06.092. [DOI] [PubMed] [Google Scholar]
- 54.Ribacka C, Pesonen S, Hemminki A. Cancer, stem cells and oncolytic viruses. Ann Med. 2008:1–10. doi: 10.1080/07853890802021342. [DOI] [PubMed] [Google Scholar]
- 55.Trumpp A, Wiestler OD. Mechanisms of Disease: cancer stem cells—targeting the evil twin. Nat Clin Pract Oncol. 2008;5:337–47. doi: 10.1038/ncponc1110. [DOI] [PubMed] [Google Scholar]
- 56.Johannessen TC, Bjerkvig R, Tysnes BB. DNA repair and cancer stem-like cells—Potential partners in glioma drug resistance? Cancer Treat Rev. 2008;34:558–67. doi: 10.1016/j.ctrv.2008.03.125. [DOI] [PubMed] [Google Scholar]
- 57.Yang XJ. The diverse superfamily of lysine acetyltransferases and their roles in leukemia and other diseases. Nucleic Acids Res. 2004;32:959–76. doi: 10.1093/nar/gkh252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Chen CC, Carson JJ, Feser J, Tamburini B, Zabaronick S, Linger J, et al. Acetylated lysine 56 on histone H3 drives chromatin assembly after repair and signals for the completion of repair. Cell. 2008;134:231–43. doi: 10.1016/j.cell.2008.06.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Chen CC, Tyler J. Chromatin reassembly signals the end of DNA repair. Cell Cycle. 2008;7:3792–7. doi: 10.4161/cc.7.24.7188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Dumuis-Kervabon A, Encontre I, Etienne G, Jauregui-Adell J, Mery J, Mesnier D, et al. A chromatin core particle obtained by selective cleavage of histones by clostripain. EMBO J. 1986;5:1735–42. doi: 10.1002/j.1460-2075.1986.tb04418.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Estepa I, Pestana A. Activation by polyamines of the acetylation of endogenous histones in isolated chromatin and nuclei from Artemia. Eur J Biochem. 1981;119:431–6. doi: 10.1111/j.1432-1033.1981.tb05626.x. [DOI] [PubMed] [Google Scholar]
- 62.Dod B, Kervabon A, Parello J. Effect of cations on the acetylation of chromatin in vitro. Eur J Biochem. 1982;121:401–5. doi: 10.1111/j.1432-1033.1982.tb05801.x. [DOI] [PubMed] [Google Scholar]
- 63.Wei G, Hobbs CA, Defeo K, Hayes CS, Gilmour SK. Polyamine-mediated regulation of protein acetylation in murine skin and tumors. Mol Carcinog. 2007;46:611–7. doi: 10.1002/mc.20350. [DOI] [PubMed] [Google Scholar]
- 64.Bhaskara S, Chyla BJ, Amann JM, Knutson SK, Cortez D, Sun ZW, et al. Deletion of histone deacetylase 3 reveals critical roles in S phase progression and DNA damage control. Mol Cell. 2008;30:61–72. doi: 10.1016/j.molcel.2008.02.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Cerna D, Camphausen K, Tofilon PJ. Histone deacetylation as a target for radiosensitization. Curr Top Dev Biol. 2006;73:173–204. doi: 10.1016/S0070-2153(05)73006-4. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.