Abstract
Objective
To determine factors differentiating LPA3 from other nine lysophospholipid (LP) receptors for its role in embryo implantation.
Design
Experimental mouse models.
Setting
Institute/University research laboratories.
Animal(s)
Wild type, Lpar3(−/−), Lpar1(−/−)Lpar2(−/−), and S1pr2(−/−)S1pr3(−/−) mice.
Intervention(s)
Ovariectomy.
Main Outcome Measure(s)
Blue dye injection for determining implantation sites on gestation day 4.5. Realtime PCR for measuring gene expression in whole uterus and separated uterine layers. In situ hybridization for detecting progesterone (P)-induced Lpar3 expression in the uterine luminal epithelium (LE).
Result(s)
Normal implantation was observed in Lpar1(−/−)Lpar2(−/−) and S1pr2(−/−)S1pr3(−/−) females. Temporal expression showed peak expression of Lpar3 in the preimplantation uterus and constitutive expression of the other nine LP receptors in the peri-implantation uterus. Spatial localization revealed main expression of Lpar3 in the LE and broad expression of the remaining LP receptors in all three main uterine layers: LE, stromal, and myometrial layers. Hormonal regulation in ovariectomized uterus indicated upregulation of Lpar3 but downregulation or no effect of the remaining nine LP receptors by P, and downregulation of most LP receptors, including Lpar3, by 17β-estradiol (E2).
Conclusion(s)
LE localization and upregulation by progesterone differentiates LPA3 from the other nine LP receptors and may underlie its essential role in embryo implantation.
Keywords: lysophospholipid receptors, uterine luminal epithelium, progesterone, embryo implantation
Introduction
Lysophospholipids (LPs) are quantitatively minor lipid species that are well known as components in the biosynthesis of membrane phospholipids and as metabolic intermediates (1). A few LPs, mainly lysophosphatidic acid (LPA) and sphingosine 1-phosphate (S1P), have been proven to function as extracellular signaling molecules through cell surface G protein-coupled receptors (2), including LPA1-5 (LPA1-5) and S1P1-5 (S1P1-5) (3–5). Other potential LP receptors have also been reported (6–9) . Several LP receptor-specific in vivo functions have been identified in receptor specific knockout mice, e.g., LPA1 in proper craniofacial formation, neural development, and neuropathic pain (10–12); S1P1 in vascular development (13); S1P2 in auditory and vestibular function (14, 15); S1P4 in shaping the terminal differentiation of megakaryocytes (16); LPA1-3 in spermatogenesis (17); and LPA3 in embryo implantation (18–21).
Embryo implantation is a crucial step for the successful establishment of pregnancy in mammals. It requires effective reciprocal signaling between a competent blastocyst and a receptive uterus during a discrete “implantation window”. The “implantation window” is a limited time during which the uterine environment favors blastocyst growth, attachment, and implantation into the uterine wall (22–25). We have previously shown that deletion of Lpar3 in mice delays embryo implantation and alters embryo spacing. These defects are caused by uterine rather than embryonic loss of Lpar3, indicating uterine defects (18). Defective embryo implantation has not been reported in other available LP receptor-deficient females (10, 13, 16, 26–29) and in this study we confirm normal embryo implantation in mice lacking LPA1, LPA2, S1P2, or S1P3. Some LP receptors, e.g., LPA1, LPA2, and LPA3 (30), share high sequence homology and have comparable expression levels in the preimplantation uterus, yet only deletion of Lpar3 but not of Lpar1 and Lpar2 affects embryo implantation. What factors contribute to LPA3 receptor-specific role in embryo implantation Here we evaluate the ten bona fide LP receptors for their temporal expression patterns in the peri-implantation uterus, their localization in the preimplantation day 3.5 uterus, as well as their uterine regulation by ovarian hormones to identify the uniqueness of Lpar3 among the LP receptors, which may underscore LPA3 receptor-specificity in embryo implantation, especially in the establishment of uterine receptivity. The information from this study can also provide guidance for identifying candidate genes that are potentially involved in the establishment of uterine receptivity.
Material and Methods
Animals
Wild type (WT) and Lpar1, Lpar2, Lpar3, S1pr2, and S1pr3 knockout mice (129/SvJ and C57BL/6 mixed background) were generated and genotyped as described (10, 18, 26, 27, 31). The animal facility is on a 12-hour light/dark cycle (6:00 AM to 6:00 PM) at 23±1°C with 30–50 relative humidity. All methods used in this study were approved by the Animal Subjects Programs of The Scripps Research Institute and the University of Georgia and conform to National Institutes of Health guidelines and public law.
Localization of implantation sites and uterine sample collection
Implantation sites were localized as previously described (18). Uterine tissues from gestation days 0.5, 3.5, and 4.5 WT females between 11:00 AM and 12:00 PM were flash-frozen. Pregnancy status was determined by the presence of eggs and sperms in the oviduct (day 0.5), or blastocysts in the uterus (day 3.5), or implantation sites (day 4.5). Uteri from pregnant WT females were included in the study.
Isolation of three uterine layers
Previously reported procedures were modified to separate luminal epithelium (LE), stromal layer, and myometrium from three gestation day 3.5 WT uteri (32, 33). Briefly, sliced open uterine horns were submerged in 0.5% dispase in calcium- and magnesium-free Hanks balanced salt solution (HBSS, Invitrogen) and digested for 2 hours at room temperature. LE sheets were gently scraped. The stromal layer (Str) with glandular epithelium was isolated with a stronger scraping force. LE and stromal layer were collected for RNA isolation. The remaining myometrium was pulverized in liquid nitrogen for RNA isolation.
Hormonal treatment
Hormonal treatment was administered as previously described (34). Each group included 4–6 ovariectomized females. Uterine horns were flash-frozen.
Realtime reverse transcriptase-PCR (RT-PCR)
RNA isolation and realtime PCR were done as previously described (18, 35). To quantify the relative expression levels of each LP receptor, cDNA product from each primer pair (Supplementary Table S1) was subcloned into pGEM vector (Promega). A standard curve was prepared for each primer pair from serial dilution of the pGEM-cDNA plasmid (10−1~10−6 fmol). The relative transcript number of each gene was quantified and then normalized to β-actin as a loading control.
In situ hybridization
Statistical analysis
Data were expressed as mean ± SD. Statistical analyses were done using Student’s unequal variance t-test. The significance level was set at p<0.05.
Results
Expression levels of LP receptors in the day 3.5 uterus
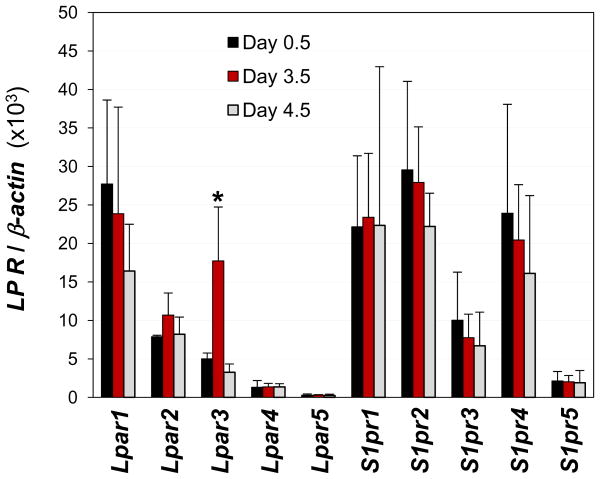
Lpar3 shows the highest expression level in the WT uterus at gestation day 3.5 (18). To compare the relative expression of other LP receptors in the uterus, day 3.5 WT uterus was chosen to quantify the expression levels of Lpar1-5 and S1pr1-5. Figure 1 showed that Lpar1, Lpar2, S1pr1, S1pr2, and S1pr4 mRNA levels were comparable to Lpar3 level in gestation day 3.5 WT uterus. The expression level of S1pr3 was about 40% of Lpar3. The other three LP receptors, Lpar4, Lpar5, and S1pr5, had significantly lower expression levels (Fig. 1).
Figure 1.
Expression of LP receptors in gestation days 0.5, 3.5, and 4.5 mouse uterus by realtime PCR. *, P<0.01, compared to day 0.5 or day 4.5. N=3–6. Error bars represent standard deviation.
Embryo implantation in different LP receptor knockout mice
Since other LP receptors are also highly expressed in the preimplantation WT uterus, to identify other LP receptor subtypes (besides LPA3) that might specifically influence implantation, we evaluated embryo implantation on the available and relevant receptor knockout mice. Of the ten LP receptors examined, seven (Lpar1-3 and S1pr1-4) were expressed at notably higher levels than the other three (Lpar4-5 and S1pr5) in the day 3.5 preimplantation uterus (Fig. 1). These seven highly expressed LPs have been genetically deleted and mice deficient of six of them (except S1pr1) have produced viable offspring: Lpar1, Lpar2, S1pr2, S1pr3, S1pr4and the previously analyzed Lpar3. Embryo implantation was evaluated in females deficient of these LP receptors except S1P4.
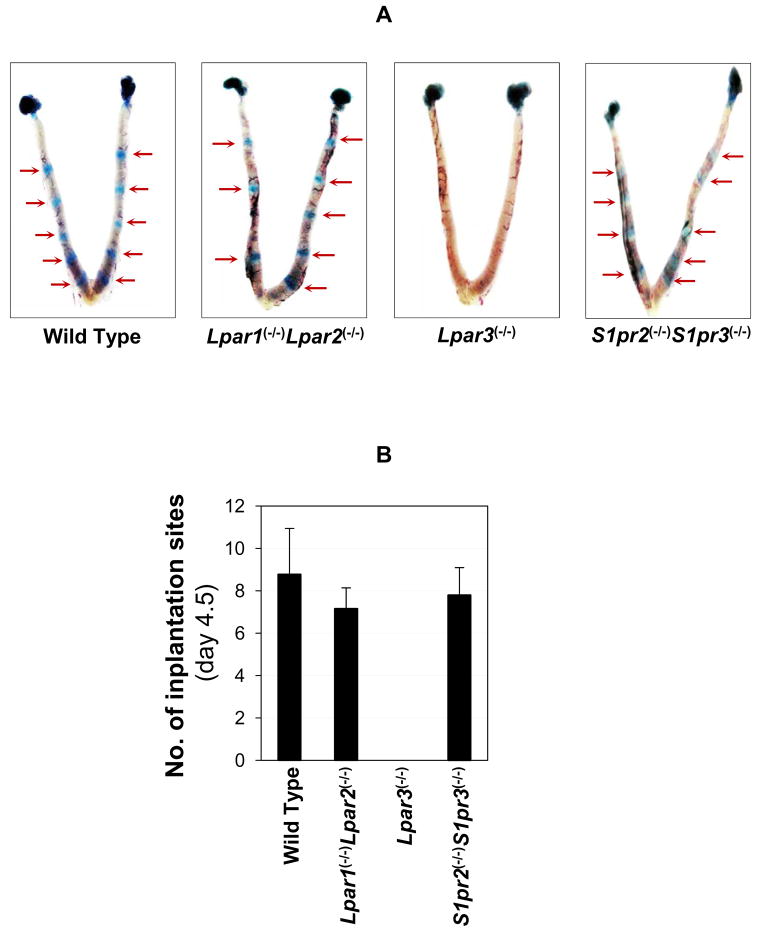
Implantation sites were detected at day 4.5 by Evans blue dye in Lpar1(−/−), Lpar2(−/−), S1pr2(−/−), S1pr3(−/−) females, as well as Lpar1(−/−)Lpar2(−/−) and S1pr2(−/−)S1pr3(−/−). On-time implantation was detected in the single knockout mice (data not shown) as well as the double knockout mice (Fig. 2A). The numbers of implantation sites from Lpar1(−/−)Lpar2(−/−) and S1pr2(−/−)S1pr3(−/−) double knockout mice detected at gestation day 4.5 were comparable to that from the WT (Fig. 2B). In addition, normal implantation spacing was observed in these mice, suggesting that these four LP receptors, Lpar1, Lpar2, S1pr2, and S1pr3, are not critical for embryo spacing either (Fig. 2A). These observations are contrasting with delayed implantation and embryo crowding in Lpar3(−/−) females (Fig. 2A) (18). These results suggest that LPA1, LPA2, S1P2, and S1P3 are not critical for embryo implantation timing and spacing, in spite of their significant expression levels in the preimplantation day 3.5 uterus.
Figure 2.
Detection of implantation sites in wild type, Lpar1(−/−)Lpar2(−/−), Lpar3(−/−), and S1pr2(−/−)S1pr3(−/−) females on gestation day 4.5 by Evans blue dye injection. A. Uterine images. Red arrows indicate implantation sites. No implantation sites were detected in the Lpar3(−/−) uterus but healthy-looking blastocysts were flushed from the uterine horns (data not shown). B. Number of implantation sites. N=5–9. Error bars represent standard deviation.
Receptor mRNA localization in the day 3.5 uterus
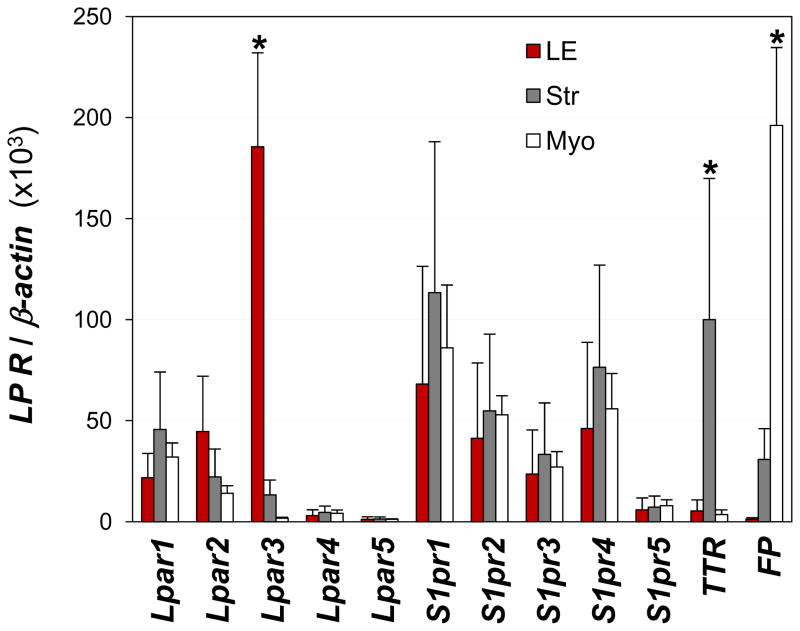
Lpar3 mRNA in the uterus is mainly expressed in the LE (18). The LE is the first layer of cells with which a blastocyst communicates prior to implanting in the uterine wall. This LE-specific expression pattern underscores the importance of LPA3 in embryo implantation. To determine if any other LP receptors share this expression pattern, we quantified mRNA levels of the ten LP receptors in the three uterine layers: LE, stromal, and myometrial layers. In addition to Lpar3, all of the remaining nine LP receptors were also detectable to some extent in the LE. However, while Lpar3 was mainly detected in the LE, the remaining nine LP receptors had comparable levels of expression in all three layers (Fig. 3). Transthyretin (TTR) is mainly expressed in the glandular epithelium in the stromal layer (37). TTR was used as a marker for stromal layer. Prostaglandin F2α receptor (FP) is mainly expressed in the myometrium of day 3.5 uterus and it served as a marker for myometrium (Fig. 3) (38). These results indicate that Lpar3 has a unique LE localization that is different from the rest LP receptors examined.
Figure 3.
Localization of LP receptors in isolated uterine luminal epithelium (LE), stromal layer (Str) with glandular epithelium, and myometrium (Myo) from preimplantation day 3.5 wild type uterus by realtime PCR. LE-specific Lpar3 (18), glandular epithelium-specific TTR (transthyretin) (37), and myometrium-specific FP (prostaglandin F2α receptor) (38) were served as a marker for LE, stromal layer, and myometrium, respectively. * P<0.05 compared to “Str” and “Myo” for Lpar3, “LE” and “Myo” for TTR, “LE” and “Str” for FP. N=3. Error bars represent standard deviation.
Receptor regulation by ovarian hormones progesterone and estrogen
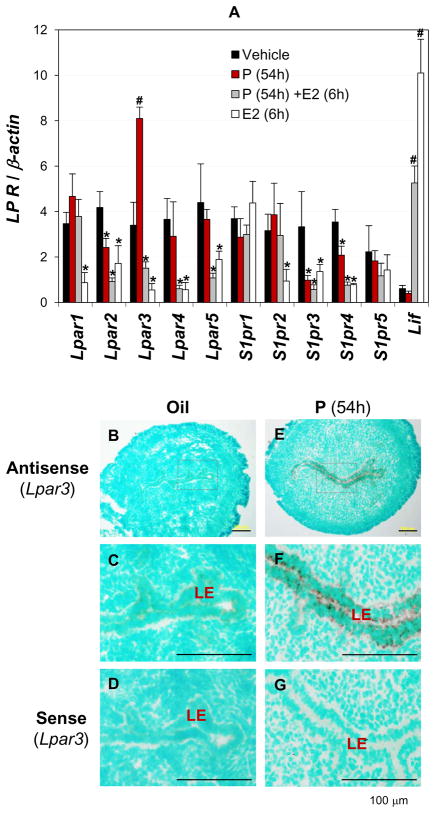
Ovarian hormones progesterone (P) and estrogen are the master controls of embryo implantation process in mice (39). LP receptor gene expression was therefore assessed for ovarian hormonal influences. It had been demonstrated that Lpar3 is regulated by both P and 17β-estrodial (E2) (40). To determine the regulation of Lpar1-5 and S1pr1-5 by ovarian hormones, prior published hormonal treatment approaches (34) were combined with realtime PCR to quantify the expression levels of LP receptors. Lpar1 expression was not changed by P (54h) or P (54h)+E2. It was transiently downregulated by E2 only (1h and 6h) (Figs. 4A, S1A). Lpar2 expression was downregulated by both P and E2 (6h and 24h) (Figs. 4A, S1B). Lpar3 expression was upregulated by P and downregulated by E2 (6h and 24h). The P-induced upregulation of Lpar3 could be reversed by E2 treatment for 6h and 24h (Figs. 4A, S1C). Lpar3 was also upregulated by P after 24h of treatment and this upregulation could be abolished by progesterone receptor (PR) antagonist RU486 (data not shown), indicating the involvement of PR in regulating P-induced uterine Lpar3 expression. P induced Lpar3 expression mainly in the LE (Figs. 4B~E). Lpar4 expression was downregulated by P+E2 (6h) and E2 (6h and 24h), but not P alone (Figs. 4A, S1D). Lpar5 expression was transiently downregulated by P+E2 and E2 treatments for 6h only (Figs. 4A, S1E). S1pr1 expression was decreased only by P+E2 and E2 treatments for 24h, but not affected by P treatment, indicating a main role of E2 in this regulation (Figs. 4A, S2A). S1pr2 expression was suppressed by E2 treatment at all the time points examined (1h, 6h, and 24h) but not changed by P or P+E2 treatments, indicating that although P did not affect the expression of S1pr2, it could block the effect of E2 on S1pr2 expression (Figs. 4A, S2B). S1pr3 expression was downregulated by P, P+E2, or E2 (6h and 24h) treatments, indicating that both P and E2 affected S1pr3 expression (Figs. 4A, S2C). S1pr4 had a similar response to P and E2 treatments as that of S1pr3 and Lpar2 (Figs. 4A, S1B, S2C, S2D). S1pr5 expression was not affected by either P or E2 (Figs. 4A, S2E). Leukaemia inhibitory factor (Lif) was upregulated by E2 and served as a control to demonstrate that the downregulation of LP receptors by E2 was specific (Figs. 4A, S2F) (41). Although LP receptor mRNA levels can be differentially regulated by P and E2, Lpar3 was the only one upregulated by P treatment (Figs. 4A, S1, S2). The upregulation of uterine Lpar3 by P is another unique feature among the ten LP receptors.
Figure 4.
Regulation of LP receptors by progesterone (P) and 17β-estrodiol (E2) in ovariectomized mouse uterus. A. Effects of P and E2 on LP receptor expression determined by realtime PCR. Wild type virgin females (6 weeks old) were ovariectomized and allowed to recover for two weeks. “Oil”: daily injection of vehicle (0.1 ml sesame oil) for three days, sacrificed 6h after the final injection (54h from the first injection). “P”: daily injection of P (2 mg/mouse, Sigma) for 3 days, sacrificed 6h after the final injection (54h from the first injection). “P+E2”: daily injection of P for 3 days, plus a single dose of E2 (100 ng/mouse, Sigma) on day 3, sacrificed 1h, 6h, and 24h later. “E2”: daily injection of vehicle for two days, plus a single dose of E2 on day three, sacrificed 1h, 6h, and 24h later. A complete set of data is in Supplementary Figures S1 and S2. Arbitrary scale. Leukaemia inhibitory factor (Lif), as a control for regulation of LP receptors by E2; β-actin as a loading control. * P<0.05 (downregulation) and # P<0.05 (upregulation), compared to control “oil”. N=4–6. Error bars represent standard deviation. “54h” indicates the duration of P treatment (54 hours); “6h” indicates 6 hours post E2 injection. In situ hybridization on ovariectomized uteri are shown in B~G. B. Lpar3 antisense probe, oil-injected. C. Enlarged view of the rectangle area in B. D. Lpar3 sense probe, oil-injected. E. Lpar3 antisense probe, P-injected. F. Enlarged view of the rectangle area in E. G. Lpar3 sense probe, P-injected. The duration of treatment was 54h. The sections were counterstained with 1% methyl green. Specific signal (dark brown) is detected in the uterine luminal epithelium (LE). No specific signals were detected in the negative control using a sense Lpar3 probe (D, G). Positive control (day 3.5 uterus, Lpar3 antisense probe) is shown in Supplementary Figure S3. N=3. Scale bar: 100 μm.
Temporal expression of LP receptors in the peri-implantation uterus
Lpar3 has its peak expression in the preimplantation day 3.5 WT uterus during pregnancy (18). The expression of Lpar1-5 and S1pr1-5 at gestation days 0.5, 3.5 (preimplantation), and 4.5 (post-implantation) in WT uterus was also assessed. Interestingly, only Lpar3 showed a dynamic expression pattern whereas the other nine LP receptors maintained comparable expression levels in the uterus during peri-implantation (Fig. 1).
Discussion
Laser-capture microdissection coupled with realtime PCR and in situ hybridization were previously used to localize Lpar3 in uterus (18). Both methods gave precise gene localization, but were inefficient for screening large number of genes and were not amenable to simultaneously determining their relative expression levels. The simple method used in this study for separating the three main uterine layers has the advantages of roughly localizing uterine gene expression and quantifying relative expression levels of different genes in the same uterine layers. This approach can be validated using Lpar3, TTR, and FP as markers for LE, stromal layer, and myometrium, respectively (Fig. 3) (18, 37, 38). In addition, about 0.5~1.5 μg of total RNA from gestation day 3.5 LE can be obtained, greatly exceeding that from laser-capture microdissection, which is in 10–150 ng range (42). The precise mRNA localization of candidate genes could be further analyzed using the two previously mentioned techniques (18).
Evidence suggests that the LE plays important roles in embryo implantation, especially the establishment of uterine receptivity: it is the first layer of cells that a blastocyst communicates with during the initial stage of implantation; LE cells develop mid-secretory uterodomes or pinopodes, an attribute of receptive uterine tissue; and the LE acts as a transducer of the embryo’s presence to elicit underlying stromal responses (33, 42–45). Therefore, the localization of Lpar3 in the LE underscores its important role in the establishment of uterine receptivity. All ten LP receptors are detectable in the LE, however, only Lpar3 shows nearly exclusive expression in the LE. Thus, while LP receptors may have roles in the uterus, only LPA3 has the identified role in the establishment of uterine receptivity (18).
Lpar3 is the only LP receptor upregulated by P-PR, and specifically in the LE (Fig. 4 and data not shown), which may be another critical determinant of its function in the establishment of uterine receptivity. P is a master control of embryo implantation and PR-mediated P signaling is indispensable for embryo implantation (39, 46–48). Results from indian hedgehog (Ihh, P-PR target gene) uterine epithelium conditional knockout mice reinforce the critical role of P-PR signaling in LE for embryo implantation (49–51). However, sustained PR expression in the uterine epithelium beyond the expected “implantation window” is detrimental to embryo implantation (46, 47, 52–55). We have demonstrated that PR remains expressed in the LE before decidualization becomes manifested and disappears from LE afterwards (56), suggesting an active role of PR in the LE during the initial implantation process. The observations that Lpar3 expression peaks in the preimplantation LE and returns to basal level in the post-implantation uterus (Fig. 1) (18) and Lpar3 is upregulated by P-PR signaling in the LE (Fig. 4 and data not shown) suggest that these two receptors, PR and LPA3, may communicate with each other in the LE for the establishment of uterine receptivity.
P and E2 differentially regulate LP receptors (Figs. 4A, S1, S2). Surprisingly, the downregulation of LP receptors by P and/or E2 seen in ovariectomized uterus was not observed in the peri-implantation uterus, especially at day 3.5 when the P level is high in addition to an estrogen surge (57). One possible explanation is that the uterine local levels and ratios of P and E2 received in the ovariectomized mice did not mimic the physiological conditions in the peri-implantation uterus. However, uterine Lpar3 peaks at day 3.5 (Fig. 1) (18), which agrees with the upregulation of Lpar3 by P in the ovariectomized uterus (Fig. 4). The information obtained from this study, e.g., differential spatiotemporal uterine expression and hormonal regulation, can potentially be used as a guide to identify uterine genes critical for embryo implantation.
LPA1, LPA2, S1P2, and S1P3 are demonstrated to be not individually critical for embryo implantation (Fig. 2). Other reports indicate no implantation defects due to loss of LPA4 and S1P5 (28, 29). There was no mention of implantation defects in the S1pr4(−/−) mice either (16). The in vivo role of S1P1 in embryo implantation could not be assessed because of embryonic lethality of S1pr1(−/−) mice and the lack of uterine-specific S1pr1(−/−) mice (13). Although Lpar5(−/−)mice are not available yet, the lack of distinct spatiotemporal expression patterns in the peri-implantation uterus and its low levels of expression in the peri-implantation uterus (Figs. 1, 3) suggest that LPA5 may not be critical for embryo implantation. Similar implantation defects between Lpar1(−/−)Lpar2(−/−)Lpar3(−/−) and Lpar3(−/−) females (20) reinforces the critical role of LPA3 in the establishment of uterine receptivity. However, these observations do not exclude other potential uterine functions of LP receptors, e.g., S1P1, S1P2, and S1P3 may play a role in decidualization (58).
Two interesting studies suggest that human LPA3 may play a role in the establishment of uterine receptivity (59, 60). Further analyses of human uterine microarray data (identifier GSE6364) (59) indicate that LPAR3 (EDG7) mRNA peaks in the normal early secretory phase endometrium (ESE, preimplantation stage), paralleling data obtained from mouse (18). LPAR3 mRNA levels are significantly downregulated in the ESE of endometriosis patients, while LPA3 protein levels are marginally downregulated in the ESE and significantly downregulated in the middle and later secretory phase endometrium (59, 60). The differential expression patterns of LPAR3 in normal and endometriosis patients are not observed in other LP receptors that were included in the database (59). The dysregulation of uterine LPA3 in endometriosis patients suggests that LPA3 may be involved in endometriosis-associated defective uterine receptivity.
Supplementary Material
Supplementary Figures S1. Regulation of LPA receptors by progesterone (P) and 17β-estrodiol (E2) in ovariectomized mouse uterus determined by realtime PCR. A. Lpar1; B. Lpar2; C. Lpar3; D. Lpar4; E. Lpar5. * P<0.05, downregulation; ** P<0.01, downregulation; # P<0.05, upregulation; ## P<0.01, upregulation, compared to control “oil”. N=4–6. Error bars represent standard deviation. “1h”, “6h”, and “24h” indicate hours post E2 injection.
Supplementary Figures S2. Regulation of S1P receptors by progesterone (P) and 17β-estrodiol (E2) in ovariectomized mouse uterus determined by realtime PCR. A. S1pr1; B. S1pr2; C. S1pr3; D. S1pr4; E. S1pr5; F. Leukaemia inhibitory factor (Lif), as a control for regulation of LP receptors by E2, arbitrary scale. * P<0.05, downregulation; ** P<0.01, downregulation; # P<0.05, upregulation; ## P<0.01, upregulation, compared to control “oil”. N=4–6. Error bars represent standard deviation. “1h”, “6h”, and “24h” indicate hours post E2 injection.
Supplementary Figures S3. Positive control for in situ hybridization. A. Lpar3 antisense probe, gestation day 3.5 wild type uterus. B. Enlarged view of the rectangle area in A. The section was counterstained with 1% methyl green. Specific signal (dark brown) is detected in the uterine luminal epithelium (LE). Scale bar: 100 μm.
Acknowledgments
We thank Ms. Grace Kennedy for technical assistance and Ms. Danielle Letourneau at The Scripps Research Institute for editorial assistance. We also thank Dr. James N. Moore and Dr. Zhen Fu at the College of Veterinary Medicine, University of Georgia for access to the ABI 7900 Realtime PCR machine and the imaging system, respectively. This work was supported by funding from the Office of the Vice President for Research at the University of Georgia and NIH R15HD066301 (XY) and NIH R01HD050685 (JC).
Funding: Office of the Vice President for Research at University of Georgia and National Institute of Health R15HD066301 (to X.Y.) and R01HD050685 (to J.C.).
Footnotes
All authors, X.Y., D.R.H, H.D., R.R., and J.C., have nothing to disclose.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Pieringer RA, Bonner H, Jr, Kunnes RS. Biosynthesis of phosphatidic acid, lysophosphatidic acid, diglyceride, and triglyceride by fatty acyltransferase pathways in Escherichia coli. J Biol Chem. 1967;242:2719–24. [PubMed] [Google Scholar]
- 2.Ishii I, Fukushima N, Ye X, Chun J. LYSOPHOSPHOLIPID RECEPTORS: Signaling and Biology. Annu Rev Biochem. 2004;73:321–54. doi: 10.1146/annurev.biochem.73.011303.073731. [DOI] [PubMed] [Google Scholar]
- 3.Hecht JH, Weiner JA, Post SR, Chun J. Ventricular zone gene-1 (vzg-1) encodes a lysophosphatidic acid receptor expressed in neurogenic regions of the developing cerebral cortex. J Cell Biol. 1996;135:1071–83. doi: 10.1083/jcb.135.4.1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mutoh T, Chun J. Lysophospholipid activation of G protein-coupled receptors. Subcell Biochem. 2008;49:269–97. doi: 10.1007/978-1-4020-8831-5_10. [DOI] [PubMed] [Google Scholar]
- 5.Choi JW, Herr DR, Noguchi K, Yung YC, Lee CW, Mutoh T, et al. LPA receptors: subtypes and biological actions. Annu Rev Pharmacol Toxicol. 2010;50:157–86. doi: 10.1146/annurev.pharmtox.010909.105753. [DOI] [PubMed] [Google Scholar]
- 6.Pasternack SM, von Kugelgen I, Aboud KA, Lee YA, Ruschendorf F, Voss K, et al. G protein-coupled receptor P2Y5 and its ligand LPA are involved in maintenance of human hair growth. Nat Genet. 2008;40:329–34. doi: 10.1038/ng.84. [DOI] [PubMed] [Google Scholar]
- 7.Yanagida K, Masago K, Nakanishi H, Kihara Y, Hamano F, Tajima Y, et al. Identification and characterization of a novel lysophosphatidic acid receptor, p2y5/LPA6. J Biol Chem. 2009;284:17731–41. doi: 10.1074/jbc.M808506200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tabata K, Baba K, Shiraishi A, Ito M, Fujita N. The orphan GPCR GPR87 was deorphanized and shown to be a lysophosphatidic acid receptor. Biochem Biophys Res Commun. 2007;363:861–6. doi: 10.1016/j.bbrc.2007.09.063. [DOI] [PubMed] [Google Scholar]
- 9.Murakami M, Shiraishi A, Tabata K, Fujita N. Identification of the orphan GPCR, P2Y(10) receptor as the sphingosine-1-phosphate and lysophosphatidic acid receptor. Biochem Biophys Res Commun. 2008;371:707–12. doi: 10.1016/j.bbrc.2008.04.145. [DOI] [PubMed] [Google Scholar]
- 10.Contos JJ, Fukushima N, Weiner JA, Kaushal D, Chun J. Requirement for the lpA1 lysophosphatidic acid receptor gene in normal suckling behavior. Proc Natl Acad Sci U S A. 2000;97:13384–9. doi: 10.1073/pnas.97.24.13384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kingsbury MA, Rehen SK, Contos JJ, Higgins CM, Chun J. Non-proliferative effects of lysophosphatidic acid enhance cortical growth and folding. Nat Neurosci. 2003;6:1292–9. doi: 10.1038/nn1157. [DOI] [PubMed] [Google Scholar]
- 12.Inoue M, Rashid MH, Fujita R, Contos JJ, Chun J, Ueda H. Initiation of neuropathic pain requires lysophosphatidic acid receptor signaling. Nat Med. 2004;10:712–8. doi: 10.1038/nm1060. [DOI] [PubMed] [Google Scholar]
- 13.Liu Y, Wada R, Yamashita T, Mi Y, Deng CX, Hobson JP, et al. Edg-1, the G protein-coupled receptor for sphingosine-1-phosphate, is essential for vascular maturation. J Clin Invest. 2000;106:951–61. doi: 10.1172/JCI10905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Herr DR, Grillet N, Schwander M, Rivera R, Muller U, Chun J. Sphingosine 1-phosphate (S1P) signaling is required for maintenance of hair cells mainly via activation of S1P2. J Neurosci. 2007;27:1474–8. doi: 10.1523/JNEUROSCI.4245-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.MacLennan AJ, Benner SJ, Andringa A, Chaves AH, Rosing JL, Vesey R, et al. The S1P2 sphingosine 1-phosphate receptor is essential for auditory and vestibular function. Hear Res. 2006;220:38–48. doi: 10.1016/j.heares.2006.06.016. [DOI] [PubMed] [Google Scholar]
- 16.Golfier S, Kondo S, Schulze T, Takeuchi T, Vassileva G, Achtman AH, et al. Shaping of terminal megakaryocyte differentiation and proplatelet development by sphingosine-1-phosphate receptor S1P4. FASEB J. 2010;24:4701–10. doi: 10.1096/fj.09-141473. [DOI] [PubMed] [Google Scholar]
- 17.Ye X, Skinner MK, Kennedy G, Chun J. Age-dependent loss of sperm production in mice via impaired lysophosphatidic acid signaling. Biol Reprod. 2008;79:328–36. doi: 10.1095/biolreprod.108.068783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ye X, Hama K, Contos JJ, Anliker B, Inoue A, Skinner MK, et al. LPA3-mediated lysophosphatidic acid signalling in embryo implantation and spacing. Nature. 2005;435:104–8. doi: 10.1038/nature03505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hama K, Aoki J, Inoue A, Endo T, Amano T, Motoki R, et al. Embryo Spacing and Implantation Timing Are Differentially Regulated by LPA3-Mediated Lysophosphatidic Acid Signaling in Mice. Biol Reprod. 2007;77:954–9. doi: 10.1095/biolreprod.107.060293. [DOI] [PubMed] [Google Scholar]
- 20.Ye X. Lysophospholipid signaling in the function and pathology of the reproductive system. Hum Reprod Update. 2008;14:519–36. doi: 10.1093/humupd/dmn023. [DOI] [PubMed] [Google Scholar]
- 21.Ye X, Chun J. Lysophosphatidic acid (LPA) signaling in vertebrate reproduction. Trends Endocrinol Metab. 2010;21:17–24. doi: 10.1016/j.tem.2009.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Carson DD, Bagchi I, Dey SK, Enders AC, Fazleabas AT, Lessey BA, et al. Embryo implantation. Dev Biol. 2000;223:217–37. doi: 10.1006/dbio.2000.9767. [DOI] [PubMed] [Google Scholar]
- 23.Dey SK, Lim H, Das SK, Reese J, Paria BC, Daikoku T, et al. Molecular cues to implantation. Endocr Rev. 2004;25:341–73. doi: 10.1210/er.2003-0020. [DOI] [PubMed] [Google Scholar]
- 24.Sharkey AM, Smith SK. The endometrium as a cause of implantation failure. Best Pract Res Clin Obstet Gynaecol. 2003;17:289–307. doi: 10.1016/s1521-6934(02)00130-x. [DOI] [PubMed] [Google Scholar]
- 25.Paria BC, Reese J, Das SK, Dey SK. Deciphering the cross-talk of implantation: advances and challenges. Science. 2002;296:2185–8. doi: 10.1126/science.1071601. [DOI] [PubMed] [Google Scholar]
- 26.Contos JJ, Ishii I, Fukushima N, Kingsbury MA, Ye X, Kawamura S, et al. Characterization of lpa(2) (Edg4) and lpa(1)/lpa(2) (Edg2/Edg4) lysophosphatidic acid receptor knockout mice: signaling deficits without obvious phenotypic abnormality attributable to lpa(2) Mol Cell Biol. 2002;22:6921–9. doi: 10.1128/MCB.22.19.6921-6929.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ishii I, Ye X, Friedman B, Kawamura S, Contos JJ, Kingsbury MA, et al. Marked perinatal lethality and cellular signaling deficits in mice null for the two sphingosine 1-phosphate (S1P) receptors, S1P(2)/LP(B2)/EDG-5 and S1P(3)/LP(B3)/EDG-3. J Biol Chem. 2002;277:25152–9. doi: 10.1074/jbc.M200137200. [DOI] [PubMed] [Google Scholar]
- 28.Jaillard C, Harrison S, Stankoff B, Aigrot MS, Calver AR, Duddy G, et al. Edg8/S1P5: an oligodendroglial receptor with dual function on process retraction and cell survival. J Neurosci. 2005;25:1459–69. doi: 10.1523/JNEUROSCI.4645-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lee Z, Cheng CT, Zhang H, Subler MA, Wu J, Mukherjee A, et al. Role of LPA4/p2y9/GPR23 in negative regulation of cell motility. Mol Biol Cell. 2008;19:5435–45. doi: 10.1091/mbc.E08-03-0316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Contos JJ, Chun J. The mouse lp(A3)/Edg7 lysophosphatidic acid receptor gene: genomic structure, chromosomal localization, and expression pattern. Gene. 2001;267:243–53. doi: 10.1016/s0378-1119(01)00410-3. [DOI] [PubMed] [Google Scholar]
- 31.Ishii I, Friedman B, Ye X, Kawamura S, McGiffert C, Contos JJ, et al. Selective loss of sphingosine 1-phosphate signaling with no obvious phenotypic abnormality in mice lacking its G protein-coupled receptor, LP(B3)/EDG-3. J Biol Chem. 2001;276:33697–704. doi: 10.1074/jbc.M104441200. [DOI] [PubMed] [Google Scholar]
- 32.Sidhu SS, Kimber SJ. Hormonal control of H-type alpha(1–2)fucosyltransferase messenger ribonucleic acid in the mouse uterus. Biol Reprod. 1999;60:147–57. doi: 10.1095/biolreprod60.1.147. [DOI] [PubMed] [Google Scholar]
- 33.Campbell EA, O'Hara L, Catalano RD, Sharkey AM, Freeman TC, Johnson MH. Temporal expression profiling of the uterine luminal epithelium of the pseudo-pregnant mouse suggests receptivity to the fertilized egg is associated with complex transcriptional changes. Hum Reprod. 2006;21:2495–513. doi: 10.1093/humrep/del195. [DOI] [PubMed] [Google Scholar]
- 34.Brown N, Deb K, Paria BC, Das SK, Reese J. Embryo-uterine interactions via the neuregulin family of growth factors during implantation in the mouse. Biol Reprod. 2004;71:2003–11. doi: 10.1095/biolreprod.104.031864. [DOI] [PubMed] [Google Scholar]
- 35.Diao H, Aplin JD, Xiao S, Chun J, Li Z, Chen S, et al. Altered Spatiotemporal Expression of Collagen Types I, III, IV, and VI in Lpar3-Deficient Peri-Implantation Mouse Uterus. Biol Reprod. 2011;84:255–65. doi: 10.1095/biolreprod.110.086942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Diao H, Xiao S, Zhao F, Ye X. Uterine luminal epithelium-specific proline-rich acidic protein 1 (PRAP1) as a marker for successful embryo implantation. Fertil Steril. 2010;94:2808–11. e1. doi: 10.1016/j.fertnstert.2010.06.034. [DOI] [PubMed] [Google Scholar]
- 37.Diao H, Xiao S, Cui J, Chun J, Xu Y, Ye X. Progesterone receptor-mediated up-regulation of transthyretin in preimplantation mouse uterus. Fertil Steril. 2010;93:2750–3. doi: 10.1016/j.fertnstert.2010.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yang ZM, Das SK, Wang J, Sugimoto Y, Ichikawa A, Dey SK. Potential sites of prostaglandin actions in the periimplantation mouse uterus: differential expression and regulation of prostaglandin receptor genes. Biol Reprod. 1997;56:368–79. doi: 10.1095/biolreprod56.2.368. [DOI] [PubMed] [Google Scholar]
- 39.Wang H, Dey SK. Roadmap to embryo implantation: clues from mouse models. Nat Rev Genet. 2006;7:185–99. doi: 10.1038/nrg1808. [DOI] [PubMed] [Google Scholar]
- 40.Hama K, Aoki J, Bandoh K, Inoue A, Endo T, Amano T, et al. Lysophosphatidic receptor, LPA3, is positively and negatively regulated by progesterone and estrogen in the mouse uterus. Life Sci. 2006;79:1736–40. doi: 10.1016/j.lfs.2006.06.002. [DOI] [PubMed] [Google Scholar]
- 41.Yang ZM, Chen DB, Le SP, Harper MJ. Differential hormonal regulation of leukemia inhibitory factor (LIF) in rabbit and mouse uterus. Mol Reprod Dev. 1996;43:470–6. doi: 10.1002/(SICI)1098-2795(199604)43:4<470::AID-MRD9>3.0.CO;2-M. [DOI] [PubMed] [Google Scholar]
- 42.Niklaus AL, Pollard JW. Mining the mouse transcriptome of receptive endometrium reveals distinct molecular signatures for the luminal and glandular epithelium. Endocrinology. 2006;147:3375–90. doi: 10.1210/en.2005-1665. [DOI] [PubMed] [Google Scholar]
- 43.Aplin JD. Embryo implantation: the molecular mechanism remains elusive. Reprod Biomed Online. 2006;13:833–9. doi: 10.1016/s1472-6483(10)61032-2. [DOI] [PubMed] [Google Scholar]
- 44.Murphy CR. Understanding the apical surface markers of uterine receptivity: pinopodsor uterodomes? Hum Reprod. 2000;15:2451–4. doi: 10.1093/humrep/15.12.2451. [DOI] [PubMed] [Google Scholar]
- 45.Carver J, Martin K, Spyropoulou I, Barlow D, Sargent I, Mardon H. An in-vitro model for stromal invasion during implantation of the human blastocyst. Hum Reprod. 2003;18:283–90. doi: 10.1093/humrep/deg072. [DOI] [PubMed] [Google Scholar]
- 46.Bazer FW, Slayden OD. Progesterone-induced gene expression in uterine epithelia: a myth perpetuated by conventional wisdom. Biol Reprod. 2008;79:1008–9. doi: 10.1095/biolreprod.108.072702. [DOI] [PubMed] [Google Scholar]
- 47.Mulac-Jericevic B, Conneely OM. Reproductive tissue selective actions of progesterone receptors. Reproduction. 2004;128:139–46. doi: 10.1530/rep.1.00189. [DOI] [PubMed] [Google Scholar]
- 48.Lydon JP, DeMayo FJ, Funk CR, Mani SK, Hughes AR, Montgomery CA, Jr, et al. Mice lacking progesterone receptor exhibit pleiotropic reproductive abnormalities. Genes Dev. 1995;9:2266–78. doi: 10.1101/gad.9.18.2266. [DOI] [PubMed] [Google Scholar]
- 49.Takamoto N, Zhao B, Tsai SY, DeMayo FJ. Identification of Indian hedgehog as a progesterone-responsive gene in the murine uterus. Mol Endocrinol. 2002;16:2338–48. doi: 10.1210/me.2001-0154. [DOI] [PubMed] [Google Scholar]
- 50.Jeong JW, Lee KY, Kwak I, White LD, Hilsenbeck SG, Lydon JP, et al. Identification of murine uterine genes regulated in a ligand-dependent manner by the progesterone receptor. Endocrinology. 2005;146:3490–505. doi: 10.1210/en.2005-0016. [DOI] [PubMed] [Google Scholar]
- 51.Lee K, Jeong J, Kwak I, Yu CT, Lanske B, Soegiarto DW, et al. Indian hedgehog is a major mediator of progesterone signaling in the mouse uterus. Nat Genet. 2006;38:1204–9. doi: 10.1038/ng1874. [DOI] [PubMed] [Google Scholar]
- 52.Palomino WA, Fuentes A, Gonzalez RR, Gabler F, Boric MA, Vega M, et al. Differential expression of endometrial integrins and progesterone receptor during the window of implantation in normo-ovulatory women treated with clomiphene citrate. Fertil Steril. 2005;83:587–93. doi: 10.1016/j.fertnstert.2004.11.020. [DOI] [PubMed] [Google Scholar]
- 53.Lessey BA, Killam AP, Metzger DA, Haney AF, Greene GL, McCarty KS., Jr Immunohistochemical analysis of human uterine estrogen and progesterone receptors throughout the menstrual cycle. J Clin Endocrinol Metab. 1988;67:334–40. doi: 10.1210/jcem-67-2-334. [DOI] [PubMed] [Google Scholar]
- 54.Wakitani S, Hondo E, Phichitraslip T, Stewart CL, Kiso Y. Upregulation of Indian hedgehog gene in the uterine epithelium by leukemia inhibitory factor during mouse implantation. J Reprod Dev. 2008;54:113–6. doi: 10.1262/jrd.19120. [DOI] [PubMed] [Google Scholar]
- 55.Bazer FW, Wu G, Spencer TE, Johnson GA, Burghardt RC, Bayless K. Novel pathways for implantation and establishment and maintenance of pregnancy in mammals. Mol Hum Reprod. 2010;16:135–52. doi: 10.1093/molehr/gap095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Diao H, Paria BC, Xiao S, Ye X. Temporal expression pattern of progesterone receptor in the uterine luminal epithelium suggests its requirement during early events of implantation. Fertil Steril. 2011 doi: 10.1016/j.fertnstert.2011.01.160. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.McCormack JT, Greenwald GS. Evidence for a preimplantation rise in oestradiol-17beta levels on day 4 of pregnancy in the mouse. J Reprod Fertil. 1974;41:297–301. doi: 10.1530/jrf.0.0410297. [DOI] [PubMed] [Google Scholar]
- 58.Skaznik-Wikiel ME, Kaneko-Tarui T, Kashiwagi A, Pru JK. Sphingosine-1-phosphate receptor expression and signaling correlate with uterine prostaglandin-endoperoxide synthase 2 expression and angiogenesis during early pregnancy. Biol Reprod. 2006;74:569–76. doi: 10.1095/biolreprod.105.046714. [DOI] [PubMed] [Google Scholar]
- 59.Burney RO, Talbi S, Hamilton AE, Vo KC, Nyegaard M, Nezhat CR, et al. Gene expression analysis of endometrium reveals progesterone resistance and candidate susceptibility genes in women with endometriosis. Endocrinology. 2007;148:3814–26. doi: 10.1210/en.2006-1692. [DOI] [PubMed] [Google Scholar]
- 60.Wei Q, St Clair JB, Fu T, Stratton P, Nieman LK. Reduced expression of biomarkers associated with the implantation window in women with endometriosis. Fertil Steril. 2009;91:1686–91. doi: 10.1016/j.fertnstert.2008.02.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figures S1. Regulation of LPA receptors by progesterone (P) and 17β-estrodiol (E2) in ovariectomized mouse uterus determined by realtime PCR. A. Lpar1; B. Lpar2; C. Lpar3; D. Lpar4; E. Lpar5. * P<0.05, downregulation; ** P<0.01, downregulation; # P<0.05, upregulation; ## P<0.01, upregulation, compared to control “oil”. N=4–6. Error bars represent standard deviation. “1h”, “6h”, and “24h” indicate hours post E2 injection.
Supplementary Figures S2. Regulation of S1P receptors by progesterone (P) and 17β-estrodiol (E2) in ovariectomized mouse uterus determined by realtime PCR. A. S1pr1; B. S1pr2; C. S1pr3; D. S1pr4; E. S1pr5; F. Leukaemia inhibitory factor (Lif), as a control for regulation of LP receptors by E2, arbitrary scale. * P<0.05, downregulation; ** P<0.01, downregulation; # P<0.05, upregulation; ## P<0.01, upregulation, compared to control “oil”. N=4–6. Error bars represent standard deviation. “1h”, “6h”, and “24h” indicate hours post E2 injection.
Supplementary Figures S3. Positive control for in situ hybridization. A. Lpar3 antisense probe, gestation day 3.5 wild type uterus. B. Enlarged view of the rectangle area in A. The section was counterstained with 1% methyl green. Specific signal (dark brown) is detected in the uterine luminal epithelium (LE). Scale bar: 100 μm.