Abstract
Amonafide is a DNA intercalator in clinical development for the treatment of cancer. The drug has a 5-position amine that is variably acetylated to form a toxic metabolite in humans, increasing adverse effects and complicating the dosing of amonafide. Numonafides, 6-amino derivatives of amonafide that avoid the toxic acetylation, also show in vitro anticancer activity, as we have previously described. Here, we report the in vitro and in vivo activities of two numonafides, 6-methoxyethylamino-numonafide (MEAN) and 6-amino-numonafide (AN) with comparisons to amonafide. The in vitro potencies and cellular anticancer mechanisms are similar for the two numonafides and amonafide. Results from several mouse models of human cancer demonstrate that AN and MEAN require slightly higher doses than amonafide for equal efficacy in short-term dosing models, but the same dose of all three compounds in long-term dosing models are equally efficacious. MEAN is tolerated much better than amonafide and AN at equally efficacious doses based on weight change, activity, stool consistency, and dose tolerance with survival as the end point. The studies presented here demonstrate that MEAN is much less toxic than amonafide or AN in mouse models of human liver and gastric cancers while being equally efficacious in vivo and inhibiting cancer cells through similar mechanisms. These findings demonstrate that numonafides can be less toxic than amonafide and support further preclinical development and novel anticancer agents or as replacements or amonafide.
Introduction
Naphthalamides are a class of anticancer compounds that have been the focus of considerable development during the last 25 years [1,2]. One naphthalamide drug, amonafide (AMN), which is a DNA intercalator and a possible inhibitor of topoisomerase II [1,2], has proceeded to clinical development for the treatment of neoplastic diseases. AMN has shown good activity against advanced breast cancer [3] and as a second-line therapy for AML [4]; however, the 5-position amine of AMN is acetylated in humans by N-acetyl-transferase 2 (NAT2), converting the parental molecule to a toxic 5-amino-acetyl metabolite [1,2]. Polymorphisms in the NAT2 gene cause varying enzymatic activity of NAT2 among individuals, which thereby requires phenotyping of acetylation status or genotyping of NAT2 in each patient before AMN treatment [1,2], a process that is costly and delays treatment initiation.
We have previously described the synthesis of 6-position amino derivatives of AMN called numonafides (Figure 1A) [5]. The numonafide with a free amine in the 6-position and one with a substituted amine in the 6-position are not acetylated, whereas the parental compound, AMN, is extensively acetylated as determined by an NAT2 biochemical assay [5]. Our previous characterization of numonafides showed that 6-amino-numonafide (AN) and 6-methoxyethylamino-numonafide (MEAN) (Figure 1A) had the best antitumor properties in vitro [5]. In this report, we have further characterized the in vitro mechanisms, in vivo antitumor efficacy, and in vivo toxicities of AN and MEAN in murine tumor models of human cancer using AMN as a comparative control throughout.
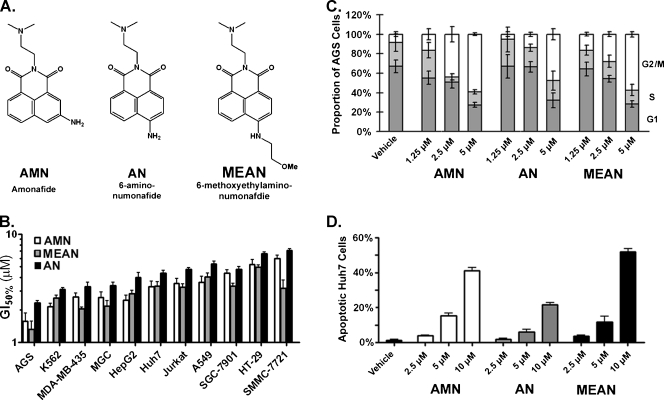
Figure 1.
Numonafides behave similar to AMN in vitro. (A) Structures of AMN and the two numonafides examined herein. (B) Concentrations of AMN, MEAN, and AN that cause 50% growth inhibition in human gastric carcinoma (AGS, MGC, and SGC-7901), leukemia (K562 and Jurkat), breast carcinoma (MDA-MB-435), hepatoma (HepG2, Huh7, and SMMC-7721), lung carcinoma (A549), and colon carcinoma (HT-29) cell lines as determined by the MTT assay. (C) Cell cycle analysis of AGS gastric cancer cells after 24 hours of treatment with AMN, AN, or MEAN as determined by flow cytometry for DNA content (bottom dark gray bars = cells in G1 phase, middle light gray bars = S phase, and white bars = G2/M phases). (D) Results from flow cytometry analysis of apoptosis in Huh7 hepatoma cells after 24 hours of treatment with AMN or numonafides. All error bars = SD from three or more independent experiments and vehicle = 1% DMSO.
Materials and Methods
Cell Culture and In Vitro Assays
All the cells were cultured in Dulbecco modified Eagle medium (Gibco, Carlsbad, CA) supplemented with 10% fetal bovine serum (Gibco).
MTT Assay
Briefly, cells were seeded into 96-well plates and treated with AMN, AN, or MEAN for 72 hours. The medium was removed, and the cells were incubated with a solution containing 0.5 mg MTT/ml phosphate-buffered saline at 37°C for 4 hours. The MTT solution was removed, and the cells were lysed with 100 µl/well dimethylsulfoxide (DMSO) for 15 minutes at 37°C. The optical density was measured using a Bio-Rad microplate reader at 570 nm with DMSO as blank. Triplicate wells were assayed for each condition. Data were analyzed by GraphPad Prism 5 software (GraphPad, La Jolla, CA) to get the 50% inhibitory concentration (IC50). For DNA content assays, 1 x 106 treated cells were collected, stained using Coulter DNA Prep Reagents Kit (Beckman Coulter) according to the manufacturer's protocol, and then analyzed by FACS (Beckman Coulter FC500 MPL). Apoptosis assays were performed on 2 x 105 treated cells stained with using annexin V-fluorescein isothiocyanate Kit (BD Biosciences, San Jose, CA), and then FACS was performed.
Gene Expression Array
RNA was isolated from 106 HepG2 cells with QIAGENs RNeasy Mini Kit after an overnight treatment with 2 µM of AMN, AN, MEAN, or vehicle (0.2% DMSO). RNA expression analysis was performed Illumina Human HT-12 Expression Beadchips, which provides coverage of 48,802 genes and expressed sequence tags. Raw signal intensities of each probe were obtained using data analysis software (Beadstudio; Illumina, San Diego, CA) and imported to the Lumi package of Bioconductor for data analysis. Before transformation and normalization [6–8], A/P call detection was performed based on detection of P value. Of 48,802 probes with less than 0.01, 18,678 were considered as valid signals. For each pair of five comparisons (AMN and vehicle, AN and vehicle, MEAN and vehicle, MEAN and AN, and MEAN and AMN), differentially expressed genes were identified using an analysis of variance (ANOVA) model with empirical Bayesian variance estimation [9]. Initially, genes were identified as being differentially expressed on the basis of a statistical significance (raw, P < .01; false discovery rate-adjusted, P < .05) and 1.5-fold change (up or down) in expression level in each comparison.
In Vivo Xenograft Models
Huh7-luc cell line
pGL3-control (Promega, Madison, WI) was first digested with XbaI (Takara, Shiga, Japan) and then blunted with DNA polymerase Klenow fragment (Takara). The resulting DNA was then digested with BglII (Takara) and the DNA fragment (1902 bp) encoding luciferase (luc). This fragment was then ligated to the BamHI/SmaI (Takara) digested backbone of pWPXL (Addgene, Cambridge, MA) to create pWPXL-luc.Next, 2.5 x 106 of HEK-293T cells were plated in a 10-cm diameter plate. The following day, 20 µg of pWPXL-Luc, 15 µg of psPAX2 (Addgene), and 6 µg of pMD2.G (Addgene) were diluted in 1 ml Hank's buffered saline with 50 µl of 2.5 M CaCl2 and mixed gently. After 20 minutes of incubation at room temperature, the plasmid solution was added to the HEK-293T (Invitrogen) medium, and after 6 hours, the medium was replaced with medium containing no plasmids. Four days later, the medium was collected, and the lentivirus was purified with 0.45-µm filters. Then, Huh7 cells were infected with pWPXL-luc lentivirus virus at a multiplicity if infection of 1000:1 in the presence of polybrene. After 3 days of infection, single cells were plated into the wells of a 96-well plate and allowed to grow for 3 weeks, at which point the highest expressing clone was expanded and used for the studies described here.
AGS-luc cell line
Lentiviral vectors expressing firefly luciferase (luc) were generated using a four-plasmid system. Briefly, a lentiviral expression construct encoding luciferase and green fluorescent protein, each under the control of an individual CAG-enhanced CMV promoter (pLenti CMV Puro LUC; Addgene), was cotransfected with lentiviral packaging plasmids (pMD2.G, psPAX2) and a VSV-G envelope expressing plasmid (CVG) into HEK-293T cells using Lipofectamine 2000 (Invitrogen, Carlsbad, CA). The medium was collected every 24 hours and replaced with fresh media for 3 days. Virus-containing medium was filtered with 0.45-µm filters, and then the viral particles were concentrated with sucrose ultracentrifugation. The viral pellet was resuspended in the medium with polybrene and added to AGS cells for 12 hours. After infection, the virus-containing medium was replaced with fresh medium for 24 hours. Cells expressing high levels of green fluorescent protein were isolated by fluorescence-activated cell sorting, and the pooled population was expanded to create the AGS-luc cell line.
Mice and xenografts
Male nu/nu (nude) mice (18–20 g at experiment initiation) were maintained at the vivarium of First Affiliated Hospital, College of Medicine, in a pathogen-free unit, under a 12-hour light/dark cycle, and were provided with food and water ad libitum. Mice were inoculated subcutaneously at the right axilla or the peritoneal cavity with HepG2 (106 cells), AGS-luc (106 cells), or Huh7-luc cells (106 cells for subcutaneous inoculation and 2 x 106 cells for intraperitoneal inoculation). For the experiments using AGS-luc and Huh7-luc cells, in vivo bioluminescent imaging was performed with a Lumina imaging system (Nippon Roper, I.C.E., Tokyo, Japan). Fifteen minutes before imaging, mice were injected with 150-mg/kg luciferin through an intraperitoneal route. Images were collected and analyzed with Living Image software (Slidebook 4.1, Denver, CO). Vehicle control was 20% DMSO.
Results
AMN, AN, and MEAN Share Similar In Vitro Growth Inhibition and Apoptotic Properties
Our previous studies showed that the numonafides AN and MEAN (Figure 1A) inhibit the growth of three cancer cell lines with potencies similar to AMN and demonstrated similar selectivity for growth inhibition of cancer cells over normal cells [5]. Here, we systematically investigated the growth inhibition of numonafides and AMN in 11 cell lines derived from various cancers. The results show that the numonafides, AN and MEAN, inhibit cancer cell growth with a similar potency as AMN, although AN tends to be slightly less potent (Figure 1B). Because AN and MEAN are potent inhibitors of gastric and liver cancer cell lines and because these cancers are prevalent malignancies with relatively few pharmacologically viable treatment options, here we evaluate their antitumor properties using AGS (gastric cancer), Huh7, and HepG2 (hepatomas) cell lines.
We evaluated the in vitro effect of these compounds on cell proliferation and apoptosis. First, AGS cells were treated with varying doses of AMN, AN, and MEAN and stained to determine the DNA content. AMN, AN, and MEAN all cause AGS cells to increase their DNA content in a dose-dependent manner, and all compounds significantly (P < .05 by one-way ANOVA with Dunnett test) increase DNA content at 5 µM, indicating that these compounds cause G2 arrest (Figure 1C), which is likely the result of DNA damage through intercalation or topoisomerase II inhibition by these compounds [10]. Next, Huh7 cells were treated with the numonafides and AMN for 24 hours then stained to determine the apoptosis index. The results show that AMN, AN, and MEAN all cause significant (P < .05 by one-way ANOVA with Dunnett test) increases in apoptosis at 5 and 10 µM with AMN and MEAN being significantly (P < .05 by unpaired t-test with Welch correction) more potent than AN at both doses (Figure 1D).
AMN, AN, and MEAN Similarly Influence Gene Expression Pattern in Cancer Cells
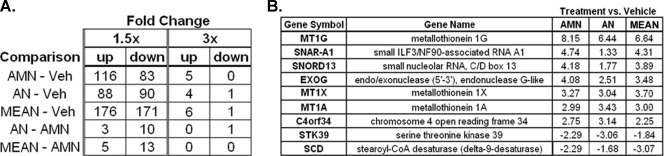
Gene array analyses on cancer cells treated with numonafides, MEAN, and AMN were performed using to identify and compare the molecular mechanism and cellular pathways that are affected by the treatment of these compounds. HepG2 cells were treated with AMN, MEAN, or AN at 2 µM (a pharmacologically achievable concentration) overnight, and the changes in the level of approximately 25,000 transcripts were determined with the gene array. MEAN, AMN, and AN significantly (P < .05 by t-test) changed the level of 347, 199, and 178 transcripts, respectively, by greater than 1.5-fold (Figure 2A). The number of transcripts changed is positively correlated with the in vitro DNA intercalation potencies of these compounds [5], suggesting that the change in gene expression is due to the differential efficiency of DNA intercalation by each compound in the cellular milieu. There is a lack of differences in gene expression when each treatment group is compared to one another instead of vehicle treatment (Figure 2A), indicating that all three compounds change the expression of similar genes and are thereby acting through similar mechanisms. Supporting this theory is the finding that transcripts changed greater than three-fold are all similarly altered in the three treatment groups compared to the vehicle group, with a few exceptions where AN does not modulate the transcript level to the extent of AMN and MEAN (Figure 2B), likely due to the lower DNA intercalation efficiency or cellular potency of AN [5].
Figure 2.
Amonafide and numonafides alter gene expression with similar patterns. (A) Number of transcripts that are significantly (P < .05 by t-test, n = 3) changed in HepG2 cells determined by Illumina's BeadArray after an overnight treatment with 2 µM of each compound compared to vehicle-treated cells and compared to AMN-treated cells as. (B) Change in levels of transcripts that are significantly upregulated or downregulated by greater than three-fold in cells treated with AMN or numonafides compared to vehicle (average values from of three independent experiments and P < .05 for fold changes more than three-fold).
Numonafides Are Efficacious in a Hepatoma Xenograft Model, but MEAN Is Better Tolerated than AMN and AN
The in vivo tolerance and anticancer properties of AMN, AN, or MEAN were initially tested in a xenograft model, in which nude immunocom promised mice were implanted with the human HepG2 hepatoma cells subcutaneously under the front right axilla (armpit). Mice were treated with vehicle, 50 µmol/kg, or 100 µmol/kg of each compound or 200 µmol/kg of MEAN (this dose of AMN or AN rapidly killed mice). The compounds were administered through the intraperitoneal route once per day for 14 days, 2 weeks after the implantation of the xenografts. After treatment, the mice were sacrificed, and the tumors were resected and weighed. AMN inhibited tumor growth most potently at the 50- and 100-µmol/kg doses (Table 1). MEAN was less efficacious than AMN at 50 and 100 µmol/kg; however, the 200-µmol/kg dose of MEAN was equally efficacious as the 100-µmol/kg dose of AMN. This initial end point tumor measurement in this study suggested that MEAN and AN are less potent than AMN, but based on the lack of mice that died in the MEAN-treated groups, up to 200-µmol/kg MEAN is tolerated much better than AMN and AN and can be equally efficacious due to its lower toxicity.
Table 1.
Numonafides Are Efficacious in the HepG2 Human Xenograft Model.
| Treatment | Dosage (µmol/kg) once a day x 14 d | Final Number of Mice (/10) | Tumor Weight (g) | % Growth Inhibition | P, t-Test |
| Vehicle | 0 | 10 | 2.185 ± 0.242 | — | — |
| AN | 100 | 8 | 1.455 ± 0.288 | 33.4 | <.01 |
| AN | 50 | 10 | 1.969 ± 0.274 | 9.9 | <.05 |
| MEAN | 200 | 10 | 0.427 ± 0.212 | 80.5 | <.01 |
| MEAN | 100 | 10 | 0.869 ± 0.301 | 60.2 | <.01 |
| MEAN | 50 | 10 | 1.889 ± 0.181 | 13.5 | <.01 |
| AMN | 100 | 9 | 0.509 ± 0.199 | 76.7 | <.01 |
| AMN | 50 | 10 | 1.141 ± 0.216 | 47.8 | <.01 |
Nude mice were implanted with HepG2 cells in the front axilla and treated through intraperitoneal injection once per day for 14 days, 2 weeks after implantation of tumor. After treatment, tumors were resected and weighed (each group started with 10 mice). Statistics (unpaired t-test) are for each group compared to vehicle-treated mice.
Numonafides Can Inhibit Tumor Growth and Reduce the Size of Established Tumors
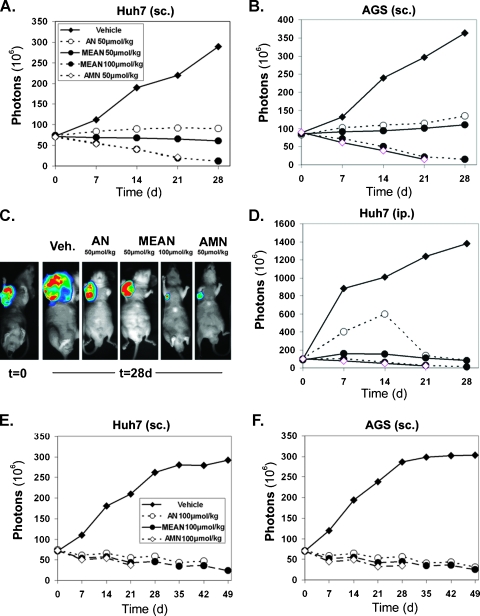
AGS and Huh7 cells expressing luciferase were used to evaluate tumor inhibition properties of numonafides and AMN in a time-dependent manner. In this model, mice were imaged every 7 days to quantify tumor growth through luminescent output of the tumor. First, mice were implanted with the tumors (subcutaneous AGS-luc [Figure 3, B and C] and Huh7-luc [Figure 3A] or intraperitoneal Huh7-luc [Figure 3D]) and were treated continuously for 28 days with 50 µmol/kg per day each compound and 100 µmol/kg MEAN through intraperitoneal administration [11]. The treatment was initiated 2 weeks after implantation of the subcutaneous xenograft and 1 week after the intraperitoneal xenograft. In all three tumor xenograft models, 50 µmol/kg AN is the least effective and 50 µmol/kg MEAN is slightly more effective than AN, both halting tumor growth. AMN at the 50-µmol/kg dose and MEAN at the 100-µmol/kg dose were equally effective (P > .05 by unpaired t-test at day 21), actually causing a significant (P < .05 by unpaired t-test with Welch correction at day 21) decrease in tumor size from treatment start (Figure 3, A–D).
Figure 3.
Numonafides are efficacious in xenograft models of liver and gastric cancer with various dosing schedules. Subcutaneous (sc.) axilla xenografts of (A) Huh7 and (B and C) AGS cells and (D) intraperitoneal xenograft of Huh7 cells expressing luciferase-implanted nude mice. Two weeks after implantation of subcutaneous xenografts and 1 week after implantation of the intraperitoneal (ip.) xenografts, mice were treated with 50 µmol/kg each of AMN, AN, and MEAN and 100 µmol/kg of MEAN once per day for 28 consecutive days. Tumor burden is determined by the total number of photons emitted from the luminescent tumors. Once the number of live mice in a treatment group dropped below three of six, the analysis is not shown on the plots for the subsequent days. For subcutaneous xenografts (A and B), tumor size was inhibited significantly (P < .05 by one-way ANOVA with Dunnett test) in mice treated with compound compared to vehicle-treated mice by day 14. AMN inhibited tumor growth significantly more (P < .05) than 50 µmol/kg AN and MEAN but not more (P > .05) than 100 µmol/kg MEAN (by one-way ANOVA) on day 21. For the intraperitoneal xenograft (D), all treatments significantly inhibited tumor growth compared to vehicle treatment from day 7 to 21 (P < .05 by one-way ANOVA with Dunnett test). (E and F) Mice were implanted with Huh7 or AGS subcutaneous xenografts and treated with 100 µmol/kg AMN, AN, and MEAN (six mice per group) with a cycle of once per day for 7 days and no treatment for 7 days (four cycles). All treatments significantly (P < .05 by one-way ANOVA with Dunnett test) inhibited the growth of the tumor compared to the control after 7 days, but the tumor size among the treatment groups was not significantly different (P > .05 by one-way ANOVA) at day 21.
Owing to the high toxicity observed with AMN and AN and the need to compare equivalent doses over a longer treatment time, a different dosing strategy was used in the same Huh7 and AGS xenograft models. About 100 µmol/kg of each compound was administered through the intraperitoneal route on a 7-day-on/7-day-off schedule for a total of four treatment courses. At 100 µmol/kg, all three drugs inhibit the growth of the tumor significantly (P < .05 by one-way ANOVA with Dunnett test) after day 7 compared to vehicle, but the tumor size in the treatment groups was not significantly different (Figure 3, E and F; P > .05 by one-way ANOVA with Dunnett) from one another at day 21. All three compounds shrank the tumor volume compared to day 0 using this dosing schedule.
MEAN Is Less Toxic In Vivo Compared to AMN and AN
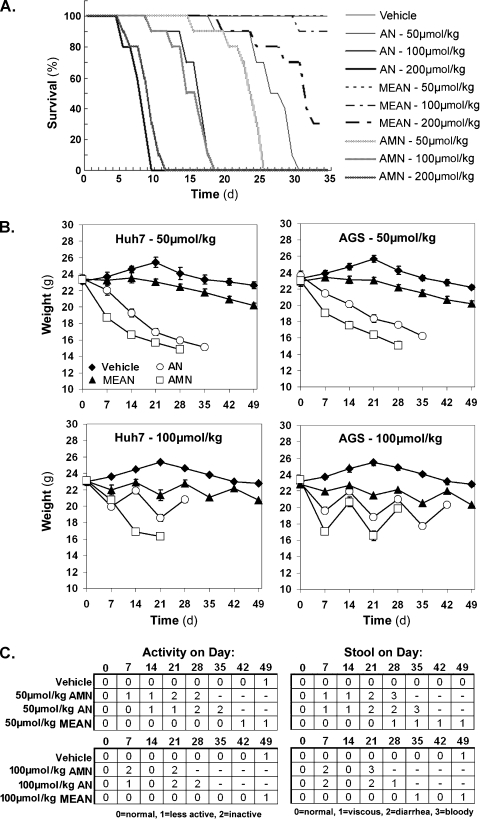
The toxicity of these compounds was examined by survival analysis on mice treated with 50, 100, or 200 µmol/kg of AMN, AN, and MEAN for up to 35 consecutive days. The results show that AMN and AN were similarly toxic, whereas the MEAN was tolerated much better (Figure 4A). Only 1 in 10 mice died by the 35th day at the 100-µmol/kg dose of MEAN compared with the 100-µmol/kg dose of AMN or AN, which killed all mice by day 20 (Figure 4A). In addition, the median survival time for mice treated with 200 µmol/kg MEAN was more than 30 days compared to a median survival of 10 days for mice treated with equivalent doses of AMN and AN (Figure 4A).
Figure 4.
MEAN is much less toxic than AMN or AN. (A) Survival of mice treated with intraperitoneal injection of vehicle or 50, 100, or 200 µmol/kg AMN, AN, or MEAN once a day for 35 continuous days (n = 10 mice per treatment group). (B) Weight of mice implanted with subcutaneous tumors treated with 50 µmol/kg once per day for 49 consecutive days or with 100 µmol/kg once per day on a cycle of 7 days on and 7 days off (n = 6 mice per treatment group). (C) Activity and stool consistency in mice implanted with Huh7 xenografts (same mice represented in the left half of B). Once the number of mice alive in a treatment group dropped below three of six, the analysis is not shown for the subsequent days.
The body weight, activity, and stool consistency were recorded during the treatments described in Figure 3 to further assess the toxicities of these compounds. When dosed at 50 µmol/kg every day for up to 49 days, AMN and AN caused approximately 30% decrease in weight by days 28 and 35, respectively (Figure 4B). In contrast, MEAN caused approximately a 10% decrease in weight at day 49 compared to vehicle-treated mice (Figure 4B). On the other hand, all mice in the AMN and AN groups died by day 49 and no mice died in the MEAN group (not shown). In treatment with 100 µmol/kg on a 7-day-on/7-day-off dosing cycles, the final weight of MEAN-treated mice was only 10% to 15% lower than vehicle-treated mice after four treatment courses (Figure 4B). Given the large size of the AGS and Huh7 tumors (Figure 2D), the differences in final weights between MEAN- and vehicle-treated mice may be partially attributed to the smaller tumors in MEAN-treated mice. Furthermore, a rebound to a healthy weight is observed on removal of MEAN during the 7-day-off portion of the dosing (Figure 4B), and there is a minimal alteration of activity and stool consistency (Figure 4C). In comparison, AN and AMN treatments demonstrated less body weight recovery during the 7-day-off in weight and caused decreased activity and worse stool consistency. These findings demonstrate that MEAN is much less toxic than AMN and AN in nude mice, in terms of weight loss, activity levels, gastrointestinal toxicities, and survival, suggesting that MEAN is a good candidate to be developed as a novel antigastric and hepatic cancer drug or as a replacement for AMN.
Discussion
Here, we demonstrate that two numonafides, AN and MEAN, inhibit tumor cell growth, induce G2 arrest, and apoptosis in vitro with potencies similar to the parental drug, AMN (Figure 1), indicating these three compounds inhibit tumor cell growth through similar cellular mechanisms. In addition, the numonafides alter the transcriptome in cancer cells in a similar pattern to AMN (Figure 2), as has been reported with other derivatives of AMN containing substituted 5-position aryl amines [12], further indicating that this class of drugs act on cancer cells with similar mechanisms, independent of alterations in aryl amine substitution. Although the association between most transcripts altered over three-fold (Figure 2B) by these compounds and cancer is unknown; however, two genes have been associated with cancer. First, metallothionein 1G, which is upregulated greater than six-fold in cells treated by all three compounds, has been described as a tumor suppressor in hepatocellular carcinoma [13] and other carcinomas [14]. This finding suggests that up-regulation of metallothionein 1G could be a potential anticancer mechanism of numonafides and AMN. The second gene, stearoyl-CoA desaturase, downregulated by all three compounds, plays a critical role in fatty acid metabolism that increases cancer cell proliferation and malignant transformation and decreases apoptosis [15]. The down-regulation of this gene by numonafides and AMN may contribute to the growth inhibition and apoptosis induction properties of these compounds. The identification of changes in gene expression patterns by these compounds not only helps confirm the common cellular targets between the numonafides and AMN but also provides potentially new mechanisms for tumor cell inhibition by AMN and numonafides, such as the changes in expression of known and unknown genes and noncoding RNA (Figure 2B) and provides potential clinical biomarkers for response. Future studies will explore the additional mode of action for these compounds in cells that contribute to their antitumor properties in vitro and in vivo.
In three human cancer cell line xenograft models using short-term daily doses, we found that AN and MEAN are slightly less potent in vivo, but MEAN can be equally efficacious as AMN at higher doses (Figure 3, A–D). A long-term periodic dosing regimen showed that all three compounds could be equally efficacious at the same dose, actually shrinking established tumors, in two different xenograft models (Figure 3, E and F). The xenograft models indicate that numonafides are efficacious in vivo and that MEAN is more effective than AN. Numonafides were developed as potentially less toxic derivatives of AMN because they avoid acetylation of the arylamine, which causes toxicities associated with AMN [2]. Mice were injected with 50, 100, or 200 µmol/kg AN, MEAN, and AMN once daily for up to 35 days to initially determine the toxicities of numonafides. AN is about equally toxic as AMN in nude mice, suggesting that the free amine of AN is being metabolized in vivo similar to AMN, but MEAN is much less toxic and better tolerated by mice. MEAN treatment at the dose of 200 µmol/kg kill less mice than the 50-µmol/kg dose of AN and AMN (Figure 4A). Further evaluation as judged by weight, activity, and stool consistency in the two different dosing regimens used for the tumor efficacy studies confirmed that AN and AMN are equally toxic, whereas MEAN is much less toxic than both of these compounds (Figure 4, B and C). The similar in vivo potencies and in vitro mechanisms suggest that these compounds inhibit tumor cell growth by similar mechanisms; however, the large difference in toxicity in vivo between MEAN and AMN/AN may be due to differential pharmacokinetics, biodistribution, metabolism, or a combination thereof. Although this remains to be elucidated, here we have provided proof of principle that numonafides can be developed as less toxic counterparts to AMN and have identified MEAN as the first numonafide for future development as an anticancer drug.
Acknowledgment
The authors thank the Genomic Core facility at Northwestern University for performing the array studies.
Abbreviations
- AMN
amonafide
- AN
6-amino-numonafide
- MEAN
6-methoxyethylamino-numonafide
Footnotes
The authors thank the partial funding from the Robert H. Lurie Comprehensive Cancer Center and grant to S.H. from the National Institutes of Health (R01 GM078555).
References
- 1.Lv M, Xu H. Overview of naphthalimide analogs as anticancer agents. Curr Med Chem. 2009;16(36):4797–4813. doi: 10.2174/092986709789909576. [DOI] [PubMed] [Google Scholar]
- 2.Ingrassia L, Lefranc F, Kiss R, Mijatovic T. Naphthalimides and azonafides as promising anti-cancer agents. Curr Med Chem. 2009;16(10):1192–1213. doi: 10.2174/092986709787846659. [DOI] [PubMed] [Google Scholar]
- 3.Costanza ME, Berry D, Henderson IC, Ratain MJ, Wu K, Shapiro C, Duggan D, Kalra J, Berkowitz I, Lyss AP. Amonafide: an active agent in the treatment of previously untreated advanced breast cancer—a Cancer and Leukemia Group B study (CALGB 8642) Clin Cancer Res. 1995;1(7):699–704. [PubMed] [Google Scholar]
- 4.Kolitz JE, Lundberg AS, Bennett JM, Capizzi RL, Budman DR. Phase I trials of amonafide as monotherapy and in combination with cytarabine in patients with poor-risk acute myeloid leukemia. Leuk Res. 2010;34(4):487–491. doi: 10.1016/j.leukres.2009.07.038. [DOI] [PubMed] [Google Scholar]
- 5.Witschi MA, Luong L, Kawamura A, Ghosh S, Stack MS, Sim E, Avram MJ, Appella DH, Huang S. Synthesis and anticancer activities of 6-amino amonafide derivatives. Anticancer Drugs. 2008;19(1):23–36. doi: 10.1097/CAD.0b013e3282f00e17. [DOI] [PubMed] [Google Scholar]
- 6.Du P, Kibbe WA, Lin SM. lumi: a pipeline for processing Illumina microarray. Bioinformatics. 2008;24(13):1547–1548. doi: 10.1093/bioinformatics/btn224. [DOI] [PubMed] [Google Scholar]
- 7.Lin SM, Du P, Huber W, Kibbe WA. Model-based variance-stabilizing transformation for Illumina microarray data. Nucleic Acids Res. 2008;36(2):e11. doi: 10.1093/nar/gkm1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Du P, Kibbe WA, Lin SM. nuID: a universal naming scheme of oligonucleotides for Illumina, Affymetrix, and other microarrays. Biol Direct. 2007;2:16. doi: 10.1186/1745-6150-2-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wettenhall JM, Smyth GK. limmaGUI: a graphical user interface for linear modeling of microarray data. Bioinformatics. 2004;20(18):3705–3706. doi: 10.1093/bioinformatics/bth449. [DOI] [PubMed] [Google Scholar]
- 10.Clifford B, Beljin M, Stark GR, Taylor WR. G2 arrest in response to topoisomerase II inhibitors: the role of p53. Cancer Res. 2003;63(14):4074–4081. [PubMed] [Google Scholar]
- 11.Van Quaquebeke E, Mahieu T, Dumont P, Dewelle J, Ribaucour F, Simon G, Sauvage S, Gaussin JF, Tuti J, El Yazidi M, et al. 2,2,2-Trichloro-N-({2-[2-(dimethylamino)ethyl]-1,3-dioxo-2,3-dihydro-1H-benzo[de]isoquinolin-5-yl}carbamoyl)acetamide (UNBS3157), a novel nonhematotoxic naphthalimide derivative with potent antitumor activity. J Med Chem. 2007;50(17):4122–4134. doi: 10.1021/jm070315q. [DOI] [PubMed] [Google Scholar]
- 12.Mijatovic T, Mahieu T, Bruyere C, De Neve N, Dewelle J, Simon G, Dehoux MJ, van der Aar E, Haibe-Kains B, Bontempi G. UNBS5162, a novel naphthalimide that decreases CXCL chemokine expression in experimental prostate cancers. Neoplasia. 2008;10(6):573–586. doi: 10.1593/neo.08290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kanda M, Nomoto S, Okamura Y, Nishikawa Y, Sugimoto H, Kanazumi N, Takeda S, Nakao A. Detection of metallothionein 1G as a methylated tumor suppressor gene in human hepatocellular carcinoma using a novel method of double combination array analysis. Int J Oncol. 2009;35(3):477–483. doi: 10.3892/ijo_00000359. [DOI] [PubMed] [Google Scholar]
- 14.Ferrario C, Lavagni P, Gariboldi M, Miranda C, Losa M, Cleris L, Formelli F, Pilotti S, Pierotti MA, Greco A. Metallothionein 1G acts as an oncosupressor in papillary thyroid carcinoma. Lab Invest. 2008;88(5):474–481. doi: 10.1038/labinvest.2008.17. [DOI] [PubMed] [Google Scholar]
- 15.Igal RA. Stearoyl-CoA desaturase-1: a novel key player in the mechanisms of cell proliferation, programmed cell death and transformation to cancer. Carcinogenesis. 2010;31(9):1509–1515. doi: 10.1093/carcin/bgq131. [DOI] [PubMed] [Google Scholar]