Abstract
Several types of epidermal growth factor receptor (EGFR) gene alternations have been observed in human tumors. Here we present a novel EGFR variant with aberrant splicing of exon 4 (named as de4 EGFR). Variant-specific polymerase chain reaction showed that de4 EGFR was expressed in some glioma (4/40), prostate cancer (3/11), and ovarian cancer (3/9) tissues but not in tissues adjacent to tumors or normal tissues. de4 EGFR displayed an enhanced transformation and a higher metastasis-promoting capacity in comparison to wild-type EGFR. With minimal EGF-binding activity, de4 EGFR underwent ligand-independent autophosphorylation and self-dimerization. Moreover, in serum-starved condition, de4 EGFR expression in U87 MG cells significantly upregulated the extracellular signal-regulated kinase and AKT phosphorylation and expression of JUN and Src. Importantly, E-cadherin expression was barely detectable in the U87 MG cells expressing de4 EGFR and restored expression of E-cadherin in these cells inhibited their metastatic behaviors. Taken together, we identified a novel EGFR variant with increased metastasis-promoting activity that may become a promising new target for cancer therapy.
Introduction
The epidermal growth factor receptor (EGFR) is a 170-kDa transmembrane glycoprotein that belongs to the receptor tyrosine kinase family of growth factor receptors. Owing to its important contributions to tumor cell survival, proliferation, and motility, EGFR has been associated with a large number of human malignancies such as breast cancer, lung cancer, brain cancer, prostate cancer, and liver cancer [1–7]. Overexpression, deletion, and mutation of the EGFR gene are the most common mechanisms by which EGFR exerts influence on tumorigenesis [8–10].
Coding sequence alterations of EGFR are frequently found in many types of human tumors [11–15]. In most cases, the EGFR variants are likely to be generated through genomic deletion. Conversely, in some instances involving the deletion or rearrangement of the intact exon(s), variants may occur as a consequence of alternative splicing [16]. Most variants with deletions in the extracellular domain correlate with a poor prognosis. These variants, although sometimes having reduced ligand-binding capacity, generally are constitutively active and mediate different signaling transduction pathways, thus giving the tumor cells a growth advantage and increased malignant potential [17,18].
The most common EGFR variant is the type III EGFR deletion mutant EGFRvIII (also called Δ801EGFR or de2-7 EGFR), which has an in-frame deletion of exons 2 to 7. EGFRvIII has been detected in 16% of non-small cell lung carcinoma cells, 57% of high-grade gliomas, 24% to 67% of glioblastomas, and 42% of head and neck squamous cell carcinomas [19–21]. Despite its frequent occurrence, the expression of EGFRvIII is restricted to tumor cells, thus making EGFRvIII an ideal target for anticancer therapy. Currently, several monoclonal antibodies and vaccines directed against EGFRvIII are undergoing preclinical and clinical trials [22–24]. Recently, a phase 2 multicenter trial assessing an EGFRvIII-targeted peptide vaccine was undertaken; patients who received this vaccine had significantly longer overall survival than the control group [25].
With the exception of EGFRvIII, the occurrence of variants in the extracellular domain of EGFR has not been thoroughly studied because most of these variants only occur in exclusive tumor types with a very low frequency of appearance [26]. The aim of this study was to find a relatively common EGFR variant in human cancers, such as gliomas, which often accompany amplification and alternation of EGFR [14]. The association between the variant and tumorigenesis would be established, and hence, the novel variant could provide a promising cancer therapeutic target.
Materials and Methods
Cells
Mouse embryonic fibroblast cells (NIH/3T3) and human glioblastoma-astrocytoma epithelial-like cells (U87MG) were obtained from ATCC (Manassas, VA): NIH/3T3 cells were cultured in a Dulbecco modified Eagle medium (DMEM; Gibco, Invitrogen, Carlsbad, CA) and supplemented with 10% bovine calf serum (PAA Laboratories, Dartmouth, MA) and antibiotics (Gibco, Invitrogen). U87MG cells were cultured in a DMEM supplemented with 10% fetal bovine serum (Gibco, Invitrogen) and antibiotics.
Clinical Samples
Human cancer tissues were obtained along with a written informed consent and pathology reports from hospitals and institutes as follows: Shanghai Ninth People's Hospital Affiliated with the Shanghai JiaoTong University School of Medicine (ovarian cancer), Huashan Hospital (glioma), and Changhai Hospital of Shanghai (prostate cancer). Among the 40 gliomas, 36 cases with available clinical data are clarified as grade 1 (n = 3), grade 2 (n = 12), grade 3 (n = 6), and grade 4 (n = 15). This study, including the use of all clinical materials, was approved by the institutional ethics review committee of Shanghai Cancer Institute.
Reverse Transcription-Polymerase Chain Reaction
Total RNA was extracted with TRIzol reagent (Invitrogen, Carlsbad, CA) from tumors or from normal/adjacent tissues. Reverse transcription-polymerase chain reaction (PCR) was performed using nested primers. The sequences of the primers for the first round of reverse transcription-PCR were as follows: 5′-GTATTGATCGGGAGAGCCG-3′ (forward primer, de4-S1) and 5′-GTGGAGATCGCCACTGATG-3′ (reverse primer, de4-AS1). EGFRvIII detection was performed using forward primer 5′-ATGCGACCCTCCGGGACG-3′ and reverse primer 5′-ATTCCGTTACACACTTTGCGGC-3′.
For amplification of de4 EGFR in gliomas, an aliquot from the first round of production was used as a template for the second round of PCR amplification using the following primers: 5′-ATGCGACCCTCCGGGACG-3′ (forward primer, de4-S2) and 5′-GTGGTGGGGTTGTAGAGCATG-3′ (reverse primer, de4-AS2).
To selectively eliminate wild-type genes, specific PCR amplifications were used. The principle behind this assay is shown in Figure 2A. In short, an aliquot of first-round product was used as the template for the second round of PCR amplification using the following primers: 5′-CCCATGAGAAATTTACAGGGC-3′ (forward primer, de4-S3) and 5′-GTGGTGGGGTTGTAGAGCATG-3′ (reverse primer, de4-AS2). Primer de4-S3, which spans exons 3 and 5, can selectively bind to de4 EGFR.
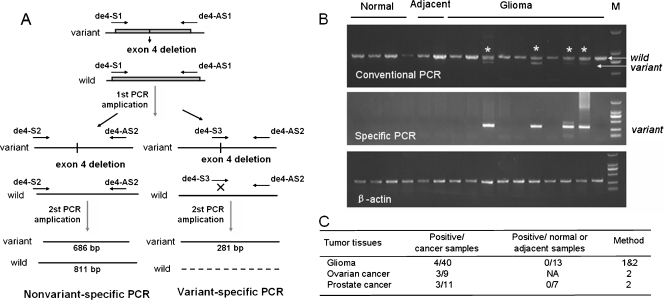
Figure 2.
de4 EGFR is observed in multiple cancer tissues. (A) Schematic of the PCR method used to detect the deletion of exon 4 in the EGFR gene. (B) Detection of EGFRwt and de4 EGFR in gliomas by conventional and variant-specific PCR. (Partial results) The variant amplicons shown here were sequenced. β-Actin served as an internal control. (C) Summary of the occurrence of de4 EGFR in several cancer tissues. Method 1: detection by conventional PCR. Method 2: detection by variant-specific PCR. NA indicates not applicable.
All amplifications were performed with the following parameters: 25 cycles (15 seconds at 94°C, 15 seconds at 58°C, 60 seconds at 68°C) using 5 to 100 ng of DNA template, 125 µM deoxynucleotide triphosphate, 2 pmol of each primer, and 0.25 U of KOD-Plus DNA polymerase (Toyobo, Inc., Osaka, Japan). Amplification of β-actin served as an internal control.
Lentivirus Production and Transduction of Target Cells
The EGFRwt and de4 EGFR sequences were amplified by PCR (long strand amplification), confirmed by sequencing, and then inserted into a pWPT vector (a generous gift from Dr T. Didier, University of Geneva, Geneva, Switzerland) by replacing GFP to generate pWPT-EGFRwt and pWPT-de4 EGFR. To produce virus particles, 20 µg of pWPT-EGFRwt or pWPT-de4 EGFR was transfected with the 15 µg of packaging plasmid psPAX2 and 5 µg of G protein of the vesicular stomatitis virus (VSV-G) envelope plasmid pMD2.G (generous gifts from Dr T. Didier) into 293T cells using a calcium phosphate transfection system. NIH/3T3 cells and U87MG cell were transduced with recombinant lentiviral particles to produce polyclonal cells with stable expression of EGFRwt and de4 EGFR. After being confirmed by immunoblot, polyclonal cells were seeded into 96-well plates at almost onecell per well. The monoclonal cell lines with the same expression level of EGFRwt and de4 EGFR were then selected and identified by fluorescence-activated cell sorter.
Immunoblot Analysis
Cells were serum-starved for 8 hours or treated with EGF (100 ng/ml, unless stated otherwise) for 8 minutes. Proteins were separated on 10% SDS-PAGE gels and transferred to nitrocellulose membranes (BioRad, Hercules, CA). The following antibodies were purchased from Santa Cruz Biotechnology (Santa Cruz, CA): rabbit anti-human EGFR (SC-03), rabbit anti-human pEGFR (Tyr1173) (SC-101668), rabbit anti-human pEGFR (Tyr845) (SC-57542), rabbit anti-human extracellular signal-regulated kinase (ERK; SC-93), rabbit anti-human AKT (SC-8312), mouse anti-human pERK (SC-7383), rabbit anti-human JUN (SC-1684), mouse anti-human Src (SC-8056), and mouse anti-human pEGFR (Tyr1086) (SC-81490). Rabbit anti-human pEGFR (Tyr1068) antibody (ab40815) and mouse anti-human pβ-catenin (Y654) antibody (ab24925) were purchased from Abcam (Abcam, Cambridge, UK). Rabbit anti-human pEGFR (Tyr992) antibody (07-821) was purchased from Millipore (Millipore, Temecula, CA). Rabbit anti-human pAKT (Ser473) (4060) and rabbit anti-human pSrc (Tyr416) (2101) were purchased from Cell Signaling (Danvers, MA). The mouse anti-human E-cadherin antibody (610182) and mouse anti-human β-catenin antibody (610154) were purchased from BD Biosciences (San Jose, CA). The mouse anti-GAPDH antibody was purchased from KangChen Bio-tech (Shanghai, China). All experiments were replicated at least two times.
Cell Proliferation Assay
Cell proliferation was measured using a CCK-8 Kit (Dojindo Laboratories, Rockville, MD). Three hundred cells were seeded in each well in a 96-well plate. CCK-8 solution (10 µl) was added into 100 µl of culture media, and the optical density was measured at 450 nm. Three independent experiments were performed.
Tumor Formation
Tumor growth assay and spontaneous metastasis assay were performed by subcutaneous inoculation of 1 x 106 tumor cells into 6-week-old female nude mice. In the tumor growth assay, tumor volumes were recorded until mice were killed on the 26th day. The subcutaneous tumor weight of each mouse was recorded. In the spontaneous metastasis assay, mice were killed for necropsy 8 weeks later. The lung weight of each mouse was recorded.
Colony Formation in Soft Agar
DMEM (2 ml) containing 0.7% agarose (Genentech, South San Francisco, CA) and 10% fetal calf serum was poured into six-well dishes. The layer was covered with a mixture of medium, agarose (0.42%), and cells (300) in triplicates. Colonies were visualized and counted under a microscope after 3 weeks.
Transwell Migration and Invasion Assay
Cell migration and invasion were gauged using a transwell migration assay and a Matrigel invasion assay. For the transwell migration assay, 5 x 104 cells were suspended in 200 µl of DMEM without serum and placed in the cell culture insert (8 µm pore size; BD Falcon, San Jose, CA) of a companion plate (BD Falcon) with a prewarmed culture medium containing 10% fetal bovine serum in the well. Cells were incubated for 12 hours (NIH/3T3 cells) or 24 hours (U87MG cells) at 37°C in 5% CO2 and then fixed with 4% paraformaldehyde in PBS. For Matrigel invasion assay, 1 x 105 cells were suspended in 200 µl of DMEM without serum and were placed in the cell culture insert precoated with 1 µg/µl Matrigel (BD Biosciences, San Jose, CA). A prewarmed culture medium containing 10% fetal bovine serum was added to the well. Cells were incubated for 24 hours at 37°C in 5% CO2 and were then fixed with 4% paraformaldehyde in PBS. The nonmigrated or invaded cells on top of the membrane were gently removed with a cotton swab. Cell migration or invasion was determined by staining cells with 0.1% crystal violet (Sigma, St Louis, MO) and counting the cells under a light microscope (100x magnification) in eight randomly selected areas.
EGF Binding Assay
Flow cytometric binding studies were performed at 0°C in PBS containing 0.5% BSA. A total of 200 µl of cell suspension and 50 µl of EGF-Rho (Molecular Probes) were mixed so that the final cell concentration was 1 x 106/ml, and the final EGF-Rho concentration was 400 ng/ml. Autofluorescence blanks (50 µl of buffer without EGF-Rho) were also prepared and served as control. All samples were placed on ice in the dark for 2 hours before the measurement of cellular fluorescence.
Covalent Cross-linking Analyses
Cells were grown on 10-cm plates, serum-starved for 8 hours, and incubated with EGF (100 ng/ml) for 8 minutes. After being washed twice with saline, cells were incubated for 20 minutes on ice in PBS containing 2 mM BS3 (Pierce, Rockford, IL). The reaction was quenched by adding Tris (50 mM, final concentration), and then cells were harvested.
Statistical Analysis
Data are presented as means ± SD. After the Levene test was used to test for equality of the variances, data were examined using an analysis of variance (ANOVA) test and the LSD method for multi-sample comparisons or Student's t test for the two-sample comparisons A value of P < .05 was considered statistically significant.
Results
Discovery and Detection of de4 EGFR
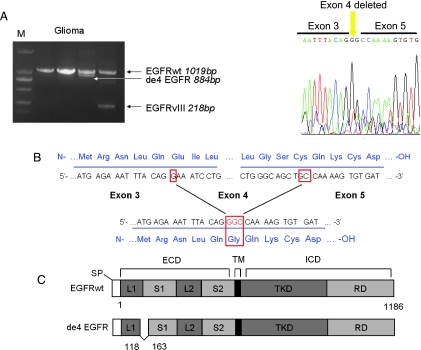
During our analysis of aberrant EGFR in gliomas, we identified an unexpected band that was slightly smaller than wild-type EGFR (Figure 1A). Sequencing of the smaller band showed that this transcript was an EGFR truncation that lacked the entire exon 4 sequence. Like EGFRvIII, this in-frame splice variant generated a novel glycine residue at the junction [19] (Figure 1B). This novel variant was named as de4 EGFR (GenBank accession number:HQ912715) to distinguish it from wild-type EGFR (Figure 1C). Further analysis of de4 EGFR mRNA expression in tumor tissues was performed. However, the length of de4 EGFR is very similar to that of EGFRwt, making it difficult to observe. Moreover, because clinical samples such as tumor biopsies may contain a small fraction of mutant genes and a large amount of wild-type genes, a highly sensitive assay for mutant detection should be applied [27]. Therefore, we designed a pair of variant-specific primers that amplified de4 EGFR but not wild-type EGFR (Figure 2A). The results shown in Figure 2B indicated that both non-variant-specific PCR and variant-specific PCR could obtain de4 EGFR amplicons, but the bands for the de4 EGFR PCR products were clearer when using the variant-specific primers. Using variant-specific PCR, we also observed the presence of de4 EGFR in ovarian cancer tissues and prostate cancer tissues but not in tissues adjacent to tumors or normal tissues (Figures 2B and W1). A summary of the expression of de4 EGFR in the examined tissues is shown in Figure 2C.
Figure 1.
Identification of de4 EGFR. (A) The discovery of de4 EGFR in glioma. The amplicon and the corresponding length of each EGFR variant are shown by an arrow. The right panel shows the determined sequencing result of de4 EGFR. (B) Salient features of de4 EGFR. The in-frame splicing removes exon 4 and creates a novel glycine residue at the splice junction. (C) Schematic representation of de4 EGFR variant. Deleted amino acid numbers are indicated. ECD indicates extracellular domain (S1, L1, S2, and L2 are subdomains); ICD, intracellular domain; RD, regulatory domain; SP, signal peptide; TKD, tyrosine kinase domain; TM, transmembrane.
de4 EGFR Owns a Proliferation-Promoting Capacity and an Enhanced Transformation Ability
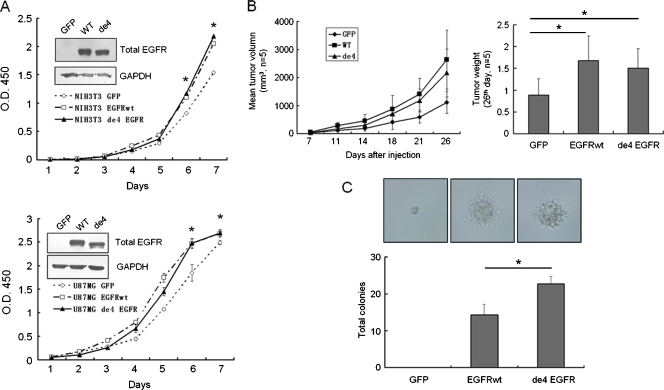
To analyze the function of de4 EGFR, two cell lines commonly used to study the function of EGFR, U87MG and NIH/3T3, were used. U87MG and NIH/3T3 were stably transfected with GFP, EGFRwt, or de4 EGFR. Growth curves demonstrated that, similar to EGFRwt, overexpression of de4 EGFR significantly increased cell proliferation (Figure 3A). Moreover, in the in vivo tumor formation assay, both EGFRwt and de4 EGFR promoted tumor growth when compared with GFP controls (Figure 3B). The effect of EGFRwt and de4 EGFR on cell transformation was further evaluated by colony formation in soft agar (Figure 3C). The colony sizes of de4 EGFR and EGFRwt transfectants were comparable, although bigger than those of GFP transfectants. However, more colony number was formed after de4 EGFR overexpression than EGFRwt-expressing cells. Collectively, these findings imply that de4 EGFR variant enhanced anchorage-independent cell growth and tumor proliferation.
Figure 3.
de4 EGFR promotes proliferation and transformation. (A) Growth curve showing the mitogenic activities of U87MG (bottom) and NIH/3T3 (top) transfectants in vitro. Inset: immunoblot analysis of whole lysates of the NIH/3T3 and U87MG transfectants using an anti-EGFR antibody. (B) Both EGFRwt and de4 EGFR promoted the proliferation of U87MG in vivo (n = 5). The right panel shows a comparison of tumor weight. (C) Colony formation in soft agar. NIH/3T3 cell derivatives (300/well) were cultured for 3 weeks. de4 EGFR transfected cells form the most colonies. *P < .05, compared with the control. Error bars, SD. Photomicrographs of colonies are representative. Magnification, x400. The normality of each data set was confirmed using the Levene test. Statistical data were evaluated using an ANOVA. Comparisons between two means were evaluated using the LSD method.
de4 EGFR Has a Higher Metastasis Potential in Comparison to Wild-type EGFR
In vitro migration and invasion assays were performed to explore whether de4 EGFR promoted the invasive capacity of tumor cells. A transwell migration and invasion assay using U87MG transfectants showed that EGFRwt induced a twofold increase in cell migration and a threefold increase in cell invasion compared with the GFP control. Intriguingly, de4 EGFR elicited even higher cell migration and invasion rates (approximately three- and eightfold higher than the GFP control, respectively) compared with EGFRwt (Figure 4A). Experiments using NIH/3T3 transfectants showed similar results (Figure W2).
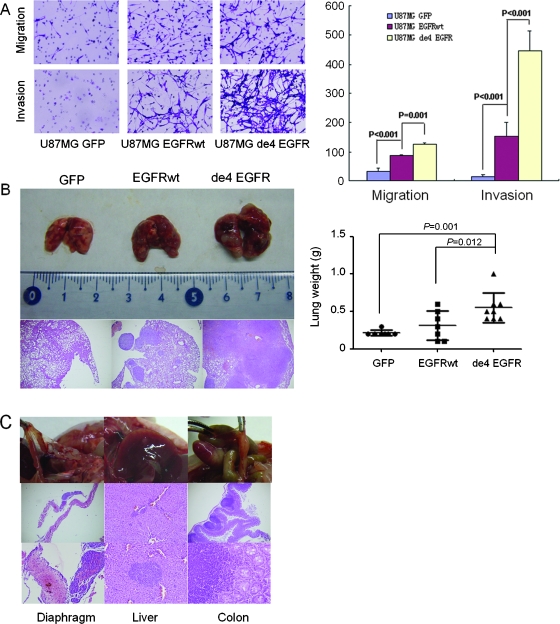
Figure 4.
de4 EGFR promotes invasion and metastasis in vitro and in vivo. (A) de4 EGFR enhances migration and invasion of U87MG in vitro (quantified on the right). Magnification, x100. Representative experiments are shown in triplicate along with SD. (B) de4 EGFR promotes more severe spontaneous metastases after subcutaneous inoculation of U87MG transfectants. Left: Comparison of lung lesions caused by three transfectants and hematoxylin and eosin staining of formalin-fixed lung sections. Magnification, x40. Right: Comparison of lung weight (tumor burden). Error bars, SD (n = 7/7/8). The normality of each data set was confirmed using the Levene test. Statistical data were evaluated using an ANOVA. Comparisons between two means were evaluated using the LSD method. (C) U87MG-de4 EGFR induced extrapulmonary metastatic foci in the diaphragm (left), liver (middle), and colon (right).
We then tried to confirm the metastasis-promoting capacity of de4 EGFR in vivo. U87MG transfectants were inoculated subcutaneously into the right hind flank of BALB/c nude mice. Eight weeks later, the mice were killed, and the lung weights were recorded. The results showed that U87MG-de4 EGFR induced the most extensive metastasis to the lungs, whereas cells bearing GFP or EGFRwt produced only occasional and less extensive pulmonary nodules. The heavier tumor burden caused by de4 EGFR in comparison to EGFRwt was readily detectable by hematoxylin and eosin staining (Figure 4B). Importantly, two of the eight mice that received U87MG de4 EGFR also exhibited extrapulmonary metastases sites, including the diaphragm, the liver, and the colon (Figure 4C). In contrast, no extrapulmonary metastases were observed in the GFP or EGFRwt groups (data not shown). These results indicated that, when compared with EGFRwt, de4 EGFR possessed significantly stronger tumor metastasis-promoting activities both in vitro and in vivo.
de4 EGFR Undergoes Basal Autophosphorylation and Constitutive Downstream Signaling
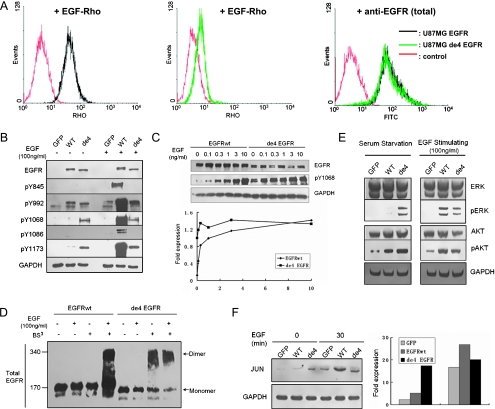
We next investigated potential mechanisms underlying the contributions of de4 EGFR to tumorigenesis and metastasis. Considering that exon 4 encodes an important part of the EGF-binding domain [28,29], it was reasonable to speculate that the binding activity between de4 EGFR and its ligands might be changed. Fluorescence-activated cell sorter analysis using tetramethylrhodamine-labeled EGF showed that, in contrast to wild-type EGFR, de4 EGFR had lost most of its EGF-binding capability (Figure 5A). Because de4 EGFR retained tumor-promoting capacity in our former experiments, we questioned whether activation modes other than EGF-dependent stimulation were present in cell lines expressing de4 EGFR.
Figure 5.
Oncogenic signaling of de4 EGFR. (A) Flow cytometry analysis for EGF-Rho binding affinity. Reactivity was tested on U87MG-EGFR (left) and U87-de4 EGFR (middle). Expression of total EGFR was assessed first (right). Autofluorescence blanks are also shown (red). (B) Tyrosine phosphorylation of EGFRwt and de4 EGFR treated with or without EGF for 8 minutes. Y1068 and Y1173 were phosphorylated in the presence of de4 EGFR in the absence of EGF activation. GAPDH served as an internal control. (C) U87MG transfectants expressing either EGFRwt or de4 EGFR were incubated with increasing EGF concentrations. The phosphorylation of Y1068 was analyzed by immunoblot analysis with the indicated antibodies. Phospho-EGFR levels were quantified according to band intensities. (D) Dimerization analysis of EGFRwt and de4 EGFR. Cells were treated with or without EGF (100 ng/ml; 8 minutes) and incubated with or without a covalent cross-linking reagent, BS3 (2mM, 20 minutes). EGFR dimers and monomers are indicated, respectively. (E) General signaling pathways relating to EGFR. ERK and AKT are activated in the presence of de4 EGFR. GAPDH served as an internal control. (F) The relative levels of JUN were determined by immunoblot analysis. The bands were quantified according to band intensities. GAPDH served as an internal control.
Previous studies reported that EGFRvIII, the most common EGFR mutant discovered to date, undergoes ligand-independent autophosphorylation, which mediates oncogenic signaling and transformation [19,30]. We hypothesized that de4 EGFR undergoes a similar autophosphorylation. We observed higher tyrosine autophosphorylation at the C-terminal of de4 EGFR (e.g., Tyr1068 and Tyr1173) in a serum-starved condition. By contrast, EGFRwt and de4 EGFR exhibited a similar degree of tyrosine phosphorylation at Tyr992 and no phosphorylation at Tyr1086 and Tyr845 (Figure 5B). When treated with EGF, the phosphorylation at all the tested tyrosine residues in EGFRwt increased dramatically, whereas in de4 EGFR, only the phosphorylation at Tyr992 changed with a slight up-regulation.
To further verify whether the basal phosphorylation of de4 EGFR variant was due to intrinsic activation or enhanced sensitivity to EGF-like ligands' stimulation, U87MG cells expressing EGFRwt or de4 EGFR were treated with increasing EGF concentrations (Figure 5C). The quantification of results illustrated that the phosphorylation of EGFRwt ascended after the increased ligand concentrations. The phosphorylation of de4 EGFR underwent a small increase when the cells were treated with 0.1 ng/ml of EGF. However, when cells were treated with EGF at a higher concentration, the phosphorylation of de4 EGFR was almost consistent. This observation indicates that intrinsic activation, rather than enhanced EGF sensitivity, may be responsible for the relatively high basal phosphorylation of de4 EGFR variant.
Because basal phosphorylation of EGFR variants can be caused by self-dimerization, dimerization of de4 EGFR was also investigated. To acquire covalently conjugated receptor dimmers for immunoblot analysis, cells were treated with the covalent cross-linking reagent bis (sulfosuccinimidyl) suberate (BS3) and harvested. Results of immunoblot assay clearly showed that dimerization of de4 EGFR occurred independent of EGF treatment. By contrast, the dimer of EGFRwt only formed in the presence of EGF (Figure 5D).
Considering the possibility that basal tyrosine activation may initiate constitutive downstream signaling, general signaling pathways related to EGFR were also examined. Activation of ERK and AKT by U87MG-de4 EGFR in the absence of serum was observed, although it was slightly weaker than EGF-induced phosphorylation in U87MG-EGFR (Figure 5E). Moreover, the induction of JUN, an oncogenic transcription factor that is mediated by EGFR activation, was detected in the serum-starved condition or under EGF treatment. In the absence of EGF stimulation, a relatively higher level of JUN was observed in de4 EGFR transfectants than that in EGFR transfectants. EGF treatment increased JUN expression in EGFR transfectants but not in de4 EGFR transfectants (Figure 5F).
Reduction of E-cadherin Expression Contributes to the Increased Invasiveness of Cell Lines Expressing de4 EGFR
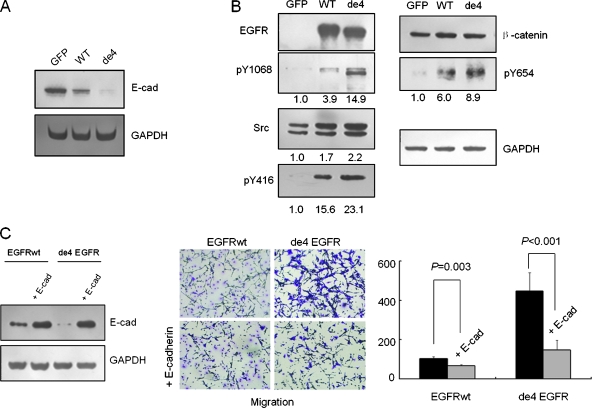
It has been reported that E-cadherin, a 120-kDa transmembrane glycoprotein located at the adherens junctions of epithelial cells, is often downregulated in highly invasive, poorly differentiated carcinomas [31]. In this study, we found that, in comparison to U87MG-GFP, U87MG-EGFR had lower E-cadherin expression and U87MG-de4 EGFR had almost no E-cadherin expression in different culture conditions (Figures 6A and W3).
Figure 6.
Reduction of E-cadherin-mediated cell-cell adhesion contributes to the increased migratory capacity of cell lines expressing de4 EGFR. (A) E-cadherin expression levels in three U87MG transfectants. (B) Activation of EGFR and overexpressed Src induced phosphorylation of β-catenin. Numbers indicate normalized intensities of bands. Cells were serum-starved for 8 hours and then harvested. (C) Exogenous E-cadherin was successfully transfected into U87MG-EGFR and U87MG-de4 EGFR. Enhanced E-cadherin expression resulted in decreased migratory capacity of U87MG-EGFR and U87MG-de4 EGFR. The normality of each data set was confirmed using the Levene test. Statistical data were conducted using the t test method.
Interestingly, the activation of EGFR generally results in the downregulation of E-cadherin-mediated cell-cell adhesion, accompanying EGFR-dependent proliferation, and migration of tumor cells. Previous studies reported that ERK activation inhibits E-cadherin expression; however, activated EGFR and overexpression of Src, a downstream target of EGFR, phosphorylate β-catenin at Y654 and then facilitate the loss of the association between E-cadherin and β-catenin [32,33]. In addition to the ERK basal activation presented previously, the phosphorylation of β-catenin was also observed in tumor cells bearing de4 EGFR. As shown in Figure 6B, there was an increase in β-catenin phosphorylation at Y654 in U87MG-de4 EGFR with basal phosphorylation of EGFR as well as enhanced Src expression and activation but no difference in total β-catenin concentration.
To further investigate the role of E-cadherin in the cell lines expressing EGFR and de4 EGFR, exogenous E-cadherin was introduced into these transfectants (Figure 6B). As shown in Figure 6C, when E-cadherin expression was rescued, the migration capacity of U87MG-EGFR and U87MG-de4 EGFR decreased.
Discussion
In this study, we report a novel variant of EGFR. This variant, which lacks the entire exon 4 sequence, is widely expressed in human tumors, and it seems to be associated with tumorigenesis. Interestingly, all the four de4 EGFR-positive gliomas belong to glioblastoma multiforme (grade 4), suggesting that de4 EGFR is positively correlated to the malignant degree of tumors. The joining of exons 3 and 5 generates a truncated EGFR transcript that is only 135 bp less than the wild-type human EGFR [34]. Because most clinical samples contain a high proportion of normal cells or genes, it is difficult to distinguish this sort of variant from the wild-type EGFR; this might be the reason why this variant was not identified in previous studies. To distinguish de4 EGFR protein from wild-type EGFR protein, we tried using the junction peptide of de4 EGFR to immunize rabbits and mice to generate polyclonal antibodies and monoclonal antibodies. Unfortunately, the antibodies we obtained could not completely discriminate between de4 EGFR and wild-type EGFR. Thus, detection of de4 EGFR at the protein level will remain problematic until an appropriate antibody is prepared.
The human EGFR gene contains 28 exons and spans approximately 200 kb. Previous studies have reported several alternatively spliced transcripts of EGFR, including the 1.8-kb transcript encoding soluble receptors [34]. The de4 EGFR variant may be produced through alternative splicing that joins exon 3 and exon 5 in some instances. Another possibility is that this variant transcript results from genomic deletion [19]. However, in our preliminary study using genomic-specific primers, amplicons of de4 EGFR were not observed in the tested tissues (data not shown), suggesting that the variant complementary DNA is more likely to be generated as a consequence of alternative splicing.
A previous study reported that EGFR gene amplification frequently occurred in gliomas [14]. We found that about half of the gliomas tested possessed EGFR amplification (data not shown). Among the four samples with de4 EGFR expression, only two samples carried EGFR amplification (1.5-folds and 7.2-folds compared with normal brains). Therefore, there seems no causal relationship between de4 EGFR expression and EGFR amplification.
It is well known that some EGFR variants with deletions in the extracellular domain cause increased malignancy in cells [15]. Our study indicates that de4 EGFR has the capacity to promote proliferation, although no growth advantage over EGFRwt was observed. The reason why de4 EGFR did not display proliferation-promotion advantage over EGFRwt is rather complicated. One possibility is that, in the presence of serum, wild-type EGFR itself can also induce strong cell proliferation. Importantly, de4 EGFR has an enhanced transformation capacity as well as a higher metastasis-promoting potential than wildtype EGFR. Because de4 EGFR was also detected in human tumors other than gliomas, the role of de4 EGFR in those tumors needs to be studied further. In addition, clinical studies on the correlation between de4 EGFR expression and prognosis in patients with such tumors should be performed.
Dimerization and tyrosine phosphorylation on EGF binding are important features of intact EGFR [35]. The extracellular domain of EGFR consists of four domains: L1, S1, L2, and S2. Domains L1 and L2 form the main part of the EGF-binding pocket, whereas S1 and S2 act as backbone structures [28,29]. A peptide composed of 45-amino-acid residues that are encoded by exon 4 functions as a hinge between the L1 and S1 domains (Figure 1C). Thus, the deletion of exon 4 should affect the conformation of the EGF binding region of EGFR and, accordingly, the receptor's binding activity to EGF. Interestingly, in the absence of ligand, de4 EGFR undergoes self-dimerization and constitutive tyrosine phosphorylation at residues Tyr1068 and Tyr1173. It seems that de4 EGFR acts as EGFRvIII in its metastasis-promoting activities as well as ligand-independent autophosphorylation and self-dimerization properties. However, unlike EGFRvIII, de4 EGFR did not show a more powerful capacity to promote proliferation than wild-type EGFR. Molecular mechanisms underlying the discrepancy on proliferation-promoting capacity between de4 EGFR and EGFRvIII need further studies.
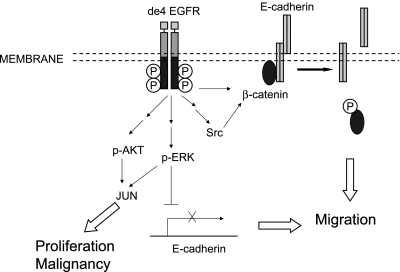
Because some tyrosine residues of de4 EGFR are constitutively phosphorylated without ligand activation, downstream signaling cascades could be activated without EGF binding. We know that through the recruitment of Grb2, Tyr1068 is associated with the activation of the mitogen-activated protein kinase (MAPK)/ERK and phosphoinositide 3-kinase/AKT pathways [19,36,37]. Thus, its constitutive phosphorylation led to a significant increase in ERK phosphorylation and a small increase in AKT phosphorylation. The expression of JUN, a downstream signaling molecule of ERK and AKT, was also upregulated in de4 EGFR-transfected U87MG cells, leading to the enhancement of transcriptional activity associated with cell proliferation and malignant transformation (Figure 7).
Figure 7.
Model showing how de4 EGFR promotes cell malignancy. Unlike EGFRwt, de4 EGFR shows basal phosphorylation independent of EGF stimulation. de4 EGFR likely enhances proliferation and transformation through constitutive activation of ERK/AKT and upregulation of the expression level of JUN in the absence of ligand stimulation. Conversely, de4 EGFR drives migration by down-regulation of E-cadherin-mediated cell-cell adhesion through two different mechanisms. One involves constitutive activation of ERK and thus inhibits E-cadherin expression. The other pathway includes β-catenin phosphorylation induced by overexpressed Src and basal activation of EGFR. This leads to the destruction of E-cadherin/catenin adhesive complexes and hence facilitating cell migration.
It has been reported that EGFR activation may cause the dismantling of cell-cell contacts by promoting the destabilization, the down-regulation, and the endocytosis or subsequent degradation of E-cadherins. Of note, MAPK activation is also involved in the negative regulation of E-cadherin [32,38]. In this study, we verified that E-cadherin is inversely related to cell invasion. Tumor cells expressing de4 EGFR had limited expression of E-cadherin protein and increased invasive activity, which was reduced by restoration of E-cadherin in these cells. Furthermore, the limited E-cadherin expression that we observed may be related to the constitutive tyrosine phosphorylation of de4 EGFR and the continuous activation of the downstream MAPK pathway. In addition, β-catenin phosphorylation at Y654 also contributes to the destabilization of E-cadherin/catenin adhesive complexes, which is potentially downregulated by EGFR and its downstream target, Src [33,38]. It is likely that these different mechanisms play a coordinated role in facilitating the loss of E-cadherin expression and hence dismantling of cell-cell contacts. Regardless, the significantly decreased activity of E-cadherin revealed a potential explanation for the potent migration-promoting ability of de4 EGFR variant (Figure 7).
EGFR is one of the most important cancer therapeutic targets discovered to date. By the end of 2008, more than 10 EGFR-targeting agents were in stages of advanced clinical development for the treatment of human cancers. Two anti-EGFR monoclonal antibodies (cetuximab and panitumumab) and two tyrosine kinase inhibitors directed against EGFR (gefitinib and erlotinib) have been approved by the US Food and Drug Administration and/or the European Medicines Agency for the treatment of various human cancer types. However, because EGFR is widely expressed in many human tissues, EGFR-targeting therapies are likely to have adverse effects, including skin toxicity and diarrhea [5,39]. Because EGFR variants are generally tumor-specific, they may be more rational targets for anticancer therapy. Furthermore, antibodies and vaccines selectively targeting these variants should have lower toxicity [40]. For example, no significant toxicity was found in the phase 1 clinical trial of mAb ch806 [23]. Therefore, de4 EGFR is a promising therapeutic target, and therapeutics targeting de4 EGFR should be developed and tested.
Supplementary Material
Acknowledgments
The authors thank the generous gift the pWPT, psPAX2, and pMD2.G lentivirus plasmids from Dr Didier Trono (University of Geneva, Switzerland). The authors also thank Dr Jian Ding from the Shanghai Institute of Materia Medica for the generous gift of the antibody recognizing phosphorylated Src.
Abbreviations
- de4 EGFR
the exon 4 deletion variant of EGFR
- EGFR
epidermal growth factor receptor
- EGFRvIII
epidermal growth factor receptor variant III
- ERK
extracellular signal-regulated kinase
- EGFRwt
wild type EGFR
Footnotes
This study is supported by National Basic Research Program (grant no. 2010CB529902), National Natural Science Foundation of China (no. 81071746), and the Key Program Project of the Shanghai Science and Technology Committee (10431903700).
This article refers to supplementary materials, which are designated by Figures W1 to W3 and are available online at http://www.neoplasia.com.
References
- 1.Xue C, Wyckoff J, Liang F, Sidani M, Violini S, Tsai K, Zhang Z, Sahai E, Condeelis J, Segall JE. Epidermal growth factor receptor overexpression results in increased tumor cell motility in vivo coordinately with enhanced intravasation and metastasis. Cancer Res. 2006;66:192–197. doi: 10.1158/0008-5472.CAN-05-1242. [DOI] [PubMed] [Google Scholar]
- 2.Pandiella A, Lehvaslaiho H, Magni M, Alitalo K, Meldolesi J. Activation of an EGFR/neu chimeric receptor: early intracellular signals and cell proliferation responses. Oncogene. 1989;4:1299–1305. [PubMed] [Google Scholar]
- 3.Liang K, Ang KK, Milas L, Hunter N, Fan Z. The epidermal growth factor receptor mediates radioresistance. Int J Radiat Oncol Biol Phys. 2003;57:246–254. doi: 10.1016/s0360-3016(03)00511-x. [DOI] [PubMed] [Google Scholar]
- 4.Ho R, Minturn JE, Hishiki T, Zhao H, Wang Q, Cnaan A, Maris J, Evans AE, Brodeur GM. Proliferation of human neuroblastomas mediated by the epidermal growth factor receptor. Cancer Res. 2005;65:9868–9875. doi: 10.1158/0008-5472.CAN-04-2426. [DOI] [PubMed] [Google Scholar]
- 5.Modjtahedi H, Essapen S. Epidermal growth factor receptor inhibitors in cancer treatment: advances, challenges and opportunities. Anticancer Drugs. 2009;20:851–855. doi: 10.1097/CAD.0b013e3283330590. [DOI] [PubMed] [Google Scholar]
- 6.Weihua Z, Tsan R, Huang W, Wu Q, Chiu C, Fidler IJ, Hung M. Survival of cancer cells is maintained by EGFR independent of its kinase activity. Cancer Cell. 2008;13:385–393. doi: 10.1016/j.ccr.2008.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang H, Jiang H, Zhou M, Xu Z, Liu S, Shi B, Yao X, Yao M, Gu J, Li Z. Epidermal growth factor receptor vIII enhances tumorigenicity and resistance to 5-fluorouracil in human hepatocellular carcinoma. Cancer Lett. 2008;279:30–38. doi: 10.1016/j.canlet.2009.01.019. [DOI] [PubMed] [Google Scholar]
- 8.Chaffanet M, Chauvin C, Lainé M, Berger F, Chédin M, Rost N, Nissou MF, Benabid AL. EGF receptor amplification and expression in human brain tumours. Eur J Cancer. 1992;28:11–17. doi: 10.1016/0959-8049(92)90374-b. [DOI] [PubMed] [Google Scholar]
- 9.Nishikawa R, Ji XD, Harmon RC, Lazar CS, Gill GN, Cavenee WK, Huang HJ. A mutant epidermal growth factor receptor common in human glioma confers enhanced tumorigenicity. Proc Natl Acad Sci USA. 1994;91:7727–7731. doi: 10.1073/pnas.91.16.7727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tang P, Steck PA, Yung WK. The autocrine loop of TGF-α/EGFR and brain tumors. J Neurooncol. 1997;35:303–314. doi: 10.1023/a:1005824802617. [DOI] [PubMed] [Google Scholar]
- 11.Lynch TJ, Bell DW, Sordella R, Gurubhagavatula S, Okimoto RA, Brannigan BW, Harris PL, Haserlat SM, Supko JG, Haluska FG, et al. Activating mutations in the epidermal growth factor receptor underlying responsiveness of non-small-cell lung cancer to gefitinib. N Engl J Med. 2004;350:2129–2139. doi: 10.1056/NEJMoa040938. [DOI] [PubMed] [Google Scholar]
- 12.Pines G, Huang PH, Zwang Y, White FM, Yarden Y. EGFRvIV: a previously uncharacterized oncogenic mutant reveals a kinase autoinhibitory mechanism. Oncogene. 2010;29:5850–5860. doi: 10.1038/onc.2010.313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lankiewicz S, Rother E, Zimmermann S, Hollmann C, Korangy F, Greten TF. Tumour-associated transcripts and EGFR deletion variants in colorectal cancer in primary tumour, metastases and circulating tumour cells. Cell Oncol. 2008;30:463–471. doi: 10.3233/CLO-2008-0432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Frederick L, Wang XY, Eley G, James CD. Diversity and frequency of epidermal growth factor receptor mutations in human glioblastomas. Cancer Res. 2000;60:1383–1387. [PubMed] [Google Scholar]
- 15.Pines G, Köstler WJ, Yarden Y. Oncogenic mutant forms of EGFR: lessons in signal transduction and targets for cancer therapy. FEBS Lett. 2010;584:2699–2706. doi: 10.1016/j.febslet.2010.04.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Moscatello DK, Montgomery RB, Sundareshan P, McDanel H, Wong MY, Wong AJ. Transformational and altered signal transduction by a naturally occurring mutant EGF receptor. Oncogene. 1996;13:85–96. [PubMed] [Google Scholar]
- 17.Antonyak MA, Moscatello DK, Wong AJ. Constitutive activation of c-Jun N-terminal kinase by a mutant epidermal growth factor receptor. J Biol Chem. 1998;273:2817–2822. doi: 10.1074/jbc.273.5.2817. [DOI] [PubMed] [Google Scholar]
- 18.Huang PH, Miraldi ER, Xu AM, Kundukulam VA, Del Rosario AM, Flynn RA, Cavenee WK, Furnari FB, White FM. Phosphotyrosine signaling analysis of site-specific mutations on EGFRvIII identifies determinants governing glioblastoma cell growth. Mol Biosyst. 2010;6:1227–1237. doi: 10.1039/c001196g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pederson MW, Meltorn M, Damstrup L, Poulsen HS. The type III epidermal growth factor receptor mutation: biological significance and potential target for anti-cancer therapy. Ann Oncol. 2001;12:745–760. doi: 10.1023/a:1011177318162. [DOI] [PubMed] [Google Scholar]
- 20.Heimberger A, Suki D, Yang D, Aldape K. The natural history of EGFR and EGFRvIII in glioblastoma patients. J Transl Med. 2005;3:38–43. doi: 10.1186/1479-5876-3-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sok JC, Coppelli FM, Thomas SM, Lango MN, Xi S, Hunt JL, Freilino ML, Graner MW, Wikstrand CJ, Bigner DD, et al. Mutant epidermal growth factor receptor (EGFRvIII) contributes to head and neck cancer growth and resistance to EGFR targeting. Clin Cancer Res. 2006;12:5064–5073. doi: 10.1158/1078-0432.CCR-06-0913. [DOI] [PubMed] [Google Scholar]
- 22.Jungbluth AA, Stockert E, Huang HJ, Collins VP, Coplan K, Iversen K, Kolb D, Johns TJ, Scott AM, Gullick WJ, et al. A monoclonal antibody recognizing human cancers with amplification/overexpression of the human epidermal growth factor receptor. Proc Natl Acad Sci USA. 2003;100:639–644. doi: 10.1073/pnas.232686499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Scott AM, Lee F, Tebbutt N, Herbertson R, Gill SS, Liu Z, Skrinos E, Murone C, Saunder TH, Chappell B, et al. A phase I clinical trial with monoclonal antibody ch806 targeting transitional state and mutant epidermal growth factor receptors. Proc Natl Acad Sci USA. 2007;104:4071–4076. doi: 10.1073/pnas.0611693104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sharma VK, Chih HW, Mrsny RJ, Daugherty AL. The formulation and delivery of monoclonal antibodies. In: An Z, editor. Therapeutic Monoclonal Antibodies: From Bench to Clinic. Hoboken, NJ: John Wiley & Sons, Inc; 2009. pp. 675–710. [Google Scholar]
- 25.Sampson JH, Heimberger AB, Archer GE, Aldape KD, Friedman AH, Friedman HS, Gilbert MR, Herndon JE, II, McLendon RE, Mitchell DA. Immunologic escape after prolonged progression-free survival with epidermal growth factor receptor variant III peptide vaccination in patients with newly diagnosed glioblastoma. J Clin Oncol. 2010;28:4722–4729. doi: 10.1200/JCO.2010.28.6963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lafky JM, Wilken JA, Baron AT, Maihle NJ. Clinical implications of the ErbB/epidermal growth factor (EGF) receptor family and its ligands in ovarian cancer. Biochim Biophys Acta. 2008;1785:232–265. doi: 10.1016/j.bbcan.2008.01.001. [DOI] [PubMed] [Google Scholar]
- 27.Asano H, Toyooka S, Tokumo M, Ichimura K, Aoe K, Ito S, Tsukuda K, Ouchida M, Aoe M, Katayama H, et al. Detection of EGFR gene mutation in lung cancer by mutant-enriched polymerase chain reaction assay. Clin Cancer Res. 2006;12:43–48. doi: 10.1158/1078-0432.CCR-05-0934. [DOI] [PubMed] [Google Scholar]
- 28.Ogiso H, Ishitani R, Nureki O, Fukai S, Yamanaka M, Kim J, Saito K, Sakamoto A, Inoue M, Shirouzu M, et al. Crystal structure of the complex of human epidermal growth factor and receptor extracellular domains. Cell. 2002;110:775–787. doi: 10.1016/s0092-8674(02)00963-7. [DOI] [PubMed] [Google Scholar]
- 29.Garrett TP, McKern NM, Lou M, Elleman TC, Adams TE, Lovrecz GO, Zhu H, Walker F, Frenkel MJ, Hoyne PA. Crystal structure of a truncated epidermal growth factor receptor extracellular domain bound to transforming growth factor α. Cell. 2002;110:763–773. doi: 10.1016/s0092-8674(02)00940-6. [DOI] [PubMed] [Google Scholar]
- 30.Ning Y, Zeineldin R, Liu Y, Rosenberg M, Stack MS, Hudson LG. Down-regulation of integrin α2 surface expression by mutant epidermal growth factor receptor (EGFRvIII) induces aberrant cell spreading and focal adhesion formation. Cancer Res. 2005;65:9280–9286. doi: 10.1158/0008-5472.CAN-05-0407. [DOI] [PubMed] [Google Scholar]
- 31.Mrgineanu E, Cotrutz CE, Cotrutz C. Correlation between E-cadherin abnormal expressions in different types of cancer and the process of metastasis. Rev Med Chir Soc Med Nat Iasi. 2008;112:432–436. [PubMed] [Google Scholar]
- 32.Conacci-Sorrell M, Simcha I, Ben-Yedidia T, Blechman J, Savagner P, Ben-Ze'ev A. Autoregulation of E-cadherin expression by cadherin-cadherin interactions: the roles of β-catenin signaling, Slug, and MAPK. J Cell Biol. 2003;163:847–857. doi: 10.1083/jcb.200308162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lilien J, Balsamo J. The regulation of cadherin-mediated adhesion by tyrosine phosphorylation/dephosphorylation of β-catenin. Curr Opin Cell Biol. 2005;17:459–465. doi: 10.1016/j.ceb.2005.08.009. [DOI] [PubMed] [Google Scholar]
- 34.Reiter JL, Maihle NJ. A 1.8 kb alternative transcript from the human epidermal growth factor receptor gene encodes a truncated form of the receptor. Nucleic Acids Res. 1996;24:4050–4056. doi: 10.1093/nar/24.20.4050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fernandez H, Cohen S, Bishayee S. Glycosylation-induced conformational modification positively regulates receptor-receptor association. J Biol Chem. 2001;276:5375–5383. doi: 10.1074/jbc.M005599200. [DOI] [PubMed] [Google Scholar]
- 36.Pernas FG, Allen CT, Winters ME, Yan B, Friedman J, Dabir B, Saigal K, Mundinger GS, Xu X, Morris JC, et al. Proteomic signatures of epidermal growth factor receptor and survival signal pathways correspond to gefitinib sensitivity in head and neck cancer. Clin Cancer Res. 2009;15:2361–2372. doi: 10.1158/1078-0432.CCR-08-1011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sebastian S, Settleman J, Reshkin SJ, Azzariti A, Bellizzi A, Paradiso A. The complexity of targeting EGFR signalling in cancer: from expression to turnover. Biochim Biophys Acta. 2006;1766:120–139. doi: 10.1016/j.bbcan.2006.06.001. [DOI] [PubMed] [Google Scholar]
- 38.Gavard J, Gutkind JS. A molecular crosstalk between E-cadherin and EGFR signaling networks. In: Haley JD, Gullick WJ, editors. EGFR Signaling Networks in Cancer Therapy. Vol. 1. New York, NY: Humana Press, Springer; 2008. pp. 131–146. [Google Scholar]
- 39.Ciardiello F, Tortora G. EGFR antagonists in cancer treatment. N Engl J Med. 2008;358:1160–1174. doi: 10.1056/NEJMra0707704. [DOI] [PubMed] [Google Scholar]
- 40.Schmittling RJ, Archer GE, Mitchell DA, Heimberger A, Pegram C, Herndon JE, Friedman HS, Bigner DD, Sampson JH. Detection of humoral response in patients with glioblastoma receiving EGFRvIII-KLH vaccines. J Immunol Methods. 2008;339:74–81. doi: 10.1016/j.jim.2008.08.004. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.