EYE2 is a key protein in connecting the positioning information of the microtubule rootlet cytoskeleton and channelrhodopsin 1 (ChR1) photoreceptor to the formation and positioning of the eyespot pigment granules in the chloroplast of Chlamydomonas. EYE3, a ser/thr kinase of the ABC1 family, is found in pigment granules and is required for their biogenesis.
Abstract
The eyespot of the biflagellate unicellular green alga Chlamydomonas reinhardtii is a complex organelle that facilitates directional responses of the cell to environmental light stimuli. The eyespot, which assembles de novo after every cell division and is associated with the daughter four-membered (D4) microtubule rootlet, comprises an elliptical patch of rhodopsin photoreceptors on the plasma membrane and stacks of carotenoid-rich pigment granule arrays in the chloroplast. Two loci, EYE2 and EYE3, define factors involved in the formation and organization of the eyespot pigment granule arrays. Whereas EYE3, a serine/threonine kinase of the ABC1 family, localizes to pigment granules, EYE2 localization corresponds to an area of the chloroplast envelope in the eyespot. EYE2 is positioned along, and adjacent to, the D4 rootlet in the absence of pigment granules. The eyespot pigment granule array is required for maintenance of the elliptical shape of both the overlying EYE2 and channelrhodopsin-1 photoreceptor patches. We propose a model of eyespot assembly wherein rootlet and photoreceptor direct EYE2 to an area of the chloroplast envelope, where it acts to facilitate assembly of pigment granule arrays, and EYE3 plays a role in the biogenesis of the pigment granules.
INTRODUCTION
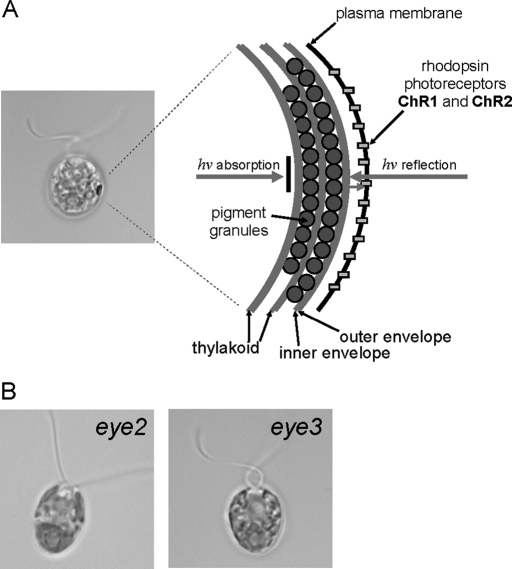
Many green algae, including the biflagellate unicell Chlamydomonas reinhardtii, possess the ability to respond to light, a behavior known as phototaxis (ptx). Phototactic cells respond to varying light levels by either swimming toward a source of low-intensity light (positive ptx) or away from high-intensity light (negative ptx), an adaptation for optimizing photosynthetic efficiency in their aqueous environment (for reviews, see Witman, 1993; Kreimer, 1994). These phototactic responses are induced by alteration of the flagellar beat pattern via Ca2+-mediated signaling mechanisms (Nultsch, 1983; Kamiya and Witman, 1984; Hegemann et al., 1990). Perception of environmental light cues is mediated by an organelle known as the eyespot apparatus, which encompasses components in both the chloroplast and plasma membrane and is observable in the light microscope as a distinct orange-red spot at the equator of the cell. The structural organization of the eyespot of C. reinhardtii is complex, comprising two to four layers of carotenoid-filled pigment granules arranged in hexagonal, closely packed arrays between layers of thylakoid membranes and tightly apposed to the chloroplast envelope (Figure 1A; Melkonian and Robenek, 1984; for reviews, see Dieckmann, 2003; Kreimer, 2009). Directly overlaying this structure is a particle-dense region of plasma membrane containing light-gated rhodopsin photoreceptors, channelrhodopsin-1 and -2 (ChR1 and ChR2) (Nagel et al., 2002, 2003; Sineshchekov et al., 2002; Berthold et al., 2008). Incoming photons not initially captured by the photoreceptors are reflected by the quarter-wave plate arrangement of the stacked eyespot pigment granule arrays, concomitantly blocking light from other directions while enhancing sensitivity of photoresponses to light stimuli (Foster and Smyth, 1980; Morel-Laurens and Feinleib, 1983).
FIGURE 1:
The eyespot apparatus is a complex multilayered organelle. (A) Bright field micrograph of a wild-type C. reinhardtii cell and diagram of the eyespot apparatus. Layers of carotenoid-filled pigment granules in the chloroplast are subtended by thylakoid membranes. The pigment granule layers are closely apposed to the chloroplast envelope and the overlying photoreceptor molecules in the plasma membrane, and act both to reflect orthogonal light onto the photoreceptors and absorb light from other directions. (B) Bright field micrographs of eyeless mutants eye2 and eye3, which lack eyespot pigment granule layers. Bars, 5 μm.
The asymmetric positioning of the eyespot in the cell is essential for the directional perception of light cues. In wild-type Chlamydomonas cells, the eyespot is invariably positioned in association with the daughter four-membered (D4) microtubule rootlet, a highly acetylated cytoskeletal component, and is offset 45° from the plane of flagellar beat and ∼90° from the anterior-posterior axis of the cell (Holmes and Dutcher, 1989). The specific characteristics of the D4 rootlet that direct positioning of the eyespot are presently undefined, although rootlet length, daughter-specific microtubule-associated proteins, or other modifications may play a role (Mittelmeier et al., unpublished data).
Assembling such an organized photosensory system in C. reinhardtii presents a complex problem: Multiple chloroplast elements (thylakoid membrane, chloroplast envelope, and pigment granules) must be coordinately assembled along with plasma membrane–localized proteins (photoreceptor molecules) in a specific area of the cell. Forward genetic approaches have been instructive in identifying several of the factors involved in this process. Loci affecting eyespot assembly and positioning include MIN1, EYE2, and EYE3 (Lamb et al., 1999). Both the eye2 and eye3 mutants lack eyespots (Figure 1B) and are unable to phototax at low light intensity, yet exhibit negative ptx in response to high light, indicative that the photosensory signaling system in these mutants remains intact (Roberts, 1999; Roberts et al., 2001). The EYE2 protein contains a LysM domain and thioredoxin motif. Because thioredoxin activity is not required for eyespot assembly (Roberts et al., 2001), EYE2 has been suggested to serve a chaperone-like function in assembly of the eyespot pigment granule arrays. The min1 mutant possesses a miniature eyespot characterized by disorganized pigment granules in the chloroplast stroma. MIN1 is a C2/LysM-domain protein, present in the eyespot proteome (Schmidt et al., 2006), and is postulated to be involved in chloroplast envelope-plasma membrane attachment (Mittelmeier et al., 2008). LysM domains were originally identified in bacterial cell-wall degrading enzymes as binding peptidoglycans (Bateman and Bycroft, 2000), and they mediate interactions with other proteins in defense responses in plants (Knogge and Scheel, 2006).
The necessity of coordinated assembly of this multicompartmental system that is both structurally intricate and precisely positioned raises important questions concerning the factors and mechanisms involved in biogenesis of the eyespot and the directional cues needed for orchestrating its asymmetric positioning. Characterization and localization of proteins involved in eyespot assembly and analysis of mutants defective in various aspects of this process have given us insight into the roles of these factors in the development of this photosensory organelle.
RESULTS
Eyespot pigment granule layers are required to maintain the shape of the ChR1 photoreceptor patch
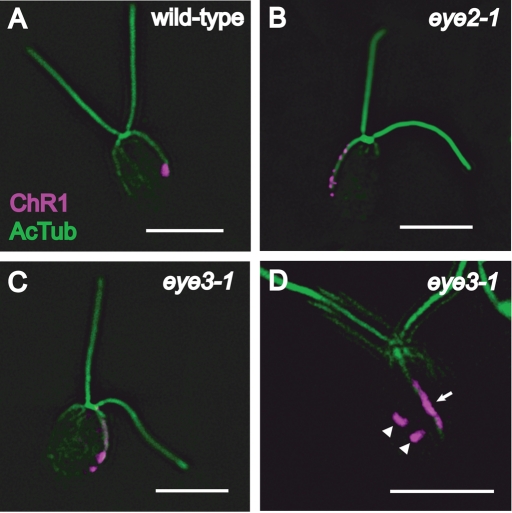
As previously noted, the ChR1 photoreceptor appears as a single elliptical patch on the plasma membrane (Berthold et al., 2008) and is associated with the D4 microtubule rootlet in wild-type C. reinhardtii cells (Figure 2A). To investigate whether the eyespot pigment granule layers affect localization of the photoreceptor molecules on the plasma membrane or their association with the D4 rootlet, wild-type, eye2, and eye3 mutant cells were stained with anti-ChR1 and anti-acetylated tubulin. In both eye2 and eye3 cells, aggregations of ChR1 are seen as multiple patches arranged in bars or stripes on the rootlet (Figure 2, B and C). A subset (23%) of cells scored in an eye3 population (n = 126) possessed distinct ChR1 patches not associated with the D4 rootlet in addition to one or more rootlet-associated patches (Table 1 and Figure 2D). In all cells of this subpopulation, off-rootlet ChR1 patches remained in one longitudinal half of the cell in proximity to the rootlet. Highly similar staining patterns were observed in photoautotrophically grown min1 cells (Table 1). These data are indicative that the presence of organized pigment granules is necessary for the maintenance of the elliptical shape of the photoreceptor patch. The D4 rootlet–associated asymmetric localization of ChR1 remains intact in the eyeless and miniature-eyed mutants.
FIGURE 2:
ChR1 photoreceptor localization pattern is altered in eyeless mutants. Combined immunofluorescence micrographs stained with antibodies against ChR1 (magenta) and acetylated α-tubulin (AcTub) (green). (A) A wild-type Chlamydomonas cell showing a single discrete patch of ChR1 near the end of the acetylated track of the D4 microtubule rootlet. (B) An eye2 cell showing ChR1 staining in multiple patches along the rootlet. (C) Photoreceptor localization in an individual cell of the eyeless mutant eye3–1. ChR1 is observed in multiple patches along the rootlet. (D) An eye3 cell showing ChR1 in a stripe associated with the rootlet (arrow). Discrete ChR1 patches not associated with the rootlet are also observed (arrowheads). Bars, 5 μm.
TABLE 1:
ChR1 localization patterns in eye3 and min1 mutants.
| Strain | No. of cells scored | No. (%) of cells with ChR1 spots only on rootlet | No. (%) of cells with off-rootlet ChR1 spots | Percentage of off-rootlet spots in proximity to rootlet |
|---|---|---|---|---|
| eye3–1 | 126 | 97 (77) | 29 (23) | 100 |
| min1–1 | 53 | 42 (79) | 11 (21) | 100 |
The EYE3 gene encodes a predicted ser/thr kinase of the ABC1 family
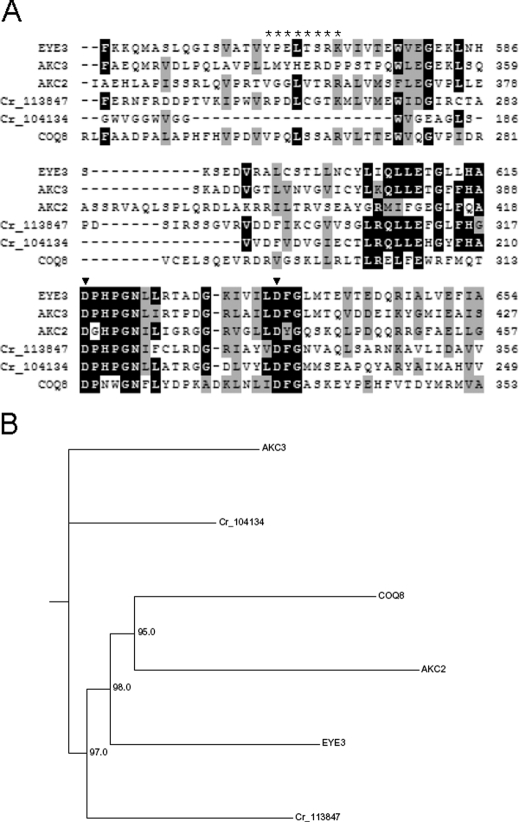
To probe how the EYE2 and EYE3 proteins function in organizing pigment granules and photoreceptors into a functional organelle, we sought to identify the EYE3 gene and use antisera to localize the gene products of both EYE3 and the previously identified EYE2 gene. C. reinhardtii strain 12–18 (eye3–1) was isolated from a ptx screen for ptx-defective strains following UV mutagenesis of wild-type strain 137c (CC-125) (Lamb et al., 1999). To enable isolation of the EYE3 gene, an NIT1 insertional allele of EYE3 was sought in a collection of ptx-defective mutants following insertional mutagenesis of strain g1 (nit1, agg1, mt+) (Pazour et al., 1995). Light microscopy revealed that cells of strain F35 were eyeless. The mutation in F35 was not complemented by eye3–1, indicating that the eyeless phenotype of F35 was most likely the result of NIT1 insertion within or close to the EYE3 gene. The genomic sequence adjacent to the NIT1 insertion was used to isolate the putative EYE3 gene in a cosmid. Sequencing the point mutation in eye3–1 and rescue of the mutant phenotype by transformation with the EYE3 cosmid verified the identity of the gene (for details, see Materials and Methods). The EYE3 gene is predicted to encode a large hydrophobic, chloroplast-localized ser/thr kinase which is a member of the ABC1 (UbiB/AarF) family of ser/thr kinases involved in regulation of quinone biosynthesis (Do et al., 2001). Six other members of this protein family are predicted from gene models in the Chlamydomonas genome (JGI version 4.0; http://www.chlamy.org), including chloroplast-targeted ABC1 kinases termed AKCs (ABC1 kinases in the chloroplast) (Merchant et al., 2007). Notably, the predicted AKC2 kinase (protein ID 205779) possesses a putative LysM domain that could be involved in protein–protein interactions. EYE3 highly resembles ABC1 kinase family members in Arabidopsis (At1g71810 and At1g79600) that are associated with plastoglobules, structures quite similar to eyespot pigment granules (Ytterberg et al., 2006). A clustal alignment of the EYE3 sequence with other ABC1 kinases in C. reinhardtii demonstrates the high degree of conservation of the characteristic kinase domain sequence of this family (Figure 3A). The characteristics of the members of this protein family contributed to the hypothesis that EYE3 localizes to the eyespot pigment granules.
FIGURE 3:
EYE3 is a member of the ABC1 kinase family. (A) Clustal alignment of the amino acid sequence of the region, including the kinase-active site of the EYE3 protein (residues 549–654) with related ABC1 kinases in C. reinhardtii. Predicted amino acid sequences of proteins supported by gene models in version 4 of the genome are shown: ABC1 kinases in the chloroplast AKC3 (protein ID 205743) and AKC2 (protein ID 205779); predicted eyespot kinases Cr_113847 and Cr_104934; and ubiquinone synthesis protein COQ8 (protein ID 126076). Background shading denotes residues that are identical (black) or similar (gray) in ≥60% of the sequences. The residues in EYE3 used for generating peptide-directed antiserum are marked with asterisks. Positions of critical aspartate residues in the kinase-active site of EYE3 are marked with arrowheads. (B) Phylogram showing inferred relation between EYE3 and related ABC1 kinases in C. reinhardtii. Bootstrap values were calculated from 100 iterations using the neighbor-joining method.
EYE3 localizes to the eyespot pigment granule layers
To determine the localization of EYE3 in wild-type Chlamydomonas cells, a polyclonal antiserum to EYE3 was used in immunofluorescence staining of methanol-fixed cells. The EYE3 signal appeared as an eyespot-shaped elliptical spot in wild-type cells that was absent in both eye3 (Figure 4A) and eye2 cells, which also lack pigment granules (Supplemental Figure S1). Blocking of anti-EYE3-labeled wild-type cells with the immunizing peptide confirmed that the immunofluorescence signal was specific to EYE3 (Supplemental Figure S2). Edge-on micrographs of wild-type cells strengthened our hypothesis that the EYE3 signal emanates from the eyespot pigment granule layers in the chloroplast below the plasma-membrane–localized ChR1 photoreceptor (Figure 4B).
FIGURE 4:
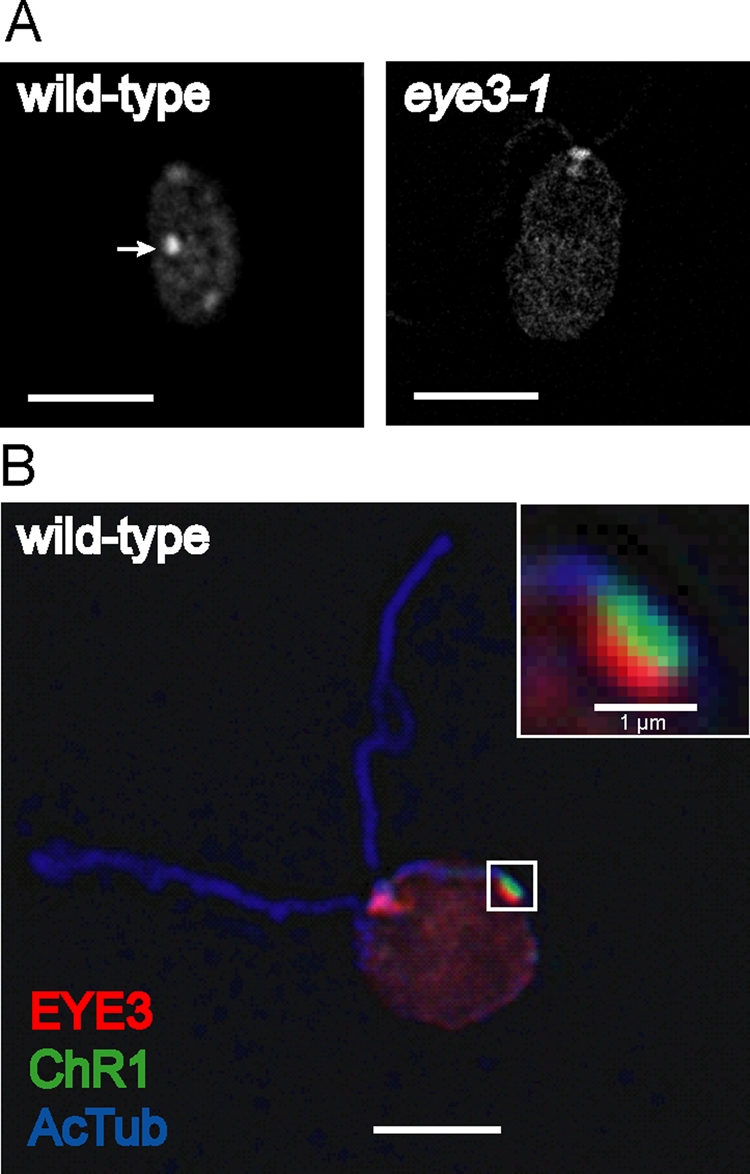
EYE3 localizes to eyespot pigment granule layers. (A) Immunofluorescence micrographs of wild-type C. reinhardtii cell (left) and eye3–1 cell (right) stained with antibody directed against EYE3. EYE3 localizes to the eyespot (arrow) in wild-type cell, whereas staining is absent in the eye3 mutant. Nonspecific staining in the basal body region is observed. (B) Combined immunofluorescence micrograph of a wild-type cell, showing localization of EYE3 to the pigment granule area of the eyespot. Inset highlights the distinct, yet closely apposed, pigment granule spot (red) and ChR1 photoreceptor patch (green) associated with the D4 microtubule rootlet (blue). Bars, 5 μm unless otherwise noted.
EYE2 localizes to the chloroplast envelope region in the eyespot
EYE2 is thought to be a chaperone-like protein required for eyespot assembly (Roberts et al., 2001). The EYE2 protein was present in a proteomic analysis of the eyespot and possesses a single predicted transmembrane domain (Schmidt et al., 2006). EYE2 has a chloroplast-targeting sequence as well as a LysM domain and thioredoxin active site motif (Supplemental Figure S3). Like the eye3 mutant, the eye2 mutant strains lack pigment granule arrays (Lamb et al., 1999). Polyclonal antiserum raised against EYE2, like that for EYE3, detected an eyespot-shaped patch in wild-type cells that was absent in an eye2 insertion mutant (Figure 5A). Given that EYE2 is required for assembly of the eyespot pigment granule layers, we expected that it, like EYE3, would be localized to this compartment. Surprisingly, immunofluorescence triple staining of wild-type cells with EYE2, EYE3, and ChR1 antisera revealed that EYE2 does not colocalize with EYE3 in the pigment granules, but is instead found in an area between the pigment granules and the plasma-membrane–localized photoreceptor patch (Figure 5B). This staining pattern suggests that EYE2 localizes to a specialized area of the chloroplast envelope in the eyespot.
FIGURE 5:
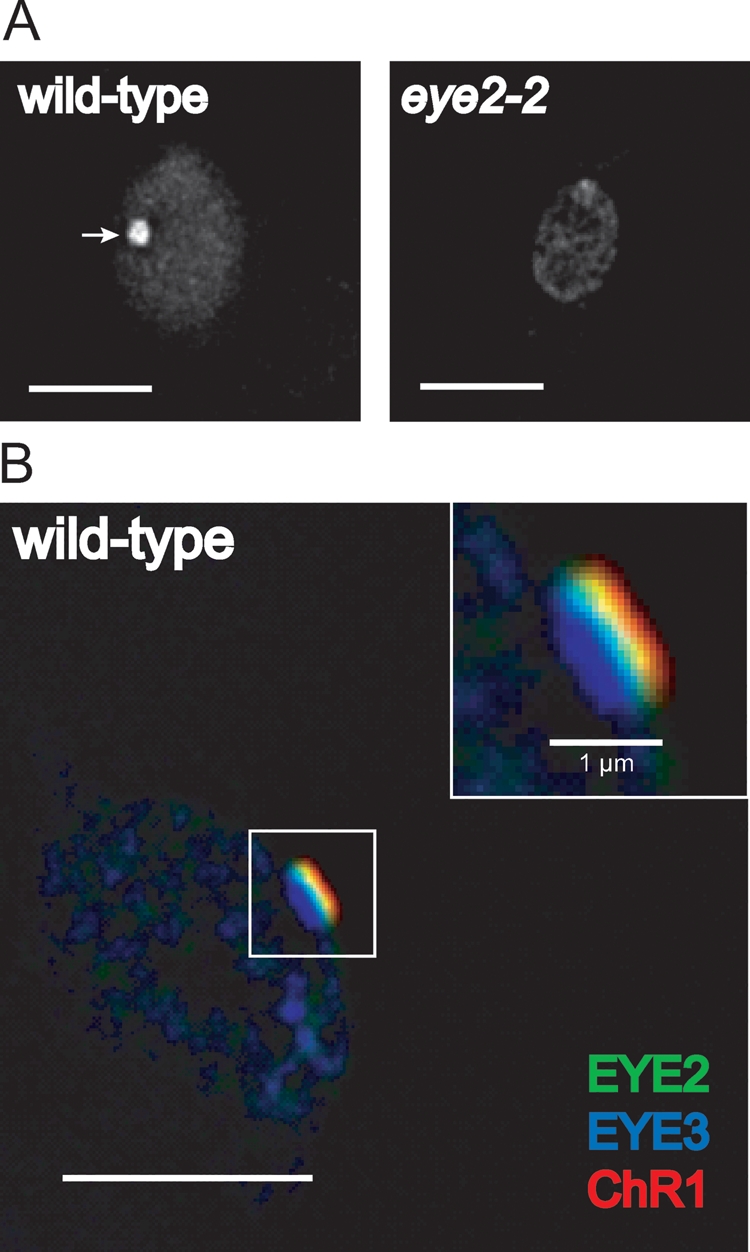
EYE2 localizes to a region corresponding to the chloroplast envelope in the eyespot. (A) Immunofluorescence micrographs of wild-type (left) and eye2–2 cells (right) stained with antibody directed against EYE2. EYE2 localizes to the eyespot (arrow) in wild-type cells, whereas staining is absent in the insertion mutant. (B) Combined immunofluorescence micrograph of wild-type C. reinhardtii cell stained with antibodies against EYE3 (blue), EYE2 (green), and ChR1 (red). Inset shows a close-up view of the eyespot area. EYE2 staining appears between the pigment granule layers and the plasma membrane–localized photoreceptor patch. Bars, 5 μm unless otherwise noted.
EYE2 copositions with ChR1 in eye3 and min1 cells
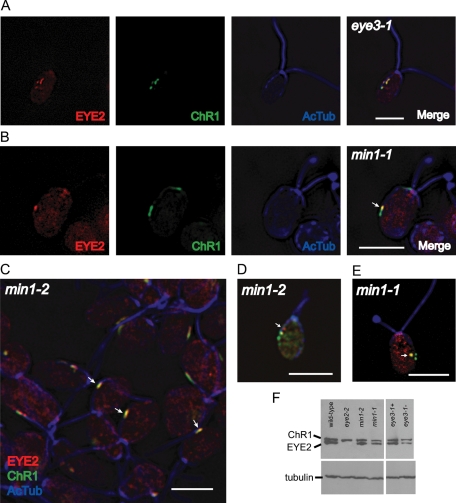
By Western blotting we discovered that anti-EYE2 antiserum detects EYE2 both in wild-type and eye3 mutant cells (Figure 6F). Because the absence of pigment granules markedly affects photoreceptor localization, we investigated whether the localization patterns of EYE2 are affected similarly in mutants lacking organized pigment granule arrays and whether EYE2 localization is dependent or independent of ChR1 localization. In both eye3 mutant cells, EYE2 consistently shared an aberrant localization pattern with the ChR1 photoreceptor on the track of, or in the proximity of, the D4 rootlet (Figure 6A), indicating that some sort of stable connection is maintained between EYE2 and ChR1.
FIGURE 6:
EYE2 protein shares aberrant localization patterns with ChR1 photoreceptor in eye3 and min1 mutant cells. (A) Immunofluorescence micrographs of a single eye3 (eyeless) cell labeled with antibodies against EYE2 (red), ChR1 (green), and acetylated α-tubulin (blue). Patches of EYE2 coposition with ChR1 patches and remain in the vicinity of the microtubule rootlet. (B) Immunofluorescence micrographs of a representative min1 cell showing EYE2-ChR1 copositioning in the area of the rootlet (arrow). EYE2-ChR1 copositioning does not require MIN1 protein. (C–E) Combined immunofluorescence micrographs of min1–2 (insertion mutant) and min1–1 cells stained for EYE2 (red), ChR1 (green), and acetylated α-tubulin (blue). (C) Combined immunofluorescence micrograph of a min1–2 field. Arrows indicate cells where EYE2 and ChR1 are copositioned (yellow). (D) Single min1–2 cell showing EYE2 spot (arrow) not copositioned with ChR1. (E) Single min1–1 cell showing EYE2 and ChR1 copositioned in a spot not associated with rootlet (arrow). Bars for all images: 5 μm. (F) Western blot of whole-cell extracts of auxotrophically grown wild-type, eye2, min1, and eye3 strains probed with anti-ChR1, anti-EYE2, and anti-tubulin. Levels of both ChR1 and EYE2 are lower in min1 and eye3 mutant strains in comparison to wild-type.
Our hypothesis had been that the MIN1 protein was required for the apposition of plasma membrane and chloroplast envelope in the eyespot during photoautotrophic growth. This hypothesis was based on electron micrographs showing lack of membrane apposition in sections containing aggregations of pigment granules in the chloroplast stroma (Lamb et al., 1999). If apposition of the chloroplast envelope and plasma membrane were required for the connection between EYE2 and photoreceptor, loss of MIN1 might be expected to abolish the copositioning of EYE2 and ChR1. Unexpectedly, in the majority of photoautotrophically grown min1 cells, EYE2 remained copositioned with ChR1 (Figure 6, B and C). Some min1 cells, however, exhibited discrete EYE2 patches that were not associated with ChR1 patches (Figure 6D). In cases where ChR1 patches were not on the rootlet track, EYE2 copositioning was still observed (Figure 6E), indicating that the copositioning phenomenon does not require interaction with the rootlet. MIN1 is known to play a role in stability of the ChR1 photoreceptor, as min1 cells exhibit reduced levels of ChR1 (Mittelmeier et al., 2008). Western blots of whole-cell extracts probed with anti-ChR1 and anti-EYE2 showed that levels of both proteins were reduced in min1 strains in comparison to wild-type cells (Figure 6F). Taken together, these results are consistent with the hypothesis that the photoreceptors or associated proteins, together with D4 microtubule rootlet–associated positioning cues, are responsible for the positioning, localization, and stability of EYE2 in the eyespot.
DISCUSSION
EYE3 localizes to and is required for formation of the eyespot pigment granule arrays
The layers of carotenoid-filled pigment granules in the eyespot function to enhance directional light perception by their shading and reflective properties. The granules are organized into regular closely packed arrays between chloroplast and thylakoid membranes. We have described the identification and initial characterization of the EYE3 gene, which encodes a serine/threonine protein kinase of the ABC1 family that localizes to, and is required for, assembly of the eyespot pigment granule arrays in C. reinhardtii. ABC1 kinases similar to EYE3 have been identified in Arabidopsis thaliana chloroplasts as components of plastoglobules, lipid-filled structures that blister from and are linked to the thylakoid membrane (Ytterberg et al., 2006; Bréhélin et al., 2007). The blistering events occur at curved margins of thylakoid membranes and are hypothesized to be induced by accumulation of carotenoids and mediated by carotenoid-binding proteins termed plastoglobulins (PAP/fibrillins) (Austin et al., 2006; Simkin et al., 2007). Eight putative proteins with a PAP/fibrillin domain were identified in the eyespot proteome (Renninger et al., 2006; Schmidt et al., 2006), seven of which have functional homologues in Arabidopsis plastoglobules (Ytterberg et al., 2006). The similarities between plastoglobules and eyespot pigment granules have led to the hypothesis that plastoglobules may be precursors of the pigment granules (Kreimer, 2009) and that ABC1 kinases may function in their biosynthesis. Analogous ABC1 kinases in Escherichia coli (Poon et al., 2000) and Saccharomyces cerevisiae (Do et al., 2001) are involved in ubiquinone biosynthesis. The enzymes of the ubiquinone biosynthesis pathway in S. cerevisiae associate to form complexes (Marbois et al., 2005, 2009), and stability of the Coq3 subunits of the Coq3/5/9 complex is dependent on the kinase Coq8 (Tauche et al., 2008). EYE3 may regulate the interactions of structural proteins via phosphorylation or may itself serve a structural role, as it is stably expressed in pigment granules. It is also possible that EYE3 functions in regulating quinone metabolism, but the absence of an observable growth defect in eye3 deletion mutants implies that such activity would be nonessential or redundant. Further studies assessing whether the kinase activity of EYE3 is required for eyespot assembly and determining the identity of kinase targets may help parse the mechanism of EYE3 function.
EYE2 localizes to an area corresponding to the chloroplast envelope in the eyespot
The EYE2 protein was found in the eyespot proteome (Schmidt et al., 2006) and possesses a LysM domain, a single transmembrane helix, and a thioredoxin active site. The role of EYE2 in eyespot assembly does not require the redox function of the EYE2 thioredoxin domain (Roberts et al., 2001). In wild-type C. reinhardtii eyespots, EYE2 localizes to an area between the pigment granule stacks and the plasma membrane–localized ChR1 photoreceptor, a region corresponding to an area of the chloroplast envelope. This localization pattern suggests a role for EYE2 in directing the site for assembly of the pigment granule arrays rather than in pigment granule biogenesis.
Our observation that EYE2 copositions with ChR1 in mutants lacking organized pigment granule arrays suggests an interaction or stable connection between these proteins. A direct connection would be supportive of a model placing EYE2 in the outer envelope, and freeze-fracture studies of C. reinhardtii eyespot membranes suggested eyespot-specific elaboration of the outer membrane (Melkonian and Robenek, 1980). EYE2, however, has a putative chloroplast transit peptide, which proteins destined for the outer membrane typically do not possess (Strittmatter et al., 2010). Additionally, in sequence alignments, EYE2 clusters with chloroplast stromal adenylyl sulfate reductases (Roberts et al., 2001), suggesting that EYE2 may be an inner envelope protein with its thioredoxin site positioned on the stromal side. Association of EYE2 with the inner envelope would be consistent with a model in which EYE2 directs the site for pigment granule array assembly. Further biochemical studies will be needed to determine the membrane association of EYE2 in the chloroplast envelope region. The N-terminal LysM domain of EYE2 likely functions to mediate protein–protein interactions necessary for the formation and stabilization of the association between the chloroplast envelope and plasma membrane in the eyespot.
Eyespot pigment granule arrays are necessary for maintaining the supramolecular organization of the eyespot
The altered localization patterns of both the ChR1 photoreceptor and EYE2 in the eyeless mutant eye3 indicate that the eyespot pigment granules, or factors associated with the granules, act to maintain the elliptical shape and integrity of both the EYE2 patch and the photoreceptor patch. Mediatory structural connections are likely responsible for keeping the specialized regions of chloroplast envelope and plasma membrane together in a single integrated unit. Surprisingly, although patches of EYE2 were sometimes observed without associated ChR1, we found that copositioning of EYE2 and ChR1 patches was maintained in the vast majority of min1 mutant cells, which were previously characterized as lacking apposition of plasma membrane with chloroplast envelope (Lamb et al., 1999; Mittelmeier et al., 2008). This conclusion was drawn from observations of eyespot pigment granules by electron microscopy in thin sections of cells grown without acetate. It is possible that the pigment granules observed in electron micrographs were not near the area of apposed eyespot membranes. Eyespot organization in min1 cells is restored during mixotrophic growth with acetate (Lamb et al., 1999), indicating that other structural proteins function redundantly with MIN1 under certain nutrient conditions. These unknown proteins may be responsible for the continued association of EYE2 and ChR1 observed in min1 cells.
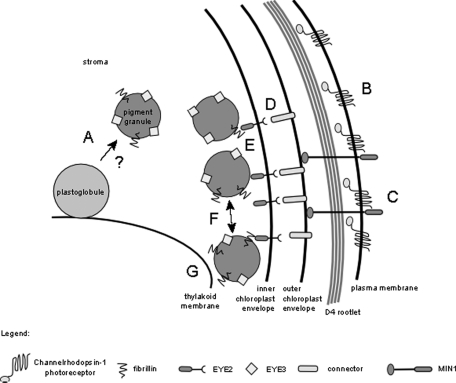
In both eye3 and min1 mutants, copositioned patches of EYE2 and ChR1 were most often observed in proximity to the D4 microtubule rootlet. The lack of pigment granules, or organized arrays of the pigment granules, does not affect the asymmetric localization of ChR1 or EYE2 to the daughter side of the cell. The rootlet association evident in staining patterns of EYE2 in eye3 and min1 mutants suggests that the localization of chloroplastic components of the eyespot is also in some way directed by the D4 rootlet. The data presented here are consistent with a working model wherein the stable ultrastructure of the eyespot is formed in a “top-down” cascade with rootlet-directed factors, likely including photoreceptor molecules, recruiting and facilitating coalescence of other components in the chloroplast (Figure 7). In this model, photoreceptor molecules are guided to the site of eyespot assembly by interaction with the D4 microtubule rootlet. MIN1 stabilizes the photoreceptors in the plasma membrane, and other membrane-spanning proteins are recruited to the area, forming a stable connection between the chloroplast envelope and plasma membrane. EYE2 forms a specialized patch on the chloroplast envelope, shown in this model associated with the inner membrane, where it nucleates formation of the pigment granule arrays, which self-assemble via interactions with associated fibrillin proteins. EYE3 plays a role in formation and stabilization of the pigment granules from plastoglobules in the chloroplast. Additional interactions are established between the arrays of pigment granules and thylakoid membrane, resulting in the regular stacked arrangement of the layers in the eyespot.
FIGURE 7:
Model of eyespot assembly. EYE3 is required for biogenesis of the carotenoid-filled pigment granules, which are possibly formed from thylakoid membrane–associated plastoglobules (A). Photoreceptor molecules are guided by the D4 microtubule rootlet to the location where the eyespot is to be assembled (B) and where they aggregate and are stabilized by proteins such as MIN1 (C). Positioning of eyespot proteins “downstream” of the photoreceptor is mediated by hypothetical connecting proteins (D) that act as a link between molecules in the plasma membrane and chloroplast envelope proteins. EYE2 is positioned in the chloroplast envelope and nucleates the pigment granule arrays (E), which then self-assemble in a closely packed arrangement (F) and associate with thylakoid membranes via other protein–protein interactions (G).
The development of the complex supramolecular structures that comprise the eyespot apparatus depends on upstream cues to direct the placement of components, ensure protein stability, and establish close contact between membrane layers in both the chloroplast and plasma membrane. It would be informative to observe the localization patterns of EYE2, EYE3, and ChR1 photoreceptor in cells early postcytokinesis to ascertain probable cause–effect relationships in the eyespot assembly process.
MATERIALS AND METHODS
Chlamydomonas strains and media
Chlamydomonas strains (Table 2) were maintained on solid Tris–acetate–phosphate (TAP) medium or TAP supplemented with 0.2 mg/ml arginine (for arginine auxotrophic strains). Liquid cultures were grown in modified Sager and Granick medium I with added Hutner’s trace elements (R medium) or without acetate (M medium) (Harris, 1989) or R medium containing 0.1% sodium acetate limited for NH4NO3 (0.125 mM) and supplemented with 0.2 mg/ml arginine (RNA medium).
TABLE 2:
Chlamydomonas strains used in this study.
| Strain name | Genotype | Reference |
|---|---|---|
| 137c mt+ | Wild-type mt+ | Harris (1989) |
| 12–18 | eye3–1 mt+ | Lamb et al. (1999) |
| H9–8 | eye2–2::ARG7 arg7–8 mt− | Roberts et al. (2001) |
| F35 | eye3–2::NIT1 mt+ | This study |
| 12–12 | min1–1 mt+ | Lamb et al. (1999) |
| H6–2 | min1–2::ARG7arg7–8 mt+ | Mittelmeier et al. (2008) |
| arg2 | arg7–8 mt+ | Harris (1989) |
| arg7 | arg7–2 mt− | Harris (1989) |
| 12–18 arg2 mt− | eye3–1 arg7–8 mt− | This study |
| 12–18 arg2 mt+ | eye3–1 arg7–8 mt+ | This study |
Chlamydomonas transformation
Chlamydomonas strain 12–18 arg2 mt+ was grown for 3 d in liquid RNA medium at 25°C under continuous light (∼20 μmol/m2) and harvested by centrifugation (5 min, 510 × g), resuspended, and grown overnight in 200 ml of low-nitrogen medium (Harris, 1989). Cells were resuspended in R medium to an approximate density of 3.0 × 107 cells/ml and then transformed with ∼10 μg of linearized plasmid DNA using the silicon carbide whisker method of Dunahy (1993) with modifications described in Mittelmeier et al. (2008). Transformants were picked after approximately 1 wk of incubation at 25°C under continuous illumination under 38 μmol/m2 fluorescent light.
Identification of EYE3 genomic sequence
Strain 12–18 (eye3–1) and NIT1 insertion strain F35 (eye3–2) were isolated from screens for ptx-defective mutants (see Results). Mating an eyeless arg7 mt+ strain (obtained from a cross of F35 to strain arg7–2 mt−, CC-1820) to the strain eye3–1 arg2 mt− yielded ptx− diploids (n = 10), whereas mating the same strain to strain arg2 mt− (CC-49) yielded ptx+ diploids (n = 10). A probe from the 5′ end of NIT1 identified a 0.9-kb fragment on a Southern blot of PvuII-digested eye3–2 genomic DNA that was not present in PvuII-digested genomic DNA from wild-type strain 137c (CC-125). To obtain a sequence from the 0.9-kb fragment, PvuII-digested eye3–2 genomic DNA was self-ligated and used as a template in PCR amplification with the nit5–675 and nit5–683 oligonucleotide primers (Supplemental Table S1). The resulting 0.9-kb product comprised 250 nucleotides of novel sequence adjacent to NIT1 sequence. Oligonucleotides F35–1A and F35–1B (Supplemental Table S1), derived from the sequence adjacent to NIT1 in strain eye3–2, were used to identify cosmid 14–1 from a pARG7.8cos library (Purton and Rochaix, 1995) for gene rescue experiments. Following transformation of strain eye3–1 arg2 mt+ with linearized 14–1, 2.5% of Arg+ transformants (n = 160) were phototactic, and light microscopy confirmed the presence of eyespots in transformed cells (Table 3). A 6.9-kb restriction fragment obtained from a partial BamHI digest of cosmid 14–1 was ligated to plasmid ARG7.8 (Debuchy et al., 1989) to create plasmid EYE3-B3. Following transformation of 12–18 arg2 mt+ with linearized pEYE3-B3, ∼4.0% (n = 288) of Arg+ transformants were phototactic and had at least partial restoration of eyespots (Table 3).
TABLE 3:
Phenotypes of transformants.
| Construct | Strain | No. of Arg+ transformants | No. of Ptx+ transformants | Ptx+ transformants with eyespots? |
|---|---|---|---|---|
| 14–1 | 12–18 arg2 | 160 | 4 | Yes |
| pEYE3-B3 | 12–18 arg2 | 288 | 12 | Yes |
A Basic Local Alignment Search Tool (BLAST) search of the Chlamydomonas genome identified the EYE3 sequence within a predicted gene on chromosome 2 (JGI Chlamydomonas version 4.0 protein ID: 205767; PubMed GeneID 10511552). The putative 1066-residue EYE3 protein has an estimated molecular weight of 113.5 kDa and a theoretical isoelectric point (pI) of 5.56. The sequence is hydrophobic overall, with a grand average of hydropathicity (GRAVY) of 0.110. EYE3 contains an N-terminal chloroplast-targeting sequence as predicted by the ChloroP 1.1 program (Emanuelsson et al., 1999; accessed at http://www.cbs.dtu.dk/services/ChloroP/). To identify the mutation in strain eye3–1, oligonucleotides throughout EYE3 were used as primers to amplify sequences from eye3–1 genomic DNA, and the PCR products were sequenced. When compared with the Chlamydomonas genome, primer pairs A3500/S4300 and A4100/F35–1A (Supplemental Table S1) yielded PCR products that contained a single nucleotide change: A G→A transition at nucleotide 3015 of the EYE3 gene predicted to change the wild-type UGG tryptophan codon to a premature UGA stop codon.
Generation of antisera
A rabbit polyclonal antiserum was raised against a TrpE-fusion construct containing residues 75–180 of the EYE2 sequence (for methods, see Weber and Dieckmann, 1990). A rabbit polyclonal antiserum was raised against a peptide epitope of EYE3 (residues 565–572) and purified by affinity chromatography (Pacific Immunology, La Jolla, CA).
Protein sequence analysis
Analysis of amino acid sequences was conducted using the ProtParam tool (Swiss Institute of Bioinformatics Expert Protein Analysis System [ExPASy]; http://us.expasy.org/tools/protparam.html). Sequences in Figure 3 were aligned using ClustalW (Thompson et al., 1994) and were shaded using the Multiple Align Show program (http://www.bioinformatics.org/sms/multi-align.html). The phylogenetic tree was constructed by the neighbor-joining method using the QuickTree program (Howe et al., 2002; accessed at http://sanger.ac.uk/resources/software/quicktree) and drawn using the Phylodendron program (accessed at http://iubio.bio.indiana.edu/treeapp). Bootstrap values were calculated with 100 iterations.
Ptx assays
Following overnight growth in M medium, cultures were screened for phototactic ability using the assay described in Lamb et al. (1999).
Bright field microscopy
Cells from overnight cultures grown in M medium were prepared and viewed according to the protocol described in Mittelmeier et al. (2008).
Immunofluorescence microscopy
Cells were fixed essentially according to the protocol described in Mittelmeier et al. (2008), except that samples were incubated with appropriate primary and secondary antibodies as noted later in the text and coverslipped with Mowiol 4–88 mounting medium (Calbiochem, La Jolla, CA). Polyclonal rabbit anti-ChR1 serum (gift of P. Berthold and P. Hegemann (Humboldt-Universität zu Berlin, Berlin, Germany) and anti-EYE2 serum were purified using a Melon Gel immunoglobulin (Ig)G spin purification kit (Pierce, Rockford, IL) at a dilution of 1:10 (vol/vol). For fluorescence microscopy, EYE2, EYE3, and ChR1 antibodies were directly conjugated to fluorophores (Alexa Fluor 488, Alexa 594, or allophycocyanin) using Zenon rabbit IgG labeling kits (Invitrogen, Carlsbad, CA) according to the protocol provided by the manufacturer. Monoclonal anti-acetylated α-tubulin (Clone 6–11B-1; Sigma, St. Louis, MO) was detected with goat anti–mouse secondary antibodies conjugated to Alexa Fluor 568 at a dilution of 1:1000 (vol/vol) (images in Figure 2) or Cy-5 at a dilution of 1:200 vol/vol (Figures 3 and 6) (Molecular Probes, Eugene, OR). Samples were viewed on a DeltaVision RT inverted epifluorescence microscope (Applied Precision, Eugene, OR) with an Olympus TH4 1.4 numerical aperture 100× objective with a 1.6 optivar at 24°C using appropriate filters. Images were captured using a CoolSnap HQ CCD camera (Photometrics, Tucson, AZ), deconvolved using the SoftWorx imaging program (Applied Precision), and combined in National Institutes of Health (NIH) ImageJ.
Peptide blocking
Solutions of affinity-purified anti-EYE3 (0.08 mg/ml in blocking buffer [1× phosphate-buffered saline + 1% bovine serum albumin]) were incubated at room temperature for 30 min with continuous agitation either with or without 1 mg/ml immunizing peptide. Wild-type cells prepared for immunofluorescence as described earlier in the text were incubated in either anti-EYE3 (0.08 mg/ml) or anti-EYE3 blocked with immunizing peptide and labeled with 1:1500 (vol/vol) donkey anti–rabbit Alexa Fluor 488 (Molecular Probes) secondary for visualization.
Immunoblotting
SDS–PAGE and immunoblotting were carried out according to the protocol described in Mittelmeier et al. (2008).
DNA sequencing
Automated DNA sequencing was performed at the DNA sequencing facility of the Laboratory of Molecular Systematics and Evolution, University of Arizona (Tucson, AZ).
Oligonucleotide synthesis
The oligonucleotides used in this study are listed in Supplemental Table S1. Oligonucleotides were synthesized by SigmaGenosys Biotechnologies (The Woodlands, TX).
Construction of figures
Figures were prepared using Adobe Illustrator (Adobe Systems, San Jose, CA) and Microsoft Word (Microsoft, Redmond, WA). Micrographs were minimally adjusted for brightness and contrast using NIH ImageJ software, cropped in Adobe Photoshop, and reduced from the original size in Adobe Illustrator. Figure 7 was drawn in Inkscape (open-source vector drawing program) and exported to Adobe Illustrator.
Supplementary Material
Acknowledgments
We thank Carl Boswell for assistance with microscopy and Kylie Swisher for comments on the manuscript. Melissa Schonauer and members of Roy Parker’s laboratory at the University of Arizona provided helpful comments and support during the course of this project. Michael Pham conducted the experiments to determine the mutation in strain eye3–1. George Witman and Greg Pazour provided the eye3–2 insertion strain. Douglas Roberts constructed the TrpE fusion for production of the EYE2 antiserum. Peter Hegemann and Peter Berthold provided the ChR1 antiserum. This work was supported by National Science Foundation Grant MCB-0843094 (to C. L. D.) and National Institutes of Health Graduate Training Grant in Biochemistry and Molecular Biology T32GM008659 (to J. S. B.).
Abbreviations used:
- AKC
ABC1 kinase in the chloroplast
- ChR
channelrhodopsin
- D4
daughter four-membered
- ptx
phototaxis
- TAP
Tris–acetate–phosphate
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E10-11-0918) on March 3, 2011.
REFERENCES
- Austin JR, Frost E, Vidi P-A, Kessler F, Staehelin LA. Plastoglobules are lipoprotein subcompartments of the chloroplast that are permanently coupled to thylakoid membranes and contain biosynthetic enzymes. Plant Cell. 2006;18:1693–1703. doi: 10.1105/tpc.105.039859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bateman A, Bycroft M. The structure of a LysM domain from E. coli membrane-bound lytic murein transglycosylase D (MltD) J Mol Biol. 2000;299:1113–1119. doi: 10.1006/jmbi.2000.3778. [DOI] [PubMed] [Google Scholar]
- Berthold P, Tsunoda SP, Ernst OP, Mages W, Gradmann D, Hegemann P. Channelrhodopsin-1 initiates phototaxis and photophobic responses in Chlamydomonas by immediate light-induced depolarization. Plant Cell. 2008;20:1665–1677. doi: 10.1105/tpc.108.057919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bréhélin C, Kessler F, van Wijk KJ. Plastoglobules: versatile lipoprotein particles in plastids. Trends Plant Sci. 2007;12:260–266. doi: 10.1016/j.tplants.2007.04.003. [DOI] [PubMed] [Google Scholar]
- Debuchy R, Purton S, Rochaix J-D. The arginosuccinate lyase gene of Chlamydomonas reinhardtii: an important tool for molecular transformation and for correlating the genetic and molecular maps of the ARG7 locus. EMBO J. 1989;8:2803–2809. doi: 10.1002/j.1460-2075.1989.tb08426.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dieckmann CL. Eyespot placement and assembly in the green alga Chlamydomonas. BioEssays. 2003;25:410–416. doi: 10.1002/bies.10259. [DOI] [PubMed] [Google Scholar]
- Dunahy TG. Transformation of Chlamydomonas reinhardtii with silicon carbide whiskers. BioTechniques. 1993;15:452–460. [PubMed] [Google Scholar]
- Do TQ, Hsu AY, Jonanssen T, Lee PT, Clarke CF. A defect in coenzyme Q biosynthesis is responsible for the respiratory deficiency in S. cerevisiae abc1 mutants. J Biol Chem. 2001;276:18161–18168. doi: 10.1074/jbc.M100952200. [DOI] [PubMed] [Google Scholar]
- Emanuelsson O, Nielsen H, von Heijne G. ChloroP, a neural network-based method for predicting chloroplast transit peptides and their cleavage sites. Protein Science. 1999;8:978–984. doi: 10.1110/ps.8.5.978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foster KW, Smyth RD. Light antennas in phototactic algae. Microbiol Rev. 1980;44:572–630. doi: 10.1128/mr.44.4.572-630.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris EH. The Chlamydomonas Sourcebook: A Comprehensive Guide to Biology and Laboratory Use. San Diego: Academic Press; 1989. [DOI] [PubMed] [Google Scholar]
- Hegemann P, Neumeier K, Hegemann U, Kuehnle E. The role of calcium in Chlamydomonas photomovement responses as analysed by calcium channel inhibitors. Photochem Photobiol. 1990;52:575–583. doi: 10.1111/j.1751-1097.1990.tb01802.x. [DOI] [PubMed] [Google Scholar]
- Holmes JA, Dutcher SK. Cellular asymmetry in Chlamydomonas reinhardtii. J Cell Sci. 1989;94:273–285. doi: 10.1242/jcs.94.2.273. [DOI] [PubMed] [Google Scholar]
- Howe K, Bateman A, Durbin R. QuickTree: building huge neighbour-joining trees of protein sequences. Bioinformatics. 2002;18:1546–1547. doi: 10.1093/bioinformatics/18.11.1546. [DOI] [PubMed] [Google Scholar]
- Kamiya R, Witman G. Submicromolar levels of calcium control the balance of beating between the two flagella in demembranated models of Chlamydomonas. J Cell Biol. 1984;98:97–107. doi: 10.1083/jcb.98.1.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knogge W, Scheel D. LysM receptors recognize friend and foe. Proc Natl Acad Sci USA. 2006;103:10829–10830. doi: 10.1073/pnas.0604601103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kreimer G. Cell biology of phototaxis in flagellated green algae. Int Rev Cytol. 1994;148:229–310. [Google Scholar]
- Kreimer G. The green algal eyespot apparatus: a primordial visual system and more? Curr Genetics. 2009;55:19–43. doi: 10.1007/s00294-008-0224-8. [DOI] [PubMed] [Google Scholar]
- Lamb MR, Dutcher SK, Worley CK, Dieckmann CL. Eyespot-assembly mutants in Chlamydomonas reinhardtii. Genetics. 1999;153:721–729. doi: 10.1093/genetics/153.2.721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marbois B, Gin P, Faull KF, Poon WW, Lee PT, Strahan J, Shepherd JN, Clarke CF. Coq3 and Coq4 define a polypeptide complex in yeast mitochondria for the biosynthesis of coenzyme Q. J Biol Chem. 2005;280:20231–20238. doi: 10.1074/jbc.M501315200. [DOI] [PubMed] [Google Scholar]
- Marbois B, Gin P, Gulmeziana M, Clarke CF. The yeast Coq4 polypeptide organizes a mitochondrial protein complex essential for coenzyme Q biosynthesis. Biochim Biophys Acta. 2009;1791:69–75. doi: 10.1016/j.bbalip.2008.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melkonian M, Robenek H. Eyespot membranes of Chlamydomonas reinhardtii: a freeze-fracture study. J Ultrastruct Res. 1980;72:90–102. doi: 10.1016/s0022-5320(80)90138-0. [DOI] [PubMed] [Google Scholar]
- Melkonian M, Robenek H. The eyespot apparatus of flagellated green algae: a critical review. In: Round FE, Chapman DJ, editors. In: Progress in Phycological Research. vol. 3. Bristol:: Biopress,; 1984. pp. 193–268. [Google Scholar]
- Mittelmeier TM, Berthold P, Danon A, Lamb MR, Levitan A, Rice M, Dieckmann CL. C2 domain protein MIN1 promotes eyespot organization in Chlamydomonas reinhardtii. Eukaryotic Cell. 2008;7:2100–2112. doi: 10.1128/EC.00118-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merchant SS. The Chlamydomonas genome reveals the evolution of key animal and plant functions. Science. 2007;318:245–251. doi: 10.1126/science.1143609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morel-Laurens NML, Feinleib ME. Photomovement in an “eyeless” mutant of Chlamydomonas. Photochem Photobiol. 1983;37:189–194. [Google Scholar]
- Nagel G, Ollig D, Fuhrmann M, Kateriya S, Musti AM, Bamberg E, Hegemann P. Channelrhodopsin-1: a light-gated proton channel in green algae. Science. 2002;296:2395–2398. doi: 10.1126/science.1072068. [DOI] [PubMed] [Google Scholar]
- Nagel G, Szellas T, Huhn W, Kateriya S, Adeishvili N, Berthold P, Ollig D, Hegemann P, Bamberg E. Channelrhodopsin-2, a directly light-gated cation-selective membrane channel. Proc Natl Acad Sci USA. 2003;100:13940–13945. doi: 10.1073/pnas.1936192100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nultsch W. The photocontrol of movement in Chlamydomonas. Symp Soc Exp Biol. 1983;36:521–539. [PubMed] [Google Scholar]
- Pazour GJ, Sineshchekov OA, Witman GB. Mutational analysis of the phototransduction pathway of Chlamydomonas reinhardtii. J Cell Biol. 1995;131:427–440. doi: 10.1083/jcb.131.2.427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poon WW, Davis DE, Ha HT, Jonassen T, Rather PN, Clarke CF. Identification of Escherichia coli ubiB, a gene required for the first monooxygenase step in ubiquinone biosynthesis. J Bacteriol. 2000;182:5139–5146. doi: 10.1128/jb.182.18.5139-5146.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purton S, Rochaix J-D. Characterization of the ARG7 gene of Chlamydomonas reinhardtii and its application to nuclear transformation. Eur J Phycol. 1995;30:141–148. [Google Scholar]
- Renninger S, Dieckmann CL, Kreimer G. Toward a protein map of the green algal eyespot: analysis of eyespot globule-associated proteins. Phycologia. 2006;45:199–212. [Google Scholar]
- Roberts DGW. 1999. Characterization of the EYE2 gene required for eyespot assembly in Chlamydomonas reinhardtii. PhD dissertation. University of Arizona, Tucson, AZ.
- Roberts DGW, Lamb MR, Dieckmann CL. Characterization of the EYE2 gene required for eyespot assembly in Chlamydomonas reinhardtii. Genetics. 2001;158:1037–1049. doi: 10.1093/genetics/158.3.1037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt M, et al. Proteomic analysis of the eyespot of Chlamydomonas reinhardtii provides novel insights into its components and tactic movements. Plant Cell. 2006;18:1908–1930. doi: 10.1105/tpc.106.041749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simkin AJ, Gaffé J, Alcaraz J-P, Carde J-P, Bramley PM, Fraser PD, Kuntz M. Fibrillin influence on plastid ultrastructure and pigment content in tomato fruit. Phytochemistry. 2007;68:1545–1556. doi: 10.1016/j.phytochem.2007.03.014. [DOI] [PubMed] [Google Scholar]
- Sineshchekov OA, Jung KH, Spudich JL. Two rhodopsins mediate phototaxis to low- and high-intensity light in Chlamydomonas reinhardtii. Proc Natl Acad Sci USA. 2002;99:8689–8694. doi: 10.1073/pnas.122243399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strittmatter P, Soll J, Bölter B. The chloroplast import machinery: a review. Methods Mol Biol. 2010;619:307–321. doi: 10.1007/978-1-60327-412-8_18. [DOI] [PubMed] [Google Scholar]
- Tauche A, Krause-Buchholz U, Rödel G. Ubiquinone biosynthesis in S. cerevisiae: the molecular organization of O-methylase Coq3p depends on Abc1p/Coq8p. FEMS Yeast Res. 2008;8:1263–1275. doi: 10.1111/j.1567-1364.2008.00436.x. [DOI] [PubMed] [Google Scholar]
- Thompson JD, Higgins DG, Gibson TJ. Improved sensitivity of profile searches through the use of sequence weights and gap extension. Comput Appl Biosci. 1994;10:19–29. doi: 10.1093/bioinformatics/10.1.19. [DOI] [PubMed] [Google Scholar]
- Weber ER, Dieckmann CL. Identification of the CBP1 polypeptide from mitochondrial extracts of S. cerevisiae. J Biol Chem. 1990;265:1594–1600. [PubMed] [Google Scholar]
- Witman GB. Chlamydomonas phototaxis. Trends Cell Biol. 1993;3:403–408. doi: 10.1016/0962-8924(93)90091-e. [DOI] [PubMed] [Google Scholar]
- Ytterberg AJ, Peltier J-B, van Wijk KJ. Protein profiling of plastoglobules in chloroplasts and chromoplasts. A surprising site for differential accumulation of metabolic enzymes. Plant Physiol. 2006;140:984–997. doi: 10.1104/pp.105.076083. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.