Members of chromosome passenger complex BIR1 and SLI15 suppress the chromosome segregation defect of bub1Δ and sgo1Δ. Neither Bub1 nor Sgo1 is required for CPC activity. This study found genetic interaction between Mps1 and Sgo1. Mps1 governs localization of Sgo1. Bub1, Sgo1, and Mps1 function in parallel to the CPC in correction of syntelic attachments.
Abstract
The conserved mitotic kinase Bub1 performs multiple functions that are only partially characterized. Besides its role in the spindle assembly checkpoint and chromosome alignment, Bub1 is crucial for the kinetochore recruitment of multiple proteins, among them Sgo1. Both Bub1 and Sgo1 are dispensable for growth of haploid and diploid budding yeast, but they become essential in cells with higher ploidy. We find that overexpression of SGO1 partially corrects the chromosome segregation defect of bub1Δ haploid cells and restores viability to bub1Δ tetraploid cells. Using an unbiased high-copy suppressor screen, we identified two members of the chromosomal passenger complex (CPC), BIR1 (survivin) and SLI15 (INCENP, inner centromere protein), as suppressors of the growth defect of both bub1Δ and sgo1Δ tetraploids, suggesting that these mutants die due to defects in chromosome biorientation. Overexpression of BIR1 or SLI15 also complements the benomyl sensitivity of haploid bub1Δ and sgo1Δ cells. Mutants lacking SGO1 fail to biorient sister chromatids attached to the same spindle pole (syntelic attachment) after nocodazole treatment. Moreover, the sgo1Δ cells accumulate syntelic attachments in unperturbed mitoses, a defect that is partially corrected by BIR1 or SLI15 overexpression. We show that in budding yeast neither Bub1 nor Sgo1 is required for CPC localization or affects Aurora B activity. Instead we identify Sgo1 as a possible partner of Mps1, a mitotic kinase suggested to have an Aurora B–independent function in establishment of biorientation. We found that Sgo1 overexpression rescues defects caused by metaphase inactivation of Mps1 and that Mps1 is required for Sgo1 localization to the kinetochore. We propose that Bub1, Sgo1, and Mps1 facilitate chromosome biorientation independently of the Aurora B–mediated pathway at the budding yeast kinetochore and that both pathways are required for the efficient turnover of syntelic attachments.
INTRODUCTION
Proper chromosome segregation is essential for successful cell division. Eukaryotic cells do not explicitly monitor the fate of individual chromosomes during mitosis; rather they have evolved elaborate surveillance mechanisms to recognize and correct errors in microtubule-kinetochore attachments. These mechanisms are so efficient that under normal conditions wild-type cells missegregate their chromosomes once in thousands of cell divisions. This is not true, however, for polyploid cells, which contain more than two chromosome sets. In animals and fungi, tetraploids (cells with four copies of each chromosome) missegregate their chromosomes at high rates, producing aneuploid cells (Mayer and Aguilera, 1990; Andalis et al., 2004; Fujiwara et al., 2005; Storchová et al., 2006; Ganem et al., 2009). Indeed, it has been suggested that cancer cells, which frequently exhibit abnormal karyotypes, might in some cases be the descendents of unstable tetraploid cells, whose elevated rate of chromosome missegregation might facilitate transformation (Shackney et al., 1989; Andalis et al., 2004; Storchova and Pellman, 2004; Fujiwara et al., 2005; Margolis, 2005; Storchová and Kuffer, 2008).
We have previously reported the genetic phenomenon of “ploidy-specific lethality” in budding yeast, in which a deletion of a particular gene kills tetraploid cells even though haploids and diploids bearing the null allele are viable (Lin et al., 2001). Using a genome-wide screening approach, we identified a set of 39 gene deletions that cause ploidy-specific lethality (Storchová et al., 2006). Interestingly, nearly all of these genes have known roles in chromosome segregation or the repair of DNA breaks, further supporting the idea that the maintenance of genetic stability is the principal cellular function compromised by polyploidization.
The proteins that are essential to tetraploids but not diploids likely participate in pathways that are either directly impaired or under increased demand following a whole-genome duplication. Because tetraploidy increases the severity of certain mutant phenotypes, we anticipated that ploidy-specific lethality could be exploited to facilitate the genetic analysis of factors with poorly understood roles in chromosome segregation. One tetraploid-essential gene, BUB1, encodes a conserved serine-threonine kinase that is required for the activity of the spindle assembly checkpoint (SAC), which delays anaphase until all kinetochores form stable attachments to microtubules (Hoyt et al., 1991). In yeast and vertebrate cells, Bub1 binds the SAC protein Bub3 (which is also essential in tetraploids [Storchová et al., 2006]), targets other checkpoint proteins to unattached kinetochores, and forms a complex with Mad1 in the presence of a checkpoint signal (Musacchio and Salmon, 2007). These interactions are essential for inactivation of the anaphase promoting complex in the presence of microtubule-attachment errors (for a review, see Musacchio and Salmon, 2007). The finding that loss of Bub1 is lethal to tetraploid yeast raised the possibility that the SAC is required for the viability of these cells, in contrast to budding yeast with normal ploidy. However, this possibility can be excluded because mad1Δ and mad2Δ tetraploids, which are similarly defective for the SAC, are viable (Storchová et al., 2006). The requirement for Bub1 in tetraploids is consistent with reports of a SAC-independent function in chromosome segregation that requires its kinase domain (Warren et al., 2002; Fernius and Hardwick, 2007).
A checkpoint-independent requirement for Bub1 in the maintenance of genetic stability may be attributable, at least in part, to its conserved role in targeting the shugoshin proteins (Sgo1 in budding yeast) to the centromere (Kitajima et al., 2005; Fernius and Hardwick, 2007). It was recently discovered that shugoshin centromere recruitment requires the phosphorylation of the histone H2A by Bub1 (Kawashima et al., 2010). Deletion of SGO1 also results in ploidy-specific lethality (Storchová et al., 2006). In organisms from yeast to man, shugoshin prevents premature separation of sister chromatids in meiosis (and mitosis in vertebrates) by recruiting protein phosphatase 2A (PP2A), which in turn promotes the retention of centromeric cohesin by antagonizing the function of mitotic kinases (Wang and Dai, 2005; Riedel et al., 2006). However, it appears that budding yeast Sgo1, despite being essential for accurate chromosome segregation, is dispensable for the protection of centromeric cohesin in mitosis (Katis et al., 2004; Indjeian et al., 2005).
Instead, Sgo1 has been shown to play a role in the establishment of sister-chromatid biorientation—that is, the tension-generating attachment of sister chromatids to microtubules emanating from opposite spindle poles (also called “bipolar attachment”). Such biorientation is a necessary condition for sister chromatids to segregate evenly between daughter cells. In normal cells, improper attachments of both sisters to the same spindle pole—syntelic attachments—are recognized by proteins of the chromosomal passenger complex (CPC) and disrupted by the activity of its effector kinase Aurora B (Ipl1 in budding yeast). This release of the syntelic attachments activates the SAC and allows bipolar attachments to form in their place (Biggins and Murray, 2001; Tanaka et al., 2002; Pinsky et al., 2006). However, sgo1Δ mutants enter anaphase with an elevated frequency of unrepaired syntelic attachments and fail to arrest under experimental conditions that abolish kinetochore tension (Indjeian et al., 2005; Indjeian and Murray, 2007). Cells that lack Bub1 kinase activity do not properly localize Sgo1 to the kinetochore and hence exhibit similar defects in biorientation (Fernius and Hardwick, 2007; Indjeian and Murray, 2007). However, the precise mechanism by which Sgo1 (and, in turn, its upstream regulator Bub1) promotes the attachment of sister kinetochores to opposing spindle poles remains unknown. Furthermore, given its role in the centromeric recruitment of PP2A (Riedel et al., 2006), it is likely that budding yeast Sgo1 functions in multiple mitotic processes. Hence, the reason for Sgo1’s importance for tetraploid viability is unclear.
In the experiments reported here, we have characterized the functions of Bub1 and Sgo1 that are essential for the growth of tetraploid budding yeast. By identifying genetic suppressors that can restore viability to bub1Δ and sgo1Δ tetraploids, we first determined the specific mitotic defects that cause the lethality of these cells and second gained insight into the function of Bub1 and Sgo1 in chromosome segregation. Our findings suggest that the ploidy-specific lethality of the bub1Δ mutation is due to a failure to properly localize Sgo1 to the kinetochore. Both bub1Δ and sgo1Δ tetraploids die due to the persistence of uncorrected syntelic attachments, which results in high rates of sister-chromatid nondisjunction. The role of Sgo1 in sister-chromatid biorientation can be bypassed—in cells of all ploidies—by overexpression of factors that activate Ipl1 kinase. We found that budding yeast Bub1 and Sgo1 are dispensable for the localization of the CPC and observed no change in Ipl1-dependent phosphorylation events in cells lacking Bub1 or Sgo1. This suggests that Sgo1 might regulate microtubule attachment without affecting the Ipl1-mediated phosphorylation of kinetochore proteins. In searching for factors that cooperate with Sgo1, we identified a reciprocal genetic interaction between SGO1 and MPS1, which encodes a mitotic checkpoint kinase that promotes the turnover of syntelic attachments (Maure et al., 2007). Notably, we found that Mps1 activity is required to maintain the kinetochore localization of Sgo1, and SGO1 overexpression allows cells to survive mitosis after Mps1 inactivation. Taken together, our data suggest that budding yeast Sgo1 is not simply a recruiting and activating factor of Aurora B. Instead, we propose that Sgo1 and Msp1 act in parallel to the CPC and that both pathways are required for the establishment of the chromosome biorientation.
RESULTS
The defect of bub1Δ and sgo1Δ cells is suppressed by the overexpression of BIR1 or SLI15
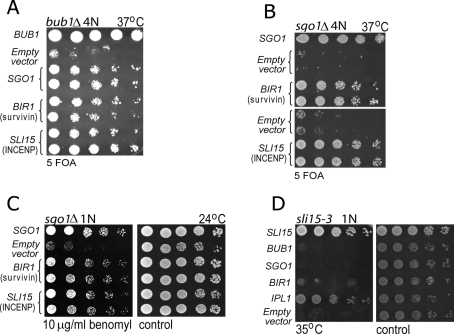
BUB1, which codes for a mitotic kinase involved in the SAC and chromosome segregation, was previously identified as 1 of the 39 genes whose deletion results in ploidy-specific lethality in budding yeast (Storchová et al., 2006). Tetraploid cells lacking BUB1 grow poorly at 24°C and are inviable at higher temperatures; this contrasts with haploid bub1Δ mutants, which propagate at any temperature, although with a slightly reduced growth rate compared with wild-type cells. We found that overexpression of SGO1 on a 2-μm plasmid rescues the growth defect of bub1Δ tetraploids (Figure 1A), consistent with the requirement of the kinase domain of Bub1 for the centromeric localization of Sgo1 (Supplemental Figure 1; Fernius and Hardwick, 2007). We reasoned that the ploidy-specific lethality of bub1Δ would facilitate its genetic analysis and thus could be exploited to elucidate the function of Bub1 in chromosome segregation. Using a high-copy number suppressor screen, we identified two suppressor genes, BIR1 and SLI15, whose overexpression bypasses the requirement for Bub1 in tetraploid cells (Figure 1A). Similar to bub1Δ tetraploids, sgo1Δ tetraploids are inviable at higher temperatures (Storchová et al., 2006), and we found that BIR1 or SLI15 overexpression also bypasses the growth defect of these cells (Figure 1B). Moreover, the overexpression of BIR1 or SLI15 suppresses the sensitivity of haploid sgo1Δ (Figure 1C) and bub1Δ cells (Supplemental Figure 5C) to the microtubule-depolymerizing drug benomyl. This verifies that the observed genetic interaction between CPC components and Bub1/Sgo1, although more easily detected in mutant tetraploid strains, is relevant to cells of any ploidy.
FIGURE 1:
The defect of sgo1Δ and bub1Δ strains is suppressed by BIR1 or SLI15 overexpression. (A) Overexpression of SGO1, BIR1, and SLI15 under control of their own promoter and carried on 2-μm plasmids suppresses the temperature sensitivity of bub1Δ tetraploids. Two independent clones were tested for each genotype, and cells were plated by fivefold serial dilution on plates lacking leucine (to select for plasmid retention) and containing 5-FOA (counterselection against plasmid containing the wild-type BUB1). The 2-μm LEU2 vector pRS425 (“empty”) and a LEU2-marked centromeric plasmid containing BUB1 were used as negative and positive controls, respectively. (B) Overexpression of BIR1 and SLI15 suppresses the lethality of sgo1Δ tetraploids at 37°C. The setup is identical to the previous experiment. (C) BIR1 and SLI15 overexpression suppresses the sensitivity of haploid sgo1Δ cells to the microtubule-depolymerizing drug benomyl. (D) The genetic interaction is not reciprocal, as overexpression of SGO1 does not improve the growth of sli15-3 mutants at the minimal restrictive temperature (35°C).
Bir1 and Sli15 facilitate accurate chromosome segregation by activating the kinase Ipl1/Aurora B, which promotes the release of microtubules from kinetochores that are not properly attached (Tanaka et al., 2002; Pinsky et al., 2006). However, we found that IPL1 overexpression does not restore the viability of the bub1Δ and sgo1Δ tetraploids (Supplemental Figure 2A), just as it cannot bypass a deficiency of either Bir1 or Sli15 (Kim et al., 1999). Presumably, this reflects the inactivity of Ipl1 kinase in the absence of binding to other CPC members.
Finally, the genetic interaction between CPC components and Sgo1 is not reciprocal. The temperature-sensitive sli15-3 mutation results in impaired Ipl1 kinase activation, and the defects of sli15-3 cells can be suppressed by IPL1 overexpression (Kim et al., 1999) (Figure 1D). By contrast, we find that SGO1 overexpression does not rescue the temperature sensitivity of the sli15-3 strain (Figure 1D). In conclusion, our results suggest that the growth defect of bub1Δ or sgo1Δ tetraploids can be ameliorated by increased expression of Sli15 or Bir1, consistent with recent evidence that Sgo1 plays a role in establishment of sister-chromatid biorientation (Indjeian et al., 2005; Kiburz et al., 2008).
Rescue of ploidy-specific lethality of bub1Δ and sgo1Δ requires the activation of Ipl1
Overexpression of Ipl1, the third subunit of the CPC, fails to cause any discernible improvement in the viability of either bub1Δ or sgo1Δ tetraploids at any temperature tested (Supplemental Figure 2A). Because Bir1 and Sli15 have an Ipl1-independent role in anaphase septin organization (Thomas and Kaplan, 2007), we tested the possibility that the Ipl1 kinase activity is not required to improve the growth of bub1Δ and sgo1Δ tetraploids. We used recently identified alleles of BIR1 that selectively abolish its physical interaction with associated kinetochore proteins (Thomas and Kaplan, 2007). In particular, the W901A point mutation in BIR1 selectively disrupts the Bir1–Ipl1 interaction but leaves the Bir1–Sli15 interaction unperturbed; by contrast, the doubly mutated Bir1A931E,I935E fails to associate with either Ipl1 or Sli15 (Thomas and Kaplan, 2007). High-copy plasmids expressing Bir1W901A and Bir1A931E,I935E failed to restore viability to bub1Δ and sgo1Δ tetraploids at the restrictive temperature (Supplemental Figure 2B). This result suggests that the mechanism by which BIR1 and SLI15 rescue the mitotic defects of bub1Δ and sgo1Δ mutants requires the kinase activity of Ipl1.
The suppressors of bub1Δ and sgo1Δ promote accurate chromosome segregation without restoring SAC function
The lack of Bub1 in haploid strains causes two different mitotic defects: The cells fail to delay anaphase in the presence of unattached kinetochores (Hoyt et al., 1991), and they exhibit sister-chromatid nondisjunction at a rate higher than that caused by checkpoint dysfunction alone (Warren et al., 2002), possibly due to a failure to localize Sgo1 at the centromere (Fernius and Hardwick, 2007). Loss-of-function mutations in SGO1 and BIR1 have been shown to compromise the ability of the SAC to respond to a lack of tension at mitotic kinetochores (Indjeian et al., 2005; Shimogawa et al., 2009) without affecting the ability to arrest mitosis after microtubule depolymerization. This finding is consistent with a role for Bir1 and Sgo1 in regulating microtubule attachment; however, it remains possible that overexpression of these proteins directly delays anaphase. Thus the overexpression of the suppressors could potentially rescue the defects of bub1Δ mutants by restoring the SAC rather than by promoting sister-chromatid biorientation.
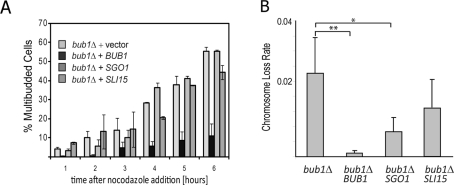
To test this possibility, we analyzed the ability of diploid bub1Δ mutants overexpressing SGO1 and SLI15 to arrest in the presence of the microtubule-depolymerizing drug nocodazole. Under these conditions, SAC-deficient cells progress through the cycle and acquire a multibudded morphology, due to concomitant bud emergence and cytokinesis failure. Over a 6-h time course, multibudded cells appeared with the same frequency and timing in bub1Δ cells overexpressing SGO1 and SLI15 as observed in cells transformed with an empty vector (Figure 2A). By contrast, cells carrying a BUB1-expressing plasmid arrested as large-budded cells for the duration of the experiment. Thus the high-copy suppression of the ploidy-specific lethality of bub1Δ does not involve mechanisms that rescue the SAC defect.
FIGURE 2:
Overexpression of SGO1 or SLI15 reduces the chromosome segregation defect in bub1Δ cells without restoring spindle checkpoint function. (A) Strains were grown in liquid culture and analyzed for their ability to arrest in the prolonged presence of the microtubule poison nocodazole. Whereas wild-type cells arrest as large-budded cells, mutants lacking a functional SAC reenter the cycle, which is indicated morphologically by the formation of a second bud. The accumulation of multibudded cells in the bub1Δ strain was unaffected by overexpression of SGO1 or SLI15, indicating that the SAC remains defective. (B) The elevated chromosome loss rate in a haploid bub1Δ strain bearing a YAC, measured by genetic fluctuation test, decreases significantly upon SGO1 or SLI15 overexpression. The experiments were performed at 30°C. *P < 0.05, **P < 0.01, Student’s unpaired t test, two-tailed.
Haploid bub1Δ cells missegregate their chromosomes at rates higher than other SAC-deficient mutants (Warren et al., 2002). We reasoned that the high-level chromosome loss of these cells might be suppressed by the overexpression of Sgo1 or CPC components. To test this hypothesis, we utilized strains carrying a yeast artificial chromosome (YAC) (Huang and Koshland, 2003), which is nonessential but contains selectable markers on both chromosome arms. Haploid bub1Δ mutants lost the YAC at a rate ∼35 times higher than the same strain expressing a single copy of BUB1 on a plasmid (Figure 2B; 2.9 × 10−2 and 8.3 × 10−4 missegregation events per division, respectively). Overexpression of SGO1 and SLI15 suppressed YAC loss in bub1Δ mutants by approximately threefold and twofold, respectively (Figure 2B; 9.2 × 10−3 and 1.56 × 10−2 missegregation events per division, respectively). We conclude that the suppressors of lethality inbub1Δ and sgo1Δ tetraploids increase survival by improving the efficiency of chromosome segregation.
The chromosome missegregation defect of sgo1Δ cells is due to the inability to establish bipolar attachments
Recent studies have shown that haploid sgo1 mutants fail to arrest the cell cycle in the presence of chromosomes that cannot form tension-generating attachments. This suggests a defect in tension sensing in these cells and is consistent with the observed high rates of sister-chromatid nondisjunction (Indjeian et al., 2005; Indjeian and Murray, 2007). We performed a series of experiments to monitor chromosome biorientation in sgo1Δ and bub1Δ tetraploids with and without overexpression of CPC components.
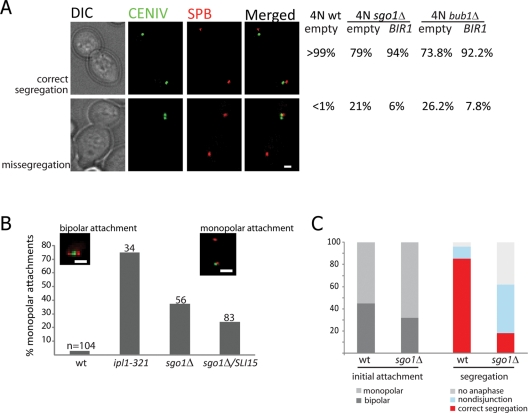
First, we followed the segregation of a single green fluorescent protein (GFP)–marked copy of chromosome IV. Whereas wild-type tetraploids missegregate chromosome IV in fewer than 1% of divisions at all temperatures, sgo1Δ tetraploids displayed modest chromosome segregation defects at 24°C (8% of the mitotic divisions) and more severe defects at 34°C (21% of divisions) (Figure 3A). Notably, 70% of nondisjunction events at 34°C were directed toward the bud; this phenotype, which has previously been reported for ipl1-321 mutants (Tanaka et al., 2002), indicates the persistence of syntelic attachments at the mother spindle pole body (SPB). Our finding, moreover, is consistent with the observation of preferential missegregation to the bud in the sgo1-100 mutant cells with unreplicated DNA (Indjeian and Murray, 2007). Overexpression of BIR1 reduced the segregation defect of sgo1Δ tetraploids more than threefold, to 6% (Figure 3A). Similarly, deletion of BUB1 in yeast tetraploids caused nondisjunction of chromosome IV in 26% of the cells grown at semipermissive temperature, and the frequency of the segregation errors dropped more than threefold after overexpression of BIR1 (Figure 3A).
FIGURE 3:
Frequent chromosome missegregation in sgo1Δ mutants is due to defects in biorientation and can be ameliorated by overexpression of BIR1 or SLI15. (A) sgo1Δ and bub1Δ tetraploid strains exhibit elevated rates of nondisjunction at chromosome IV; this defect can be suppressed by BIR1 overexpression. Chromosome IV was visualized using a TetO/TetR-GFP array, and SPBs were marked by expression of Spc29-RFP. At least 150 anaphase cells were scored for each genotype. The experiments were performed at 33°C in order to enhance the phenotype. Bar = 1 μm. (B) Haploid cells lacking SGO1 show frequent monopolar attachment of an unreplicated dicentric minichromosome, albeit less so than mutants with reduced Ipl1 kinase activity. The missegregation defect in the sgo1Δ strain can be partially suppressed by overexpression of SLI15. The experiments were performed at 33°C. The two inset images show examples of attachments: Left, the GFP-labeled attachment is positioned between the two SPBs (marked with RFP), indicating a bipolar attachment. Right, the minichromosome localizes in proximity to one of the SPBs, indicating monopolar attachment. (C) Diploid cells were released from treatment with nocodazole in order to create conditions in which a large number of syntelic attachments would be formed, even in wild-type cells. Live-cell imaging was used to monitor the initial attachment (left) and eventual segregation (right) of a GFP-marked copy of chromosome IV. sgo1Δ cells never converted monopolar attachments into bipolar attachments during the time of observation (75 min) and were consequently much more likely than wild-type cells to undergo nondisjunction in anaphase.
To directly test the ability of the sgo1Δ cells to biorient chromosomes during metaphase, we monitored the behavior of an unreplicated minichromosome containing two functional centromeres (Dewar et al., 2004). This assay allows bipolar attachments to be distinguished from monopolar attachments based on whether the minichromosome is positioned between the two SPBs or proximal to one SPB, respectively. Therefore we can directly evaluate the preference of sister kinetochores for tension-generating versus tensionless attachments, in contrast to previously published experiments in sgo1Δ cells that monitored the fate of the obligatory tensionless attachments of unreplicated chromosomes (Indjeian et al., 2005). In our experiment, cells lacking Sgo1 showed a significant accumulation of syntelic attachments, although the effect was not as severe as that observed in cells with defective Ipl1 kinase (Figure 3B). The increased frequency of monopolar attachments in sgo1Δ mutants was partially suppressed by overexpression of SLI15 (Figure 3B). Taken together, these experiments show that the primary defect in cells lacking Bub1 or Sgo1 is a high level of chromosome loss due to failed chromosome biorientation. In mutant tetraploids, the frequency of chromosome loss is too high for colony survival but can be reduced below this threshold by overexpression of the CPC components Bir1 or Sli15.
The elevated frequency of syntelic attachments in sgo1Δ mutants could be due to an increase in the formation of such attachments, a decrease in their turnover by corrective mechanisms, or both. To focus specifically on the contribution of corrective mechanisms, we performed live-cell imaging on diploid cells released from nocodazole-induced arrest and followed the movement of GFP-marked sister chromatids as they attached to the spindle. Under these conditions, the initial attachment of sister kinetochores to either the same SPB or opposing SPBs appears to be random (Indjeian and Murray, 2007). Indeed, in wild-type cells released from nocodazole (27 cells analyzed in total), we found that the rate of initial bipolar attachment at chromosome IV was similar to the rate of the monopolar attachment (44 and 56%, respectively). In the sgo1Δ mutants (54 cells analyzed in total), the chromosomes initially attached to one pole more often than to both poles (31% bipolar, 69% monopolar). However, this observed difference between sgo1Δ and wild-type cells might be biased by the fact that a correction from monopolar to bipolar attachment appears to be achieved very quickly in the wild-type strain. Thus we might have underestimated the frequency of primary monopolar attachment in these cells. Strikingly, sgo1Δ mutants that formed initial monopolar attachments failed to convert them to bipolar attachments, even in the moments when the two SPBs moved very close to each other (unpublished data). Within 75 min of release from nocodazole, 86% of the wild-type cells entered anaphase with correct bipolar attachments at chromosome IV, whereas only 18% of the cells lacking Sgo1 were able to do so (Figure 3C). Only 8% of wild-type cells, but almost 45% of sgo1Δ mutants, missegregated their chromosomes. The remaining 37% of sgo1Δ mutants did not enter anaphase within 75 min of release, in comparison to 6% of wild-type cells. The data demonstrate that cells lacking Sgo1 are either unable to recognize the tensionless attachments or cannot release them, which in turn leads to increased levels of chromosome missegregation and subsequent cell death.
The defect in biorientation is not due to PP2A-associated functions of Sgo1
Sgo1 was identified as a factor that protects centromeric cohesion in meiosis by recruiting the phosphatase PP2A to the kinetochore (Riedel et al., 2006). The phosphorylation of centromere-associated cohesins triggers their efficient removal but is antagonized by the activity of PP2A. In cells lacking Sgo1, pericentromeric cohesion in meiosis I is prematurely abolished, which results in chromosome missegregation. Thus the observed defects in chromosome segregation in bub1Δ and sgo1Δ cells could be due to premature loss of cohesion. However, budding yeast sgo1Δ mutants appear to have no discernible cohesion defects in mitosis (Kiburz et al., 2005), a finding which we have replicated here in tetraploids (Supplemental Figure 3A). Thus the frequent chromosome missegregation in sgo1Δ cells cannot be explained by the precocious separation of sister chromatids. In support of this conclusion, expression of a mutant Sgo1 protein that fails to interact with the phosphatase PP2A (which has the conserved Asn-51 residue replaced by Ile [Xu et al., 2009]) can restore viability to sgo1Δ tetraploids, although to a lesser extent than the expression of the wild-type Sgo1 protein (Supplemental Figure 3B).
Recently, it was reported that Sgo1 overexpression inhibits the nonproteolytic function of separase, resulting in delayed onset of anaphase, and that this function of Sgo1 is mediated through its recruitment of the PP2A regulatory subunit Cdc55 (Clift et al., 2009). However, overexpression of BIR1 and SLI15 has no effect on the benomyl sensitivity and cold sensitivity of haploid cdc55Δ mutants (Supplemental Figure 4A), suggesting that these mutants have a molecular defect different from that of sgo1Δ orbub1Δ cells. Moreover, we found that the defects of bub1Δ mutants can be suppressed by overexpression of SGO1 even in the absence of the PP2A regulatory subunit Cdc55: The benomyl sensitivity of a haploid bub1Δcdc55Δ strain can be rescued by SGO1 overexpression, similar to observations in the bub1Δ strain (Supplemental Figure 4B). Taken together, these results demonstrate that the function of Sgo1 in biorientation is independent of its recruitment of Cdc55/PP2A.
Bub1 and Sgo1 are dispensable for CPC localization and Ipl1 kinase activity
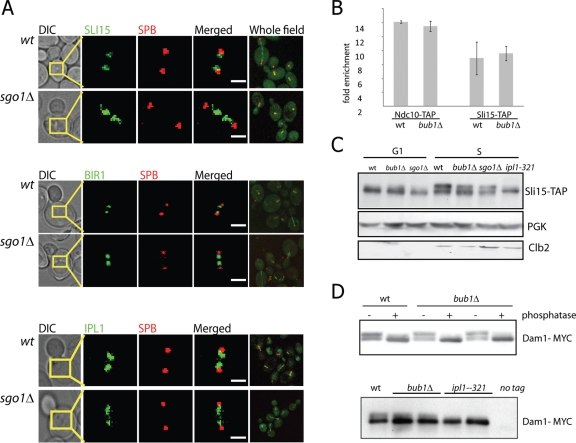
What is the underlying molecular mechanism that allows the rescue of the chromosome segregation defect in bub1Δ and sgo1Δ mutants upon Bir1 and Sli15 overexpression? We hypothesized that the absence of Sgo1 at the kinetochore might directly impair the function of the CPC in regulating sister-chromatid biorientation. One possibility is that Sgo1, like its homologue Sgo2 in fission yeast (Kawashima et al., 2007), is required to properly localize the CPC to the kinetochore. We studied the effect of the deletion of SGO1 on the localization of Bir1-GFP, Sli15-GFP, and Ipl1-GFP by quantitative fluorescence microscopy. Unlike the observations for fission yeast Sgo2, the loss of SGO1 in budding yeast has no effect on the accumulation of CPC proteins on preanaphase spindles (Figure 4A). Quantification of fluorescence intensity in mitotic cells expressing Bir1-GFP revealed no difference between wild-type and mutant strains. Values for median fluorescence intensity, averaged across 100 mitotic cells, were 48.5 ± 9.2 and 50.6 ± 12.2 arbitrary units for cells with and without SGO1, respectively.
FIGURE 4:
The localization and activity of CPC proteins are not affected by the absence of Bub1 or Sgo1. (A) The localization of Sli15-GFP, Bir1-GFP, and Ipl1-GFP to preanaphase spindles is not diminished in the diploid sgo1Δ cells. SPBs are marked with Spc29-RFP. Bar = 1 μm. (B) Sli15-directed ChIP revealed a sevenfold enrichment of centromeric DNA relative to telomeric DNA in wild-type cells. This enrichment is not affected by absence of Bub1, similar to results obtained with Ndc10-directed ChIP. (C) Phosphorylation of Sli15-TAP is not affected by the presence or absence of BUB1 and SGO1. Cells were grown at 35°C, synchronized in α-factor (G1 phase) or hydroxyurea (S phase), harvested, and analyzed by PAGE and Western blotting. Slower migrating forms of Sli15-TAP were eliminated by mutation of the Ipl1 kinase domain but not by deletion of SGO1 or BUB1. (D) The phosphorylation of Dam1, the crucial substrate of Ip1l in the release of microtubule attachments, is unaltered by the deletion of BUB1 (top, 30°C) but is abolished in the ipl1-321 mutants at 37°C (bottom). Two independent clones of both the bub1Δ and ipl1-321 mutants were tested, and the separation between phosphoforms of Dam1-myc9 was enhanced by adding 10 μM Phos-Tag AAL-107 (Kinoshita et al., 2006) to the polyacrylamide gel mixture. The slower migrating forms of Dam1-myc9 are sensitive to alkaline phosphatase treatment (top).
To more precisely measure the association of the CPC with centromeric DNA, we performed a chromatin immunoprecipitation (ChIP) of Sli15-TAP in the presence and absence of BUB1. Sli15 is enriched at budding yeast centromeres (Kang et al., 2001), albeit to a lesser extent than a constitutive kinetochore protein Ndc10, which we used as a control. Consistent with our epifluorescence data, the enrichment of CENIII DNA in Sli15-TAP immunoprecipitates was not reduced by the deletion of BUB1 (Figure 4B), further supporting the idea that the kinetochore localization of the CPC does not require Bub1/Sgo1 in budding yeast.
We next examined whether the kinase activity of Ipl1 might be impaired in the absence of Sgo1 or Bub1, even if the CPC is properly localized. We measured phosphorylation of Sli15, a direct substrate and one of the cofactors of Ipl1 kinase (Kang et al., 2001), by analyzing its mobility in SDS–PAGE. Indeed, the pattern of migration of the Sli15-TAP protein was not altered in bub1Δ and sgo1Δ mutant strains in comparison to the wild type. By contrast, the phosphorylation was almost completely abolished in cells expressing ipl1-321, a temperature-sensitive mutation that abolishes the kinase activity (Figure 4C).
Similarly, we analyzed the phosphorylation of another known substrate of Ipl1 kinase, the microtubule-binding protein Dam1 (Cheeseman et al., 2002). Catalytic interaction between Ipl1 and Dam1 is thought to be the primary mechanism by which the CPC destabilizes microtubule attachments that do not produce tension (Cheeseman et al., 2002; Gestaut et al., 2008). In asynchronous cultures, the relative abundance of the two distinctly migrating populations of Dam1-myc was unaffected by the deletion of BUB1 at 30°C or 37°C, whereas the ipl1-321 mutation eliminated the appearance of phosphorylated Dam1-myc at the restrictive temperature (Figure 4D). These findings suggest that the general activity of the Ipl1 kinase remains intact in cells that lack Bub1 or Sgo1. It should be noted that these experiments are not specific for the subset of phosphorylation events, at particular residues and particular times in the cell cycle, that directly mediate the repair of syntelic attachments. However, they show that the CPC is catalytically active in the absence of Sgo1 and Bub1.
Sgo1 supports chromosome biorientation even if Ipl1 is inactive
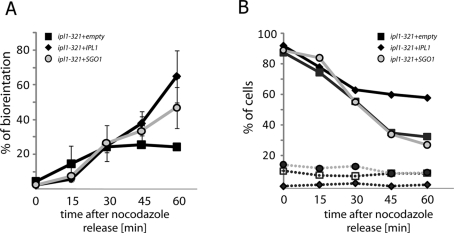
Our data suggest the possibility that Sgo1 acts in the establishment of chromosome biorientation independently of the CPC. To test this model, we used ipl1-321 strains transformed with either an empty vector control or a 2-μm vector expressing IPL1 or SGO1. One homologue of chromosome IV was marked with TetO/TetR-GFP, and SPBs were marked with Spc29–red fluorescent protein (RFP). We synchronized the cells in nocodazole, effectively abolishing microtubule-kinetochore attachment, and then microscopically analyzed chromosome segregation after release from nocodazole at the restrictive temperature. Bipolar attachments were identified by the positioning of the marked chromosome in the center between the two SPBs. As expected, up to 70% of IPL1+ cells achieved biorientation within 60 min of removal of the microtubule poison, whereas only 20% of cells with inactive Ipl1 could biorient chromosome IV (Figure 5A). Overexpression of Sgo1 improved the biorientation rate of the ipl1-321 strain to 44% (Figure 5A), suggesting that Sgo1 can alter kinetochore-microtubule attachments even in the absence of functional Ipl1. Consistently, analysis of the cells in anaphase revealed that strains overexpressing Sgo1 missegregated chromosome IV in 46% of anaphases (95% confidence interval 37.5–54.6%), whereas the ipl1-321 strain showed missegregation in 65% (58–71%) of anaphases. The IPL1+ strain missegregated chromosome IV at a rate of 8.5% (5.6–11.4%) under the identical conditions.
FIGURE 5:
Some monopolar attachments can be corrected independently of the CPC. (A) Chromosome IV was marked with a TetO/TetR-GFP array. The percentage of cells with bioriented chromosome IV (as judged by the localization of the GFP dot in relation to the marked SPBs) among all large-budded cells was assessed after release from nocodazole. All strains carry a genomic ipl1-321 allele as well as a multicopy plasmid with either the functional IPL1 gene (black diamonds), SGO1 (light circles), or marker only (gray squares). (B) Progression through anaphase after release from nocodazole is accelerated in cells lacking functional IPL1, as evidenced by the decrease of the percentage of large-budded cells in the population (full lines). These strains also accumulate bibudded cells at higher levels than wild-type (dotted lines). Overexpression of SGO1 did not alter the cell-cycle progression of ipl1-321 mutants, even though it affected biorientation (A). The experiments were performed at 35°C.
Because Ipl1 functions in the SAC, the IPL1+ and ipl1-321 strains do not respond equivalently after the release from nocodazole treatment. As expected, a higher percentage of ipl1-321 cells thanIPL1+ cells progressed through anaphase and acquired multiple buds over the course of the experiment (Figure 5B). Importantly, the decrease in the frequency of large-budded cells and increase in the frequency of multibudded cells were not altered by overexpression of Sgo1 in ipl1-321 mutants. These results indicate that Sgo1 does not slow progression through mitosis in these cells (Figure 5B). Taken together, our data suggest that some of the improper attachments created after release from nocodazole can be corrected independently of Ipl1 kinase function by an Sgo1-mediated process.
Mps1 regulates Sgo1 to establish chromosome biorientation
Another mitotic protein, the conserved checkpoint kinase Mps1, has recently been shown to promote the turnover of syntelic attachments without any apparent effect on Ipl1 localization or kinase activity (Maure et al., 2007). To test the hypothesis that Sgo1 and Mps1 act in the same pathway, we analyzed the genetic relationship between Mps1 and Sgo1. Indeed, overexpression of MPS1 (under the control of its own promoter and located on an episomal vector) weakly but reproducibly suppresses the benomyl sensitivity of sgo1Δ haploids as well as the growth defect of sgo1Δ tetraploids (Supplemental Figure 5, A and B).
However, this effect could be unrelated to Mps1’s role in regulating microtubule attachment because overexpression of Mps1 can also induce a SAC-dependent mitotic arrest. Previous reports suggest that the chromosome segregation defects of sgo1 mutants can be significantly reduced by simply delaying anaphase onset, either by weak treatment with hydroxyurea or overexpression of Mps1 at levels sufficient to activate the SAC (Indjeian et al., 2005). We therefore investigated whether this genetic interaction, the ability of Mps1 overexpression to partially compensate for the absence of Sgo1, depends upon activation of the SAC. We found that Mps1 overexpression suppresses the benomyl sensitivity of bub1ΔK cells (mutants lacking the kinase domain of Bub1 but still capable of activating the checkpoint [Fernius and Hardwick, 2007]), whereas no such effect was observed in bub1Δ cells, which lack a functional SAC (Supplemental Figure 5C). Therefore we conclude that Mps1-dependent activation of the SAC can improve the fitness of cells that lack or mislocalize Sgo1 but that Mps1 cannot bypass Sgo1’s role in regulating microtubule attachment.
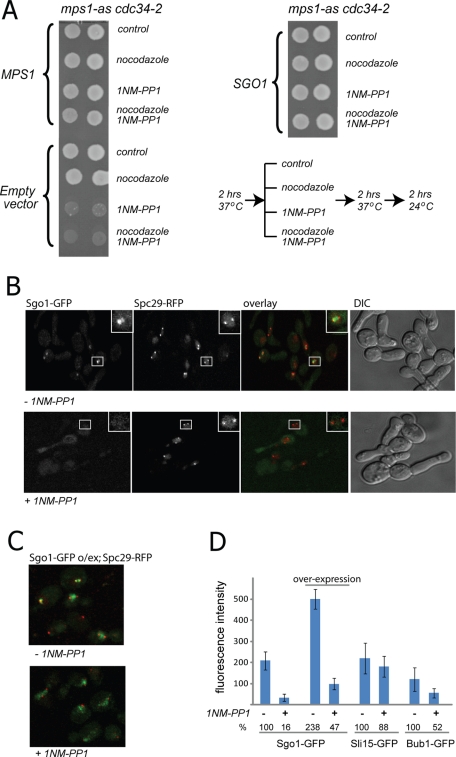
Next we analyzed whether SGO1 overexpression can reduce the mitotic defects of cells lacking Mps1 activity. In addition to its functions in the SAC and microtubule attachment, Mps1 is essential for SPB duplication during the cell cycle (Lauze et al., 1995). To be able to inactivate Mps1 only after SPB duplication is complete, we used strains expressing two mutant alleles: mps1-as (Jones et al., 2005), which enables chemical inhibition of the kinase by the ATP-analogue 1NM-PP1, and cdc34-2, which causes a temperature-sensitive arrest at the G1/S transition (after SPB duplication). The cells were arrested at 37°C, incubated for an additional 2 h at the restrictive temperature in the presence or absence of the inhibitor 1NM-PP1, and then released from the cdc34 arrest with and without the addition of nocodazole. The cells were then grown at room temperature for 2 h, which corresponds approximately to a single cell division, and subsequently plated on yeast peptone dextrose (YPD) plates. The majority of the cells with inactivated Mps1 kinase did not survive the progression through the mitosis, and addition of nocodazole did not affect the survival rates (Figure 6A). Strikingly, overexpression of Sgo1 allowed for the survival of cells after Mps1 inhibition, such that growth on the YPD plate was indistinguishable between cells treated with 1NM-PP1 and untreated cells (Figure 6A). Thus increased abundance of Sgo1 can bypass deficiency of Mps1 kinase activity in mitosis. These observations raise the possibility that Mps1’s role in chromosome biorientation—a function that appears to be independent of Ipl1 activity (Maure et al., 2007)—involves Sgo1.
FIGURE 6:
Sgo1 can partially bypass the role of Mps1 in chromosome segregation and appears to act in the same pathway. (A) Overexpression of SGO1 allows cells to survive transient pharmacological inhibition of Mps1 kinase during mitosis. The cells were synchronized in S phase (at the restrictive temperature for the cdc34-2 allele), released into mitosis with or without the mps1-as inhibitor and with or without nocodazole, and then spotted on nonselective plates. (B) Localization of Sgo1-GFP to preanaphase spindles requires active Mps1. The images were taken 10 min after adding dimethyl sulfoxide (control samples) or the ATP analogue 1NM-PP1 to inhibit Mps1. (C) Overexpression of Sgo1-GFP on a 2-μm plasmid increases retention of Sgo1 on the mitotic spindle, even in the absence of targeting by Mps1. (D). Quantification of the fluorescence signal on the spindle for Sgo1-GFP, overexpressed Sgo1-GFP, Sli15-GFP, and Bub1-GFP in cells with active or inactive Mps1 kinase.
We hypothesized that Mps1 might regulate Sgo1 function by ensuring its proper targeting to the kinetochore. To test this possibility, we again utilized the mps1-as allele in combination with the cdc34-2 temperature-sensitive allele. After release from the cdc34-2 arrest in the absence of the Mps1 inhibitor, these cells properly duplicated SPBs (marked via Spc29-RFP), and Sgo1-GFP localized within the area corresponding to the mitotic spindle. However, the localization of Sgo1-GFP to the spindle is impaired upon addition of the inhibitor 1NM-PP1 (Figure 6, B and D). In inhibitor-treated cells, accumulation of Sgo1-GFP on the spindle is partially restored (to 50% of normal levels) by overexpression of Sgo1-GFP from a multicopy vector (Figure 6, C and D), consistent with the ability of Sgo1 overexpression to rescue cell survival. The effect of Mps1 on Sgo1 localization is specific because inactivation of its kinase activity does not affect localization of Sli15-GFP (Figure 6D). In light of this evidence, we propose that Sgo1 and Mps1 act in the same pathway for the establishment of chromosome biorientation in mitosis and that Mps1 contributes to the kinetochore recruitment of Sgo1.
DISCUSSION
The chromosome segregation apparatus is compromised by increased ploidy, as demonstrated by the fact that budding yeast and mammalian tetraploids exhibit high chromosomal instability and accumulate aneuploid cells (Mayer and Aguilera, 1990; Fujiwara et al., 2005). In fact, several genes involved in the maintenance of genome stability are dispensable in haploid or diploid budding yeast but become essential in cells with increased ploidy (Storchová et al., 2006). We used this ploidy-specific lethality to analyze the function of Bub1, a conserved kinase involved in the SAC and chromosome segregation, and Sgo1, a conserved protein of unknown molecular function required for proper chromosome segregation in both mitosis and meiosis. We found that the ploidy-specific requirement for Bub1 can be bypassed by overexpression of Bir1/survivin and Sli15/INCENP, conserved proteins that are a part of the CPC. The CPC is considered to be the key protein complex involved in the establishment of bioriented attachments of sister chromatids. Bir1 and Sli15 are required for the catalytic activity of Ipl1/Aurora B, which phosphorylates several kinetochore proteins, including Dam1 and Ndc80, upon the formation of syntelic or monotelic attachments that do not produce tension. These phosphorylation events help to disrupt the unproductive attachments and enable their replacement by proper bioriented attachments (Cheeseman et al., 2002; Tanaka et al., 2002; Pinsky et al., 2006). Our finding that overexpression of CPC components restores viability to bub1Δ tetraploids suggests that these cells die due to an excess of unrepaired syntelic attachments.
The role of Bub1 in biorientation is independent of its function in the SAC. It has been shown that the C-terminal (kinase-containing) part of Bub1 is dispensable for mitotic arrest upon spindle depolymerization; instead it promotes the kinetochore recruitment of Sgo1 (Fernius and Hardwick, 2007) by phosphorylating H2A and thus creating a kinetochore mark that directs the localization of Sgo1 (Kawashima et al., 2010). Because Sgo1 overexpression bypasses the requirement for Bub1 in tetraploids, and because overexpression of the same two CPC components reverses the lethality of bub1Δ and sgo1Δ tetraploids alike, we propose that the ploidy-specific requirement for Bub1 effectively represents a requirement for Sgo1 in establishment of biorientation.
We observed that loss of Sgo1 produces a phenotype similar to that previously observed with ipl1 mutants, in which chromosome missegregation is biased toward the daughter bud and cells fail to reliably biorient an unreplicated, dicentric chromosome (Tanaka et al., 2002; Dewar et al., 2004). Sgo1 could play a role in the establishment of bioriented attachments by 1) biasing the initial attachment of kinetochores to the spindle so as to favor biorientation, 2) promoting the release of microtubule attachments that are not under tension, or 3) delaying anaphase in a SAC-independent manner and thereby allowing the CPC more time to destabilize syntelic attachments. We observed by live-cell imaging that, under experimental conditions in which cells initially form large numbers of syntelic attachments, sgo1Δ mutants were incapable of converting monopolar microtubule attachments to bioriented attachments. This observation supports the model that Sgo1 promotes the release of tensionless microtubule attachments. It should be noted that our evidence does not exclude a role for Sgo1 in cell cycle progression (Clift et al., 2009) or in biasing the initial attachment of kinetochores toward biorientation (as has been proposed for sister chromatids in meiosis [Kiburz et al., 2008]).
The defects in chromosome segregation that are observed in sgo1Δ and bub1Δ mutants are similar to those observed in CPC mutants and are suppressed by overexpression of CPC components. This suggests that the localization and/or function of CPC proteins might be impaired by the loss of Sgo1 at the kinetochore. Such a mechanism was previously observed in fission yeast: Sgo2 interacts with Bir1, and disruption of the interaction impairs the kinetochore localization of Bir1 and Aurora homologue Ark1 in metaphase (Kawashima et al., 2007). However, by quantitative fluorescence imaging we find that budding yeast lacking Sgo1 and Bub1 show normal localization of Ipl1, Bir1, and Sli15 during mitosis, which was further confirmed by ChIP of Sli15-TAP to the centromeric DNA. The intact CPC localization in Sgo1-deficient cells could have several explanations. First, Sgo1 might act independently of CPC; second, there might be a minor defect in CPC recruitment that our methods failed to detect; or, third, loss of Sgo1 triggers a compensatory response that enhances CPC targeting. However, our data clearly suggest that aberrant CPC localization could not be responsible for the striking phenotype of budding yeast lacking SGO1, which is an inability to repair syntelic attachments once they form (Figure 3C). Moreover, Western blot analysis of the phosphorylation of Dam1 and Sli15 by Ipl1 did not reveal any difference in cells with and without Bub1 and Sgo1. Previous evidence shows that the phosphorylation of Dam1 by Ipl1 is required for disassembly of incorrect chromosome attachments (Cheeseman et al., 2002; Gestaut et al., 2008). Our data show that this important catalytic interaction is preserved in the absence of Sgo1, though our experiments do not allow for the detection of more subtle changes in phosphorylation at specific residues.
If Sgo1 functions in a pathway independent of Ipl1, then at least some correction of monopolar attachments should be possible in the absence of Aurora B activity. Consistent with this idea, we observe that Sgo1 overexpression in ipl1-321 mutants increases the frequency of chromosome biorientation and successful anaphase by ∼20%. However, because these cells still missegregate chromosomes at very high rates, they are inviable. In light of this evidence, we favor a model in which multiple modifications at the kinetochore are required to eliminate a syntelic attachment. One crucial set of modifications, the phosphorylation of Dam1 and Ndc80, is catalyzed by Ipl1 and stimulated by the CPC proteins Bir1 and Sli15. However, sgo1Δ mutants present a situation in which Ipl1 kinase is active and yet syntelic attachments are hardly, if ever, repaired (Figure 3C). While we cannot rule out the possibility that Ipl1 activity is lost only in a narrow spatiotemporal window, it seems likely instead that Sgo1—or some downstream factor—provides a second signal to release tensionless microtubule attachments. This second set of kinetochore modifications would occur in parallel to Ipl1, in the sense that they are not directly catalyzed by Ipl1 kinase, although the initial “sensing” of kinetochore tension might involve a common mechanism. Analysis of ipl1 and sgo1 mutants suggests that both pathways are required for the efficient correction of syntelic attachments, though the contribution of Ipl1 is more significant: Nondisjunction of sister chromatids occurs at a rate of 85% in ipl1-321 cells (Biggins et al., 1999) but is rare enough in sgo1 mutants to permit colony growth. The viability of Sgo1-deficient strains can be explained by the observation that, under normal conditions, these cells achieve biorientation on the first try and only rarely need to repair syntelic attachments (Indjeian and Murray, 2007).
Recently, the mitotic kinase Mps1 has been implicated in the establishment of sister-chromatid biorientation in diverse organisms, in addition to its previously characterized functions in the SAC and spindle pole duplication (Winey and Huneycutt, 2002; Jones et al., 2005; Maure et al., 2007; Jelluma et al., 2008; Hewitt et al., 2010; Maciejowski et al., 2010; Santaguida et al., 2010). Budding yeast with inactivated Mps1 missegregate chromosomes at high levels (Jones et al., 2005) and are unable to release syntelic attachments (Maure et al., 2007). We observed that overexpression of SGO1 enables cells to survive transient pharmacological inhibition of Mps1 kinase activity. Furthermore, the metaphase localization of Sgo1-GFP is abolished after Mps1 inactivation. This delocalization is not due to a more general defect in kinetochore integrity because microtubule-kinetochore attachments are stable in cells with inactive Mps1 (Maure et al., 2007). Moreover, the localization of the CPC component Sli15-GFP is not altered upon Mps1 inhibition, confirming the specificity of the effect. Thus we speculate that Mps1 is an upstream regulator of Sgo1 function and that the biorientation defect caused by Mps1 inactivation can be explained, in part, by the loss of Sgo1 kinetochore function. Finally, the fact that Mps1 inhibition does not alter the activity of purified Ipl1 in an in vitro kinase assay (Maure et al., 2007), despite a marked loss of Sgo1 localization to the kinetochore, provides additional evidence consistent with the model that Sgo1 does not regulate Ipl1 function.
The proposal that budding yeast Bub1, Sgo1, and Mps1 act in parallel to the CPC implies that these pathways are wired differently in budding yeast than in fission yeast or vertebrates. Bub1 deficiency in Schizosaccharomyces pombe, Xenopus laevis, and human cells causes marked abrogation of CPC localization to the inner centromere (Boyarchuk et al., 2007; Kawashima et al., 2010), consistent with the hypothesis that Sgo2 targeting by Bub1 and subsequent Bir1–Sgo2 interaction are required for CPC recruitment (Kawashima et al., 2007; Kawashima et al., 2010). Moreover, emerging evidence suggests that Aurora B and Mps1 regulate one another in human cells: Chemical inhibition of Aurora B in mitosis causes mislocalization and hypophosphorylation of Mps1 (Santaguida et al., 2010), and depletion or inhibition of Mps1 before mitotic entry diminishes Aurora B kinase activity in metaphase-arrested cells (Jelluma et al., 2008; Kwiatkowski et al., 2010; Maciejowski et al., 2010). However, even in higher eukaryotes, the literature is consistent with the existence of a second pathway, acting independently or downstream of Ipl1, to repair syntelic attachments. Crucially, three different chemical inhibitors of Mps1, when applied to human cells after mitotic entry, cause errors in chromosome alignment and biorientation without detectable changes in Aurora B activation (Hewitt et al., 2010; Maciejowski et al., 2010; Santaguida et al., 2010; Sliedrecht et al., 2010). These studies motivate future work to identify the key downstream Mps1 substrates that are important for biorientation. The regulation of microtubule attachment in budding yeast, while distinct from that in other eukaryotes, may very well be an attractive system for manipulating conserved functions of shugoshin or Mps1 without simultaneously affecting CPC localization or activity. Our work illustrates the value of cross-species comparisons for understanding the architecture of signaling networks; different organisms appear to utilize the same players but wire the system somewhat differently. Further experiments are needed to reveal the mechanism by which budding yeast Sgo1 can alter kinetochore-microtubule attachments in the absence of functional Ipl1/Aurora B and to test whether such a function also exists for shugoshin homologues in other organisms.
MATERIALS AND METHODS
General molecular genetics methods
All strains and plasmids used in this study are listed in Table 1; details on strain construction are available upon request. The strains are derived from the BY series of S288C (leu2Δ, his3Δ, ura3Δ, lys2Δ/met15Δ) or W303 background (ade2-101, ura3-52, trp1-1, his3-1,3, leu2-1112). The tetraploid construction was performed by mating MATa/a and MATα/α strains, as previously described (Storchová et al., 2006). Yeast cultivation, α-factor synchronization, and nocodazole treatment (30 μg/ml) were performed as described previously. The SGO1, IPL1, BIR1, SLI15, and MPS1 were cloned into the plasmid pRS316 (CEN, URA3) to obtain plasmids for the shuffling experiments and into pRS425 (2 μm, LEU2) for the complementation experiments. Because the strains sgo1Δ and bub1Δ are genetically unstable, they were transformed with the URA3-marked centromeric plasmids containing a functional gene; counterselection using 5-fluoroorotic acid (5-FOA) was performed before each experiment to obtain strains of the desired genotypes.
TABLE 1:
Plasmids and strains used
| Name | Backbone/Background | Type | Relevant markers/Strain genotype | Origin |
|---|---|---|---|---|
| BZ242 | pRS316 | CEN | URA3, SGO1 | This work |
| pBS199 | pRS316 | CEN | URA3, BUB1 | A. Murray |
| BZ11 | pRS425 | 2 μm | LEU2, SGO1 | This work |
| BZ9 | pRS425 | 2 μm | LEU2, BIR1 | This work |
| BZ40 | pRS425 | 2 μm | LEU2, SLI15 | This work |
| pTR168 | pRS315 | 2 μm | LEU2, BUB1 | A. Hoyt |
| BZ278 | pRS425 | 2 μm | LEU2, IPL1 | This work |
| T431 | N/A | CEN | pGAL1-10-CEN4, TetOx112, RS-ARS-RS, CEN4 | T. Tanaka |
| BZ277 | pRS425 | 2 μm | LEU2, MPS1 | This work |
| PB2303 | pRS316 | CEN | URA3, MPS1 | This work |
| BZ279 | pRS425 | 2 μm | LEU2, BIR1W901A | This work/K. Kaplan |
| BZ280 | pRS425 | 2 μm | LEU2, BIR1A931E, I935E | This work/K. Kaplan |
| BJ25 | pRS425 | 2 μm | LEU2, BIR1ΔN | This work |
| BZ299 | pRS425 | 2 μm | LEU2, SGO1-eGFP | This work |
| YZ669 | S288C | 1N | MATa CEN4-tetO::HIS3 TetR-GFP-LEU2 | Storchová et al., 2006 |
| YZ855 | S288C | 1N | MATa sli15-3 | This work |
| YZ1063 | S288C | 1N | MATa ipl1-321, SPC42-GFP-HIS3; pMET-rec; TetR-GFP | Dewar et al., 2004 |
| YZ1064 | S288C | 1N | MATa SPC42-GFP-HIS3; pMET-rec; TetR-GFP | Dewar et al., 2004 |
| YZ640 | S288C | 4N | MATa/a/0/a CEN4 tetO::HIS3; TetR-GFP-LEU2/TetRG- FP-LEU2/leu2/leu2 sgo1/sgo1/sgo1/sgo1; SPC29-mRFP/SPC29-mRFP/SPC29/SPC29 | This work |
| YZ997 | S288C | 4N | MATa/a/a/a bub1/bub1/bub1/bub1, CEN4- tetO::HIS3 TetR-GFP-LEU2/TetR-GFP-LEU2/leu2/leu2, SPC29-mRFP/SPC29-mRFP/SPC29/SPC29 | This work |
| YZ841 | S288C | 2N | MATa/a SLI15-GFP-HIS3/SLI15, SPC29-mRFP/SPC29 | This work |
| NT745 | S288C | 2N | MATa/a SLI15-GFP-HIS3/SLI15; SPC29-mRFP/SPC29; sgo1/sgo1 | This work |
| NT712 | S288C | 2N | MATa/a BIR1-GFP-HIS3/BIR1; SPC29-mRFP/SPC29 | This work |
| NT713 | S288C | 2N | MATa/a BIR1-GFP-HIS3/BIR1; SPC29-mRFP/SPC29; sgo1/sgo1 | This work |
| NT571 | S288C | 2N | MATa/a IPL1-GFP-HIS3/IPL1; SPC29-mRFP/SPC29 | This work |
| NT608 | S288C | 2N | MATa/a IPL1-GFP-HIS3/IPL1; SPC29-mRFP/SPC29; sgo1/sgo1 | This work |
| YZ728 | S288C | 1N | MATa SLI15-TAP-HIS3 | Ghaemmaghami et al., 2003 |
| YZ741 | S288C | 1N | MATa SLI15-TAP-HIS3; bub1::kanMX | This work |
| YZ725 | S288C | 1N | MATa NDC10-TAP-HIS3 | Ghaemmaghami et al., 2003 |
| YZ735 | S288C | 1N | MATa NDC10-TAP-HIS3; bub1::kanMX | This work |
| YZ704 | S288C | 1N | MATa YAC (URA3, TRP1, ADE2, CEN) | Storchová et al., 2006 |
| YZ406 | S288C | 1N | MATa bub1::kanMX; YAC (URA3, TRP1, ADE2, CEN) | This work |
| YZ1013 | S288C | 1N | MATa sgo1::kanMX, SPC29-mRFP; pMET-rec; TetR-GFP | This work |
| JF98 | W303 | 1N | MATa bub1delK::hph | K. Hardwick |
| YZ1071 | W303 | 1N | MATa bub1delK::hph; cdc55::kanR | This work |
| YZ1003 | W303 | 1N | MATa bub1delK::hph SGO1-GFP-HIS3, SPC29-mRFP | This work |
| YZ986 | W303 | 1N | MATa bub1::kanMX; DAM1-9myc-TRP1 | This work |
| YZ1101 | W303 | 1N | MATa ipl1-321::natMX; DAM1-9myc-TRP1 | This work |
| YZ990 | W303 | 1N | MATa DAM1-9myc-TRP1 | This work |
| YZ535 | S288C | 1N | MATa IPL1-TAP-HIS3 | This work |
| YZ808 | S288C | 1N | MATa bub1::kanMXIPL1-TAP-HIS3 | This work |
| YZ991 | S288C | 1N | MATa sgo1::hph IPL1-TAP-HIS3 | This work |
| 3108 | W303 | 1N | MATa mps1-as cdc34-2 | Mark Winey |
| YZ1056 | W303 | 1N | MATa mps1-as cdc34-2 SGO1-GFP-HIS3, SPC29-mRFP | This work |
| YZ1182 | W303 | 1N | MATa mps1-as BUB1-GFP-HIS3, SPC29-mRFP | This work |
| YZ650 | S288C | 1N | MATa ipl1-321::kanMX; CEN4-tetO::HIS3, TetR-GFP-LEU2 Spc29-mRFP | |
| YZ1174 | W303 | 1N | MATa mps1-as SLI15-GFP-HIS3, SPC29-mRFP | This work |
A genome-wide suppressor screen in bub1Δ tetraploids
The high-copy suppressor screen was performed using a plasmid-shuffle strategy. In brief, a tetraploid bub1Δ deletion strain expressing BUB1 on a URA3-marked centromeric plasmid was transformed with a 2-μm genomic library (LEU2 marked). Single colonies were replica plated on media lacking leucine and containing 5-FOA and incubated at 37°C for 3 d. All robustly growing colonies (<1%) were tested by colony PCR for the presence and absence of BUB1; ∼60% of the colonies grew due to the presence of the BUB1 gene on the suppressing plasmid and were excluded from further analysis. The remaining plasmids were then extracted from the candidate clones, used for secondary verification, and sequenced.
Measurement of chromosome loss rates by fluctuation test with YAC
The fidelity of chromosome segregation in bub1Δ mutants was assessed by measuring the rate of loss of a URA3,TRP1-marked YAC (Huang and Koshland, 2003). Cells were streaked on media nonselective for the YAC and grown at 30°C for 3 d. For each strain, seven colonies were cut out of the plate and dispensed in 1 ml sterile water, diluted, and plated in duplicate on both nonselective plates (to estimate the total number of cells) and 5-FOA plates (to estimate the number of mutants). To ensure specificity for chromosome loss events, as opposed to chromosomal rearrangements, colonies on 5-FOA were replica plated to verify simultaneous loss of the TRP1 marker near the centromere. The mutation rate (the number of chromosome loss events per cell division) was calculated according to the median method (Lea and Coulson, 1949).
Western blot analysis
Protein samples were extracted, run on SDS-polyacrylamide gels, and transferred to polyvinylidene difluoride membranes. The mouse monoclonal anti–phosphoglycerate kinase antibody (Molecular Probes, Eugene, OR) was used to detect a loading control, peroxidase–anti-peroxidase rabbit antibody (Sigma-Aldrich, St. Louis, MO) for recognition of the TAP tag, and anti-Myc rabbit serum. The secondary antibodies were goat anti–rabbit and goat anti–mouse (Abcam, Cambridge, UK), both used at a dilution of 1:10,000.
Chromatin immunoprecipitation
Chromatin immunoprecipitation was performed using a protocol developed in the laboratory of Stefan Jentsch (Kalocsay et al., 2009). The proteins of interest were tagged with the TAP epitope. Then 300 ml fresh culture was fixed at room temperature by formaldehyde addition to a final concentration 1% and harvested after 16 min. After lysis (50 mM HEPES-KOH, pH 7.5, 150 mM NaCl, 1 mM EDTA, 1% Triton X-100, 0.1% sodium deoxycholate, 0.1% SDS, 1 mM phenylmethylsulfonyl fluoride) in bead beater (6 × 3 min), the chromatin was sheared to an average length of 300–500 base pairs by water bath sonication (Bioruptor UCD-200; Diagenode, Liege, Belgium) using 30 × 30–s cycles with 30-s breaks at an output of 200 W. Anti–immunoglobulin G Sepharose beads (Roche) were used for immunoprecipitation at 4°C for 120 min. The association of the TAP-tagged protein with centromeric DNA was quantified as the ratio in abundance of a 200–base pairs centromeric sequence (CEN-III) to a 200–base pairs telomeric sequence (TEL-V*) in the immunoprecipitated DNA. Quantitative PCR was performed on a Light Cycler LC480 System (Roche, Mannheim, Germany) with previously described primers (Keogh et al., 2006).
Fluorescence microscopy
Strains expressing alleles tagged with GFP (Ipl1-GFP, Sli15-GFP, Sgo1-GFP, Bir1-GFP, and Bub1-GFP) and RFP (Spc29-RFP) were visualized by a fully automated Zeiss inverted microscope (Carl Zeiss, Jena, Germany) equipped with an MS-2000 stage (Applied Scientific Instrumentation, Eugene, OR), a CSU-X1 spinning disk confocal head (Yokogawa Electric Corporation, Tokyo, Japan), and LaserStack Launch with selectable laser lines (Intelligent Imaging Innovations, Denver, CO). Images were captured at intervals of 0.5 μm in the Z focal plane using a CoolSnap HQ camera (Roper Scientific, Tuscon, AZ) under the control of the SlideBook software (Intelligent Imaging Innovations). For quantification of the fluorescence signal in metaphase cells, we analyzed all cells showing two RFP dots (SPB marker, Spc29-RFP) <2 μm apart and positioned orthogonal to the plane of the bud neck. The GFP signal localized between the spindle poles was quantified by extracting both the median and maximum fluorescence intensities over a 36-pixel mask (positioned between the RFP dots) after application of standardized signal renormalization settings.
Supplementary Material
Acknowledgments
We thank Kevin Hardwick, Kenneth Kaplan, Tomo Tanaka, and Mark Winey for plasmids and strains. We thank Karolina Peplowska, Markus Räschle, Satoshi Yoshida, and the members of the Storchová group for critical reading of the manuscript and helpful discussions. We are thankful to Susanne Gutmann and Regina Dagher for technical assistance.
Abbreviations used:
- ChIP
chromatin immunoprecipitation
- CPC
chromosomal passenger complex
- 5-FOA
5-fluoroorotic acid
- GFP
green fluorescent protein
- PP2A
protein phosphatase 2A
- RFP
red fluorescent protein
- SAC
spindle assembly checkpoint
- SPB
spindle pole body
- YAC
yeast artificial chromosome
- YPD
yeast peptone dextrose
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E10-08-0673) on March 9, 2011.
REFERENCES
- Andalis AA, Storchova Z, Styles C, Galitski T, Pellman D, Fink GR. Defects arising from whole-genome duplications in Saccharomyces cerevisiae. Genetics. 2004;167:1109–1121. doi: 10.1534/genetics.104.029256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biggins S, Murray AW. The budding yeast protein kinase Ipl1/Aurora allows the absence of tension to activate the spindle checkpoint. Genes Dev. 2001;15:3118–3129. doi: 10.1101/gad.934801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biggins S, Severin FF, Bhalla N, Sassoon I, Hyman AA, Murray AW. The conserved protein kinase Ipl1 regulates microtubule binding to kinetochores in budding yeast. Genes Dev. 1999;13:532–544. doi: 10.1101/gad.13.5.532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyarchuk Y, Salic A, Dasso M, Arnaoutov A. Bub1 is essential for assembly of the functional inner centromere. J Cell Biol. 2007;176:919–928. doi: 10.1083/jcb.200609044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheeseman IM, Anderson S, Jwa M, Green EM, Kang J, Yates JR III, Chan CS, Drubin DG, Barnes G. Phospho-regulation of kinetochore-microtubule attachments by the Aurora kinase Ipl1p. Cell. 2002;111:163–172. doi: 10.1016/s0092-8674(02)00973-x. [DOI] [PubMed] [Google Scholar]
- Clift D, Bizzari F, Marston AL. Shugoshin prevents cohesin cleavage by PP2A(Cdc55)-dependent inhibition of separase. Genes Dev. 2009;23:766–780. doi: 10.1101/gad.507509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dewar H, Tanaka K, Nasmyth K, Tanaka TU. Tension between two kinetochores suffices for their biorientation on the mitotic spindle. Nature. 2004;428:93–97. doi: 10.1038/nature02328. [DOI] [PubMed] [Google Scholar]
- Fernius J, Hardwick KG. Bub1 kinase targets Sgo1 to ensure efficient chromosome biorientation in budding yeast mitosis. PLoS Genet. 2007;3:e213. doi: 10.1371/journal.pgen.0030213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujiwara T, Bandi M, Nitta M, Ivanova EV, Bronson RT, Pellman D. Cytokinesis failure generating tetraploids promotes tumorigenesis in p53-null cells. Nature. 2005;437:1043–1047. doi: 10.1038/nature04217. [DOI] [PubMed] [Google Scholar]
- Ganem NJ, Godinho SA, Pellman D. A mechanism linking extra centrosomes to chromosomal instability. Nature. 460:278–282. doi: 10.1038/nature08136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gestaut DR, Graczyk B, Cooper J, Widlund PO, Zelter A, Wordeman L, Asbury CL, Davis TN. Phosphoregulation and depolymerization-driven movement of the Dam1 complex do not require ring formation. Nat Cell Biol. 2008;10:407–414. doi: 10.1038/ncb1702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghaemmaghami S, Huh WK, Bower K, Howson RW, Belle A, Dephoure N, O'Shea EK, Weissman JS. Global analysis of protein expression in yeast. Nature. 2003;425:737–741. doi: 10.1038/nature02046. [DOI] [PubMed] [Google Scholar]
- Hewitt L, Tighe A, Santaguida S, White AM, Jones CD, Musacchio A, Green S, Taylor SS. Sustained Mps1 activity is required in mitosis to recruit O-Mad2 to the Mad1-C-Mad2 core complex. J Cell Biol. 2010;190:25–34. doi: 10.1083/jcb.201002133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoyt MA, Totis L, Roberts BT. S. cerevisiae genes required for cell cycle arrest in response to loss of microtubule function. Cell. 1991;66:507–517. doi: 10.1016/0092-8674(81)90014-3. [DOI] [PubMed] [Google Scholar]
- Huang D, Koshland D. Chromosome integrity in Saccharomyces cerevisiae: the interplay of DNA replication initiation factors, elongation factors, and origins. Genes Dev. 2003;17:1741–1754. doi: 10.1101/gad.1089203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Indjeian VB, Murray AW. Budding yeast mitotic chromosomes have an intrinsic bias to biorient on the spindle. Curr Biol. 2007;17:1837–1846. doi: 10.1016/j.cub.2007.09.056. [DOI] [PubMed] [Google Scholar]
- Indjeian VB, Stern BM, Murray AW. The centromeric protein Sgo1 is required to sense lack of tension on mitotic chromosomes. Science. 2005;307:130–133. doi: 10.1126/science.1101366. [DOI] [PubMed] [Google Scholar]
- Jelluma N, Brenkman AB, Van Den Broek NJ, Cruijsen CW, van Osch MH, Lens SM, Medema RH, Kops GJ. Mps1 phosphorylates Borealin to control Aurora B activity and chromosome alignment. Cell. 2008;132:233–246. doi: 10.1016/j.cell.2007.11.046. [DOI] [PubMed] [Google Scholar]
- Jones MH, Huneycutt BJ, Pearson CG, Zhang C, Morgan G, Shokat K, Bloom K, Winey M. Chemical genetics reveals a role for Mps1 kinase in kinetochore attachment during mitosis. Curr Biol. 2005;15:160–165. doi: 10.1016/j.cub.2005.01.010. [DOI] [PubMed] [Google Scholar]
- Kalocsay M, Hiller NJ, Jentsch S. Chromosome-wide Rad51 spreading and SUMO-H2A.Z-dependent chromosome fixation in response to a persistent DNA double-strand break. Mol Cell. 2009;33:335–343. doi: 10.1016/j.molcel.2009.01.016. [DOI] [PubMed] [Google Scholar]
- Kang J, Cheeseman IM, Kallstrom G, Velmurugan S, Barnes G, Chan CS. Functional cooperation of Dam1, Ipl1, and the inner centromere protein (INCENP)-related protein Sli15 during chromosome segregation. J Cell Biol. 2001;155:763–774. doi: 10.1083/jcb.200105029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katis VL, Galova M, Rabitsch KP, Gregan J, Nasmyth K. Maintenance of cohesin at centromeres after meiosis I in budding yeast requires a kinetochore-associated protein related to MEI-S332. Curr Biol. 2004;14:560–572. doi: 10.1016/j.cub.2004.03.001. [DOI] [PubMed] [Google Scholar]
- Kawashima SA, Tsukahara T, Langegger M, Hauf S, Kitajima TS, Watanabe Y. Shugoshin enables tension-generating attachment of kinetochores by loading Aurora to centromeres. Genes Dev. 2007;21:420–435. doi: 10.1101/gad.1497307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawashima SA, Yamagishi Y, Honda T, Ishiguro K, Watanabe Y. Phosphorylation of H2A by Bub1 prevents chromosomal instability through localizing shugoshin. Science. 2010;327:172–177. doi: 10.1126/science.1180189. [DOI] [PubMed] [Google Scholar]
- Keogh MC, et al. The Saccharomyces cerevisiae histone H2A variant Htz1 is acetylated by NuA4. Genes Dev. 2006;20:660–665. doi: 10.1101/gad.1388106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiburz BM, Amon A, Marston AL. Shugoshin promotes sister kinetochore biorientation in Saccharomyces cerevisiae. Mol Biol Cell. 2008;19:1199–1209. doi: 10.1091/mbc.E07-06-0584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiburz BM, Reynolds DB, Megee PC, Marston AL, Lee BH, Lee TI, Levine SS, Young RA, Amon A. The core centromere and Sgo1 establish a 50-kb cohesin-protected domain around centromeres during meiosis I. Genes Dev. 2005;19:3017–3030. doi: 10.1101/gad.1373005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JH, Kang JS, Chan CS. Sli15 associates with the Ipl1 protein kinase to promote proper chromosome segregation in Saccharomyces cerevisiae. J Cell Biol. 1999;145:1381–1394. doi: 10.1083/jcb.145.7.1381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinoshita E, Kinoshita-Kikuta E, Takiyama K, Koike T. Phosphate-binding tag, a new tool to visualize phosphorylated proteins. Mol Cell Proteomics. 2006;5:749–757. doi: 10.1074/mcp.T500024-MCP200. [DOI] [PubMed] [Google Scholar]
- Kitajima TS, Hauf S, Ohsugi M, Yamamoto T, Watanabe Y. Human Bub1 defines the persistent cohesion site along the mitotic chromosome by affecting shugoshin localization. Curr Biol. 2005;15:353–359. doi: 10.1016/j.cub.2004.12.044. [DOI] [PubMed] [Google Scholar]
- Kwiatkowski N, et al. Small-molecule kinase inhibitors provide insight into Mps1 cell cycle function. Nat Chem Biol. 2010;6:359–368. doi: 10.1038/nchembio.345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lauze E, Stoelcker B, Luca FC, Weiss E, Schutz AR, Winey M. Yeast spindle pole body duplication gene MPS1 encodes an essential dual specificity protein kinase. EMBO J. 1995;14:1655–1663. doi: 10.1002/j.1460-2075.1995.tb07154.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lea D, Coulson C. The distribution of the numbers of mutants in bacterial populations. J Genet. 1949;49:264–285. doi: 10.1007/BF02986080. [DOI] [PubMed] [Google Scholar]
- Lin H, de Carvalho P, Kho D, Tai CY, Pierre P, Fink GR, Pellman D. Polyploids require Bik1 for kinetochore-microtubule attachment. J Cell Biol. 2001;155:1173–1184. doi: 10.1083/jcb.200108119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maciejowski J, George KA, Terret ME, Zhang C, Shokat KM, Jallepalli PV. Mps1 directs the assembly of Cdc20 inhibitory complexes during interphase and mitosis to control M phase timing and spindle checkpoint signaling. J Cell Biol. 2010;190:89–100. doi: 10.1083/jcb.201001050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margolis RL. Tetraploidy and tumor development. Cancer Cell. 2005;8:353–354. doi: 10.1016/j.ccr.2005.10.017. [DOI] [PubMed] [Google Scholar]
- Maure J-F, Kitamura E, Tanaka TU. Mps1 kinase promotes sister-kinetochore bi-orientation by a tension-dependent mechanism. Curr Biol. 2007;17:2175–2182. doi: 10.1016/j.cub.2007.11.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer VW, Aguilera A. High levels of chromosome instability in polyploids of Saccharomyces cerevisiae. Mutat Res. 1990;231:177–186. doi: 10.1016/0027-5107(90)90024-x. [DOI] [PubMed] [Google Scholar]
- Musacchio A, Salmon ED. The spindle-assembly checkpoint in space and time. Nat Rev. 2007;8:379–393. doi: 10.1038/nrm2163. [DOI] [PubMed] [Google Scholar]
- Pinsky BA, Kung C, Shokat KM, Biggins S. The Ipl1-Aurora protein kinase activates the spindle checkpoint by creating unattached kinetochores. Nat Cell Biol. 2006;8:78–83. doi: 10.1038/ncb1341. [DOI] [PubMed] [Google Scholar]
- Riedel CG, et al. Protein phosphatase 2A protects centromeric sister chromatid cohesion during meiosis I. Nature. 2006;441:53–61. doi: 10.1038/nature04664. [DOI] [PubMed] [Google Scholar]
- Santaguida S, Tighe A, D’Alise AM, Taylor SS, Musacchio A. Dissecting the role of MPS1 in chromosome biorientation and the spindle checkpoint through the small molecule inhibitor reversine. J Cell Biol. 2010;190:73–87. doi: 10.1083/jcb.201001036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shackney SE, Smith CA, Miller BW, Burholt DR, Murtha K, Giles HR, Ketterer DM, Pollice AA. Model for the genetic evolution of human solid tumors. Cancer Res. 1989;49:3344–3354. [PubMed] [Google Scholar]
- Shimogawa MM, Widlund PO, Riffle M, Ess M, Davis TN. Bir1 is required for the tension checkpoint. Mol Biol Cell. 2009;20:915–923. doi: 10.1091/mbc.E08-07-0723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sliedrecht T, Zhang C, Shokat KM, Kops GJ. Chemical genetic inhibition of Mps1 in stable human cell lines reveals novel aspects of Mps1 function in mitosis. PloS One. 2010;5:e10251. doi: 10.1371/journal.pone.0010251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storchová Z, Breneman A, Cande J, Dunn J, Burbank K, O’Toole E, Pellman D. Genome-wide genetic analysis of polyploidy in yeast. Nature. 2006;443:541–547. doi: 10.1038/nature05178. [DOI] [PubMed] [Google Scholar]
- Storchova Z, Kuffer C. The consequences of tetraploidy and aneuploidy. J Cell Sci. 2008;121:3859–3866. doi: 10.1242/jcs.039537. [DOI] [PubMed] [Google Scholar]
- Storchova Z, Pellman D. From polyploidy to aneuploidy, genome instability and cancer. Nat Rev. 2004;5:45–54. doi: 10.1038/nrm1276. [DOI] [PubMed] [Google Scholar]
- Tanaka TU, Rachidi N, Janke C, Pereira G, Galova M, Schiebel E, Stark MJ, Nasmyth K. Evidence that the Ipl1-Sli15 (Aurora kinase-INCENP) complex promotes chromosome biorientation by altering kinetochore-spindle pole connections. Cell. 2002;108:317–329. doi: 10.1016/s0092-8674(02)00633-5. [DOI] [PubMed] [Google Scholar]
- Thomas S, Kaplan KB. A Bir1p Sli15p kinetochore passenger complex regulates septin organization during anaphase. Mol Biol Cell. 2007;18:3820–3834. doi: 10.1091/mbc.E07-03-0201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Dai W. Shugoshin, a guardian for sister chromatid segregation. Exp Cell Res. 2005;310:1–9. doi: 10.1016/j.yexcr.2005.07.018. [DOI] [PubMed] [Google Scholar]
- Warren CD, Brady DM, Johnston RC, Hanna JS, Hardwick KG, Spencer FA. Distinct chromosome segregation roles for spindle checkpoint proteins. Mol Biol Cell. 2002;13:3029–3041. doi: 10.1091/mbc.E02-04-0203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winey M, Huneycutt BJ. Centrosomes and checkpoints: the MPS1 family of kinases. Oncogene. 2002;21:6161–6169. doi: 10.1038/sj.onc.1205712. [DOI] [PubMed] [Google Scholar]
- Xu Z, Cetin B, Anger M, Cho US, Helmhart W, Nasmyth K, Xu W. Structure and function of the PP2A-shugoshin interaction. Mol Cell. 2009;35:426–441. doi: 10.1016/j.molcel.2009.06.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.