Fas-associated factor 1 (FAF1) is identified as a negative regulator of Wnt/β-catenin signaling. In addition, its involvement in osteoblast differentiation and mineralization is shown. FAF1 was found to promote β-catenin degradation by enhancing its polyubiquitination.
Abstract
The canonical Wnt pathway plays an important role in the regulation of cell proliferation and differentiation. Activation of this signaling pathway causes disruption of the Axin/adenomatous polyposis coli/glycogen synthase kinase 3β complex, resulting in stabilization of β-catenin and its association with lymphoid enhancer factor/T-cell factor in the nucleus. Here, we identify Fas-associated factor 1 (FAF1) as a negative regulator of Wnt/β-catenin signaling. We found overexpression of FAF1 to strongly inhibit Wnt-induced transcriptional reporter activity and to counteract Wnt-induced β-catenin accumulation. Moreover, knockdown of FAF1 resulted in an increase in β-catenin levels and in activation of Wnt/β-catenin–induced transcription. FAF1 was found to interact with β-catenin upon inhibition of proteasome. Ectopic expression of FAF1 promoted β-catenin degradation by enhancing its polyubiquitination. Functional studies in C2C12 myoblasts and KS483 preosteoblastic cells showed that FAF1 depletion resulted in activation of endogenous Wnt-induced genes and enhanced osteoblast differentiation, whereas FAF1 overexpression had the opposite effect. These results identify FAF1 as a novel inhibitory factor of canonical Wnt signaling pathway.
INTRODUCTION
Wnts constitute a family of secreted proteins that regulate cell proliferation and differentiation and thereby control many biological processes, including embryonic development and tumorigenesis (Logan and Nusse, 2004; Moon et al., 2004; Reya and Clevers, 2005; MacDonald et al., 2009). One of the major components of the canonical Wnt-induced signaling pathway is β-catenin. In the absence of Wnt ligands, the majority of β-catenin is associated with E-cadherin on the plasma membrane, whereas the cytosolic β-catenin is rapidly degraded by the proteasome system due to phosphorylation by the adenomatous polyposis coli (APC)/Axin/glycogen synthase kinase (GSK)3β complex (Liu et al., 2002; Nelson and Nusse, 2004). In the presence of Wnt ligand, its receptor frizzled (Fz) and its coreceptor low-density lipoprotein receptor-related protein-5 or -6 (LRP-5/6) recruit Axin and GSK3β to the plasma membrane, together with the scaffold protein Disheveled (Dvl) (Zeng et al., 2005; Bilic et al., 2007). The membrane association of Axin and GSK3β disrupts the β-catenin destruction complex, resulting in accumulation of β-catenin in the nucleus, where it triggers target gene activation by displacing transcriptional repressors from DNA-bound lymphoid enhancer factor (LEF)/T-cell factor (TCF) (Behrens et al., 1996; Brunner et al., 1997).
Wnt/β-catenin signaling is one of the key signaling pathways required for bone formation and bone homeostasis. The canonical Wnt pathway directly stimulates Runx2 expression and promotes mineralization during osteoblastogenesis (Gaur et al., 2005). The Wnt pathway also blocks apoptosis and osteoclastogenesis by increasing the osteoprotegerin (OPG)/receptor activator of nuclear factor κB ligand (RANKL) ratio (Glass et al., 2005; Holmen et al., 2005; Spencer et al., 2006). In vivo, mutations in LRP-5 profoundly affect skeletal development and result in low bone mass (Gong et al., 2001; Little et al., 2002). The Dickkopf-1 (Dkk-1)–resistant LRP5V171 mutation leads to high bone density (Boyden et al., 2002). Conditional deletion of the β-catenin gene in osteoblasts in vivo also leads to reduced bone mass (Glass et al., 2005). Moreover, osteocyte-specific β-catenin–deficient mice develop a severe early-onset low bone mass phenotype (Kramer et al., 2010).
FAS-associated factor 1 (FAF1) was first identified as a member of the Fas death–inducing signaling complex and potentiates apoptosis in L cells and T cells (Chu et al., 1995; Yanagisawa et al., 1997; Frohlich et al., 1998; Ryu et al., 2003). Human FAF1 mRNA is abundantly expressed in testis, skeletal muscle, and heart (Ryu et al., 1999). FAF1 is also involved in the regulation of NF-κB activity, as well as in VCP (valosin containing protein)-related protein ubiquitination and proteasomal degradation (Park et al., 2004, 2007; Song et al., 2005). FAF1 has been postulated to act as a tumor suppressor. Reduced FAF1 expression was found in a high percentage of human gastric carcinomas (Bjorling-Poulsen et al., 2003), whereas genomic loss or deletion of FAF1 has been reported in uterine cervix carcinomas and mantle cell lymphoma (Hidalgo et al., 2005; Bea et al., 2009). The frequently down-regulated expression of FAF1 in malignant mesothelioma leads to aberrant NF-κB signaling (Altomare et al., 2009). Importantly, Faf1 gene-targeted mice show embryonic lethality at the two-cell stage, which demonstrates that FAF1 is also required for early embryogenesis (Adham et al., 2008).
In this study, we show that FAF1 inhibits the expression of Wnt-responsive transcriptional reporters and endogenous Wnt target genes. We further demonstrate that FAF1 is required and sufficient to decrease cytosolic β-catenin by promoting its polyubiquitination and degradation via the proteasome pathway. FAF1 depletion resulted in β-catenin accumulation and potentiation of canonical Wnt signaling, particularly in Wnt-induced osteoblast differentiation.
RESULTS
FAF1 inhibits Wnt/β-catenin signaling
We examined the effect of FAF1 on Wnt signaling because FAF1 has been postulated to act as a tumor suppressor by regulating ubiquitination and proteasomal degradation. Ectopic expression of human FAF1 in 293T cells inhibited the activation of the Wnt–transcriptional reporter constructs TopFlash–luciferase (Korinek et al., 1997) and LEF–luciferase (Hsu et al., 1998) induced by Wnt1, Wnt3a, and several other Wnt pathway components (Figure 1, A and B). The inhibition of TopFlash–luciferase by FAF1 was dose-dependent (Figure 1C) and Wnt-specific, since the control reporter Fopflash–luciferase was not inhibited (data not shown). Because β-catenin is the key factor in Wnt-induced transcription, we next tested whether increased expression of β-catenin could reverse the inhibitory effect of FAF1. Elevated levels of β-catenin indeed counteracted the inhibition by FAF1 (Figure 1D), but high doses of LEF-1 did not, indicating that FAF1 antagonizes Wnt/β-catenin signaling upstream of LEF-1.
FIGURE 1:
FAF1 inhibits Wnt/β-catenin reporter activity. HEK293T cells were cotransfected with TopFlash-Luc reporter plasmid (A) or LEF-Luc reporter plasmid (B), together with a FAF1 expression vector or the control empty vector, and Wnt1, Wnt3a, LRP6, Dvl2, or β-catenin (β-wt) expression constructs. The transfected cells were lysed for luciferase assay at 36 h posttransfection. Each experiment was performed in triplicate, and the data represent mean ± SD of three independent experiments after normalization to β-gal activity. (C) HEK293T cells were cotransfected with TopFlash-Luc reporter, Wnt3a, β-catenin (β-wt) constructs, and increasing amounts of FAF1 expression plasmid as indicated. (D) HEK293T cells were cotransfected with TopFlash-Luc reporter, FAF1 expression vector, and different amounts of β-catenin (β-wt) and LEF-1 constructs as indicated. (E) HEK293T cells infected with lentiviruses expressing control nontargeting shRNA or two independent FAF1 shRNAs were cotransfected with TopFlash-Luc and Wnt3a or β-catenin (β-wt) constructs as indicated. Forty-eight hours after transfection, cells were harvested for luciferase assay. (F) β-catenin, Axin1, GSK3β, Dapper1, and FAF1 constructs were cotransfected together with TopFlash-Luc reporter in HEK293T cells as indicated. Thirty-six hours after transfection, cells were harvested for the luciferase assay.
To examine the role of endogenous FAF1 in Wnt signaling, we used two independent human FAF1 small hairpin (sh)RNA vectors that efficiently knocked down FAF1 expression (Figure 1E). In line with the effect of FAF1 overexpression, FAF1 depletion increased the basal and Wnt3a and β-catenin–induced activity of the TopFlash–luciferase reporter (Figure 1E). Depletion of mouse FAF1 in C2C12 cells also enhanced Wnt-induced reporter activity (data not shown). Moreover, ectopic expression of FAF1 could inhibit the Wnt reporter to about the same extent as the previously identified Wnt inhibitory factors Axin, GSK3β, Chibby, and Dapper1 when their expression levels are comparable (Cheyette et al., 2002; Takemaru et al., 2003; Zhang et al., 2006) (Figure 1F and data not shown).
FAF1 reduces cytosolic β-catenin
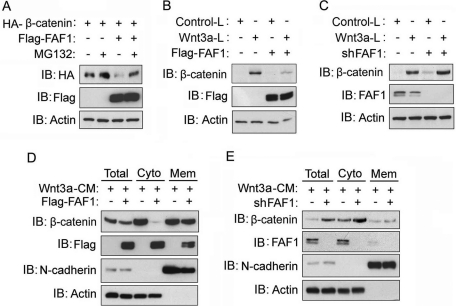
To investigate the mechanism by which FAF1 regulates canonical Wnt/β-catenin signaling, we examined its effect on β-catenin accumulation. First, we found that the levels of ectopically expressed β-catenin were reduced upon coexpression of FAF1 and that this reduction was much less after addition of the proteasome inhibitor MG132 (Figure 2A). As the majority of the ectopically expressed β-catenin accumulates in the cytoplasm, the result suggested that overexpression of FAF1 can trigger (cytosolic) β-catenin degradation. To test this, we used mouse L cells, which express almost no membrane-associated β-catenin. We stably overexpressed or knocked down FAF1 in both control and Wnt3a-expressing L cells. As shown in Figure 2 (B and C), the Wnt3a-induced β-catenin levels in these cells were indeed suppressed by FAF1 ectopic expression and increased by selective FAF1 shRNA-mediated depletion. Comparable results were obtained when we analyzed the membrane and cytosolic fractions of Wnt3a-treated HEK293T cells: forced expression of FAF1 reduced and FAF1 depletion enhanced the levels of β-catenin mainly in the cytosolic fraction (Figure 2, D and E). These results thus indicate that FAF1 controls canonical Wnt signaling by targeting β-catenin.
FIGURE 2:
FAF1 inhibits the accumulation of cytosolic β-catenin. (A) HEK293T cells were transfected with HA-tagged β-catenin and Flag-tagged FAF1 expression plasmids. Forty hours after transfection, cells were treated with MG132 (5 μM) for another 4 h. Cells were then harvested for HA-β-catenin and Flag-FAF1 immunoblot-detection (IB). Actin was included as a loading control. (B, C) Control L cells or L cells stably expressing Wnt3a were infected with lentivirus expressing Flag-tagged FAF1 (B) or FAF1 shRNA (C). Forty-eight hours after infection, cells were harvested for immunodetection of β-catenin, and FAF1 (D, E) HEK293T cells were infected with lentivirus expressing Flag-tagged FAF1 (D) or FAF1 shRNA (E). Forty hours after transfection, cells were incubated with Wnt3a conditioned medium (Wnt3a-CM) for another 6 h. Then cells were harvested, the membrane and cytosolic fractions were isolated, and β-catenin and FAF1 were detected by Western blotting. N-cadherin and actin were included as controls for the membrane and cytosolic fractions.
FAF1 promotes β-catenin polyubiquitination in vivo
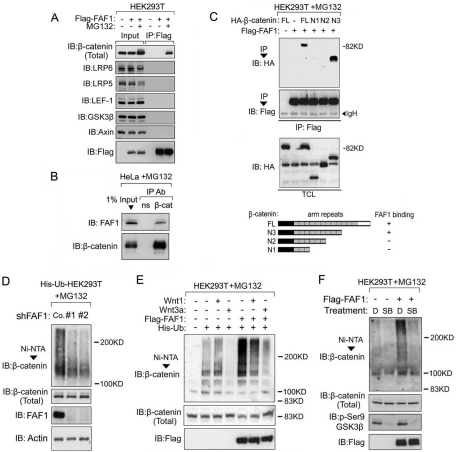
Because FAF1 is reported to be involved in ubiquitination and proteasomal degradation, we hypothesized that FAF1 may target cytosolic β-catenin for degradation through ubiquitination. We first performed coimmunoprecipitation assays to test whether FAF1 can directly interact with β-catenin. The endogenous β-catenin in HEK293T cells could indeed be coprecipitated with Flag-tagged FAF1, but only when the proteasome inhibitor MG132 was present (Figure 3A). In contrast, Flag-FAF1 did not coimmunoprecipitate with GSK3β, Axin, LRP5, LRP6, and LEF-1 (Figure 3A). These results indicate that FAF1 is not a constituent of the β-catenin destruction complex, in which Axin and GSK3β are the essential components, and show that FAF1 selectively interacts with β-catenin within the Wnt pathway. Interaction between endogenous FAF1 and β-catenin could also only be detected in MG132-treated HeLa, KS483, and C2C12 cells (Figure 3B, and Supplemental Figure S1, A and B), suggesting that, in all these cells, FAF1 promotes β-catenin degradation upon binding.
FIGURE 3:
FAF1 interacts with β-catenin and enhances its polyubiquitination. (A) Control HEK293T cells or cells transfected with Flag-tagged FAF1 were treated with MG132 (5 μM), or not, for 4 h, and cell lysates were immunoprecipitated (IP) with anti-Flag antibody. Association of the indicated Wnt pathway components was analyzed by immunoblotting (IB). Aliquots of the total cell lysates were loaded as input controls (left panel). (B) HeLa cells were treated with MG132 for 4 h. Immunoprecipitation (IP) was performed with anti-β-catenin antibody (β-cat) or a nonspecific antibody (ns). β-catenin–associated FAF1 was revealed by anti-FAF1 immunoblotting (IB). An aliquot of the total cell lysate was loaded as input control. (C) (top panel) HEK293T cells were transfected with Flag-tagged FAF1 and HA-tagged β-catenin full length (FL), or the β-catenin deletion mutants N1, N2, and N3. Forty-eight hours after transfection, cells were pretreated with MG132 for 4 h before harvesting. FAF-1 was immunoprecipitated with an anti-Flag antibody. Flag-FAF1–associated β-catenin was detected with anti-HA immunoblotting (IB). The input total cell lysate (TCL) is shown as control. (Bottom panel) Schematic representation of the β-catenin mutants and their binding ability to FAF1. (D) Stable His-ubiquitin expressing HEK293T cells were infected with lentivirus expressing control shRNA (Co.) or FAF1 shRNA (#1, #2). Sixty hours later, cells were treated with MG132 for 4 h and harvested for Nickel bead pull-down. The pulled-down polyubiquitinated endogenous β-catenin was detected by immunoblotting (IB). The total levels of β-catenin, FAF1, and actin in the input are shown in the lower panels (Total). (E) HEK293T cells were transfected with the indicated expression plasmids. Forty-eight hours later, cells were treated with MG132 for 4 h and harvested for Nickel pull-down. Polyubiquitinated endogenous β-catenin was detected as described above (D). (F) Stable His-ubiquitin expressing HEK293T cells were transfected with Flag-FAF1 plasmid or empty vector as indicated. Forty hours later, cells were treated with control dimethyl sulfoxide (D) or 10 μM SB216763 (SB) for 4 h. MG132 (5 μM) was added for another 4 h, and cells were harvested for Nickel pull-down. Polyubiquitinated endogenous β-catenin was detected by immunoblotting (IB). The levels of Flag-FAF1, β-catenin, and phosphorylated GSK3β (Ser-9) in the input are shown in the lower panels.
β-catenin contains 12 arm repeats that mediate its interaction with other proteins. To map the region required for FAF1 binding, hemagglutinin (HA)-tagged β-catenin deletion constructs were analyzed for their ability to coprecipitate with Flag-tagged FAF1. As shown in Figure 3C, construct N3, lacking arm repeat 10–12, still efficiently interacted with FAF1, but constructs N1 and N2, containing only arm repeats 1–3 and 1–6, did not. This indicates that arm repeat 7–9 mediates β-catenin binding to FAF1.
We next examined whether FAF1 has an effect on β-catenin ubiquitination. Nickel-bead pull-down in HEK293T cells stably expressing His-ubiquitin showed that knockdown of FAF1 results in reduction of the polyubiquitination of endogenous β-catenin (Figure 3D). In line with this, overexpression of FAF1 strongly increased endogenous β-catenin ubiquitination, whereas both the basal and FAF1-induced ubiquitination of β-catenin were blocked by Wnt3a stimulation (Figure 3E). Wnt3a was also found to decrease the interaction between FAF1 and β-catenin (Supplemental Figure S2). Wnt1 reduced the ubiquitination of β-catenin not or only weakly (Figure 3E), which is in line with the less efficient activation of Wnt-reporter activity by Wnt1 as compared with Wnt3a (Figure 1, A and B). This might be due to the fact that, in contrast to Wnt3a, Wnt1 can also stimulate noncanonical Wnt signaling via the Fz receptors, which can negatively affect the canonical β-catenin pathway (Kohn and Moon, 2005; Mikels and Nusse, 2006).
We subsequently analyzed whether FAF1 promotes β-catenin ubiquitination in an APC-Axin-GSK3β–dependent manner. FAF1-induced β-catenin ubiquitination was blocked by the GSK3β kinase inhibitor SB216763 (Figure 3F). However, FAF1 was not required for GSK3β-mediated β-catenin Ser-33/Ser-37/Thr-41 phosphorylation (Figure 4A) (Liu et al., 2002). This suggests that FAF1 acts downstream of GSK3β and might target phospho-β-catenin for degradation.
FIGURE 4:
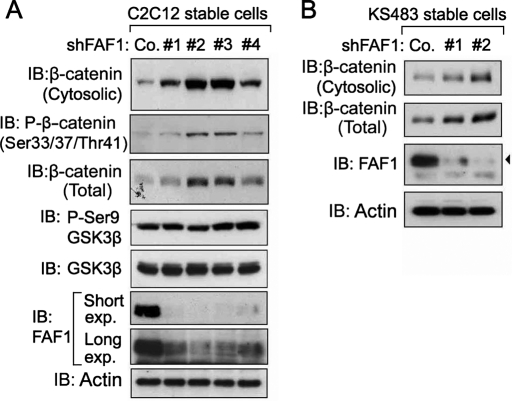
FAF1 depletion causes accumulation of cytosolic β-catenin in C2C12 and KS483 cells. (A, B) C2C12 (A) and KS483 (B) cells were infected with lentiviruses expressing independent FAF1 shRNAs (#1, #2, #3, or #4) and selected with puromycin (5 μg/ml) for 6 d. Cells were then harvested for isolation of the cytosolic fractions and analyzed by immunoblotting. Actin was included as a loading control. Co., control nontargeting shRNA.
To examine whether the inhibitory effect of FAF1 on Wnt signaling might be the result of a (relatively unspecific) effect of FAF1 on the proteasome, causing general ubiquitination and degradation of critical signaling molecules, we also examined its effect on bone morphogenic protein (BMP) signaling. However, FAF1 overexpression or knockdown did not affect BMP-induced Smad1 phosphorylation (Supplemental Figure S3). Taken together, the data indicate that FAF1 inhibits Wnt-signaling by selectively targeting β-catenin for polyubiquitination and subsequent degradation.
FAF1 depletion is sufficient for β-catenin accumulation and Wnt/β-catenin target gene activation in myoblasts and preosteoblastic cells
Due to the very efficient degradation of cytosolic β-catenin in most cell types, autocrine Wnt secretion by cells is usually not sufficient to trigger accumulation of soluble β-catenin or to activate β-catenin–dependent gene expression. However, if FAF1 is critical for β-catenin degradation in nonstimulated cells, one might detect enhanced levels of β-catenin without addition of Wnt ligands in FAF1-depleted cells. We tested this in C2C12 myoblast and KS483 preosteoblastic cells, two cell types in which Wnts induce osteoblast differentiation. As shown in Figure 4A, stable lentiviral depletion of FAF1 indeed induced the levels of cytosolic (phospho-) β-catenin in C2C12 cells, with the two strongest knockdown vectors (#2, #3) giving the highest β-catenin accumulation. Similar results were obtained in KS483 cells (Figure 4B).
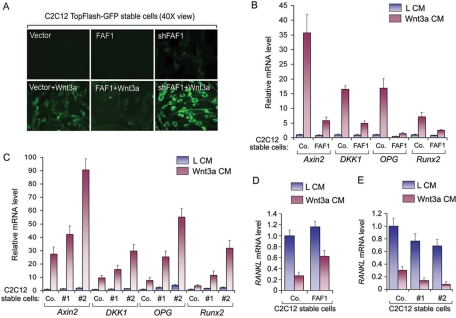
We next examined whether FAF1 depletion also results in activation of Wnt/β-catenin–induced transcription in C2C12 myoblasts. For this, we used cells stably expressing a TopFlash–green fluorescent protein (GFP) transcriptional reporter gene. In the vector control cells, GFP expression could only be detected upon Wnt3a stimulation, which, in line with the previous results, was inhibited by FAF1 overexpression and enhanced by FAF1 depletion. Importantly, FAF1 depletion also induced low levels of GFP expression in the absence of Wnt3a (Figure 5A, and Supplemental Figure S4A), and similar effects were obtained in HeLa cells stably expressing TopFlash-GFP (Supplemental Figure S4). These results show that FAF1 is critical for inhibition of β-catenin signaling in nonstimulated cells.
FIGURE 5:
FAF1 inhibits Wnt/β-catenin–induced transcription in C2C12 cells. (A) C2C12 cells stably expressing TopFlash-GFP were infected with a lentiviral vector that overexpresses FAF1 or a vector expressing FAF1 shRNA. Forty-eight hours after infection, cells were treated overnight with or without Wnt3a conditional medium and analyzed by microscopy. (B–E) C2C12 cells were stably infected with a lentiviral vector that overexpresses FAF1 (B, D) or with vectors expressing FAF1 shRNA #1 and #2 (C, E). Cells were treated with Wnt3a conditioned medium (Wnt3a CM) or control conditioned medium (L CM) for 6 h and harvested for QRT-PCR analysis. The relative mRNA levels shown were normalized to the levels in the control cells (Co.) treated with control medium. For each gene, the mRNA level in this control is set at “1.” Values and error bars represent the mean ± SD of triplicates.
To test the effect of FAF1 on endogenous Wnt/β-catenin target genes in C2C12 cells, we performed quantitative real-time (QRT)-PCR analysis. Consistent with the previous results, FAF1 overexpression strongly inhibited the Wnt3a-induced activation of Axin2, DKK1, OPG, and Runx2 (Figure 5B). Moreover, FAF1 knockdown enhanced both the basal and Wnt-induced activation of these four genes, the strongest effect being induced by the more potent knockdown vector #2 (Figures 5C and 4A). Wnt3a-mediated suppression of RANKL was also partially blocked by FAF1 overexpression and enhanced by FAF1 depletion (Figure 5, B and E). Together, these results show that FAF1 modulates Wnt/β-catenin–dependent gene expression in C2C12 myoblasts, including genes involved in osteoblast differentiation.
FAF1 negatively regulates Wnt-induced osteoblast differentiation and bone formation
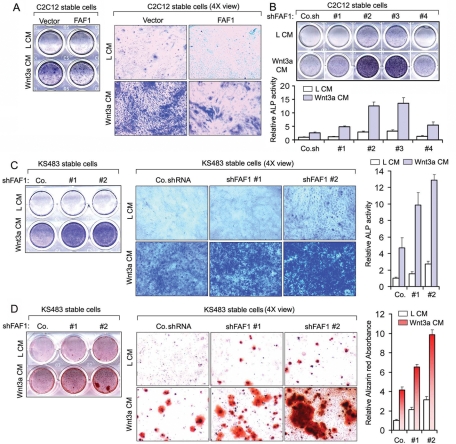
As the previous results indicated that FAF1 may regulate Wnt-induced osteoblast differentiation, we examined the effect of FAF1 modulation on the induction of two differentiation markers. For analysis of alkaline phosphatase (ALP) activity, an early differentiation marker, we incubated confluent C2C12 cells with or without Wnt3a for 4 d. Histochemical staining showed that Wnt3a-induced ALP activity was suppressed by FAF1 overexpression (Figure 6A) and highly potentiated by FAF1 depletion (Figure 6B). In particular, the stable cells expressing FAF1 shRNA constructs #2 and #3, showing the highest knockdown and highest levels of cytosolic β-catenin accumulation (Figure 4A), showed elevated ALP staining in the nontreated and Wnt3a-treated cells (Figure 6B). Stable FAF1 depletion also potentiated Wnt3a-induced ALP activity in KS483 preosteoblast cells (Figures 4B and 6C). As a (late) marker for osteogenic differentiation, we analyzed calcium deposition and formation of mineralized matrix by alizarin red S staining. As shown in Figure 6D, FAF1 knockdown also potentiated Wnt3a-induced mineralization in KS483 cells. Taken together, our results show that FAF1 negatively regulates Wnt-induced osteoblast differentiation.
FIGURE 6:
FAF1 regulates Wnt-induced osteoblast differentiation and bone formation. (A) (Left panel) C2C12 cells stably overexpressing FAF1 were treated with Wnt3a conditioned medium (Wnt3a CM) or control conditioned medium (L CM) for 4 d. Cells were then harvested for histochemical examination of ALP activity. (Right panel) Representative microscopic pictures of left panel. (B) (Top panel) C2C12 cells stably expressing FAF1 shRNA #1, #2, #3, or #4 were analyzed for Wnt-induced ALP activity as described above. (Bottom panel) The histochemically stained cell material was dissolved in 50 mM NaOH in ethanol, and absorbance was measured at 550 nm. Data show the mean and SD of triplicates. Co.sh, control nontargeting shRNA. (C) (Left panel) KS483 cells stably expressing FAF1 shRNA #1 or #2 were analyzed for Wnt-induced ALP activity as described under A. (Middle panel) Representative microscopic pictures of left panel. (Right panel) The histochemically stained cell material was dissolved in 50 mM NaOH in ethanol, and absorbance was measured at 550 nm. Data show the mean and SD of triplicates. Co, control shRNA. (D) (Left panel) KS483 cells stably expressing FAF1 shRNA #1 or #2 were stimulated with Wnt3a conditioned medium (Wnt3a CM) or control conditioned medium (L CM) containing Vitamin C (50 μg/ml) and β-glycerolphosphate (5 μM). Four days later, the medium was replaced by normal medium containing 10% FBS, Vitamin C (50 μg/ml), and β-glycerolphosphate (5 μM) for another 4 d. Cells were then fixed and stained with alizarin red S for analysis of mineralization. (Middle panel) Representative microscopic pictures of left panel. (Right panel) Quantification of alizarin red S staining. Data show the mean and SD of triplicates.
DISCUSSION
As a component of the Fas death–inducing signaling complex, FAF1 was originally identified as a negative regulator of cell survival. Moreover, the recurrent loss or down-regulation of FAF1 in certain cancers indicated a potential function in tumor suppression. FAF1-targeted mice showed embryonic lethality at the two-cell stage, indicating also an important FAF1 role in early development. Down-regulation of FAF1 in cancer has been linked to aberrant NF-κB activity, but its role in development and tumorigenesis are further unknown. The fact that FAF1 is also implicated in ubiquitination and proteasomal degradation suggested that FAF1 may also modulate additional signal transduction pathways.
The findings presented here reveal a critical role of FAF1 in the regulation of canonical Wnt signaling. By performing knockdown and ectopic expression studies, we showed that FAF1 inhibits Wnt reporter activity by increasing the degradation of cytosolic β-catenin. Moreover, we showed that FAF1 and β-catenin physically interact and that FAF1 enhances β-catenin polyubiquitination. As a consequence, FAF1 depletion results in accumulation of β-catenin and potentiation of Wnt target gene expression. Our results also indicate that, in multiple cell types, FAF1 is required for efficient degradation of (soluble) β-catenin in the absence of external Wnt ligands. FAF1 does not appear to be a constituent of the β-catenin destruction complex, because FAF1 selectively interacts with β-catenin within the Wnt pathway and is not required for GSK3β-mediated β-catenin phosphorylation. Interestingly, FAF1 contains a UBX domain involved in ubiquitin binding, which is required for FAF1-induced polyubiquitination of β-catenin (our unpublished results). As FAF1 is not an E3 ubiquitin ligase itself, the mechanism by which it promotes β-catenin ubiquitination awaits further investigation. However, as the E3 ligase β-TRCP (β-transducin repeat containing) rather than Smurf2 (SMAD specific E3 ubiquitin protein ligase 2) appears to be involved in this process, and the interaction between β-catenin and β-TRCP is reduced upon FAF1 knockdown (our unpublished results), FAF1 may facilitate the interaction between β-catenin and β-TRCP.
As an essential signaling pathway for bone formation and bone homeostasis, the Wnt/β-catenin pathway is modulated by regulators at multiple steps, the amount of (cytosolic) β-catenin being the most decisive response parameter. To investigate the functional significance of FAF1 in the modulation of canonical Wnt signaling, we examined Wnt-induced osteoblast differentiation and bone formation in C2C12 myoblasts and KS483 osteoprogenitor cells. Consistent with its effects on several critical Wnt target genes, FAF1 overexpression nearly completely blocked Wnt-induced ALP activity, whereas FAF1 depletion strongly potentiated the Wnt-induced ALP levels. Because ALP is an early marker for bone formation, we further confirmed these results by analyzing calcium deposition and the formation of mineralized matrix, occuring late in differentiation. Based on these results, we can conclude that FAF1 is a negative modulator of Wnt-induced osteoblast differentiation.
As a pathway that can promote cell growth, canonical Wnt-signaling also contributes to cancer cell proliferation. Mutation or deletion of critical genes involved in β-catenin destruction, such as APC mutations, directly contribute to cancer. Therefore the loss or down-regulation of FAF1 observed in certain cancers might be linked to the inhibitory effect of FAF1 on the Wnt/β-catenin pathway. Although much more work is needed to elucidate the exact mechanism by which FAF1 mediates β-catenin degradation, both its physiological and pathological functions in embryos, adult tissues, and in particular tumors might be mediated at least in part via the canonical Wnt signaling pathway.
MATERIALS AND METHODS
Reagents and plasmids
FAF1 (#4932), LEF-1 (#2230), GSK3β (#9315), and Phospho-GSK-3β (Ser9) (#9336) antibodies were purchased from Cell Signaling Technology (Beverly, MA). β-catenin antibody (#610153) was purchased from BD Biosciences (San Jose, CA); actin (ab8227) and LRP5 (ab36121) antibodies were purchased from Abcam (Cambridge, MA). Smad1 (sc7965), Smad4 (sc7966), and LRP6 (sc25317) antibodies were purchased from Santa Cruz Biotechnology (Santa Cruz, CA). Axin antibody was kindly provided by Shengcai Lin of Xiamen University. Mouse FAF1 shRNA (#1: TRCN0000191814; #2 TRCN0000191433; #3 TRCN0000190097; #4 TRCN0000192222) and human FAF1 shRNA (1#, TRCN0000004244; 2#, TRCN0000004246) MISSION shRNA Lentiviral Transduction Particles were purchased from Sigma Mission Library (St. Louis, MO). TopFlash-luciferase, FopFlash-luciferase, LEF-luciferase, Wnt1, Wnt3a, Axin1, Dapper1, GSK3β, β-catenin, LEF-1, and 6His-Ubiquitin constructs were described previously (Zhang et al., 2006; Zhou et al., 2008a, 2008b). FAF1 cDNA was obtained from Novartis (East Hanover, NJ). FAF1 and Flag-tagged FAF1 were cloned into plv-bc-puro lenti vector. All of the constructs were verified by DNA sequencing.
Cell culture
C2C12, KS483, HeLa, HEK293T, and L cells were cultured in DMEM supplemented with 10% fetal bovine serum (FBS) (HyClone, Logan, UT), nonessential amino acids, l-glutamine, and penicillin/streptomycin in a 5% CO2-containing atmosphere at 37°C.
Luciferase reporter assay
Cells were transfected and lysed as described (Zhang et al., 2006), and luciferase activities were measured by a luminometer (Berthold Technologies). Reporter activity was normalized to β-galactosidase (gal) activity, resulting from a cotransfected internal control plasmid. Experiments were performed in triplicate.
Transfection, immunoprecipitation, and immunoblotting
Cells were transiently transfected using calcium phosphate or Lipofectamine (Invitrogen, Carlsbad, CA). Forty hours posttransfection, cells were lysed with 1 ml of lysis buffer (20 mM Tris-HCl, pH 7.4, 2 mM EDTA, 25 mM NaF, 1% Triton X-100) plus protease inhibitors (Sigma) for 30 min at 4°C. After centrifugation at 12 × 103 g for 15 min, the lysates were immunoprecipitated with specific antibody and protein A-Sepharose (Zymed Laboratories, San Francisco, CA) for 3 h at 4°C. Thereafter the precipitants were washed three times with washing buffer (50 mM Tris-HCl, pH 8.0, 150 mM NaCl, 1% Nonidet P-40, 0.5% sodium deoxycholate, and 0.1% SDS), and the immune complexes were eluted with sample buffer containing 1% SDS for 5 min at 95°C and analyzed by SDS–PAGE. Immunoblotting was performed with specific antibody and secondary anti–mouse or anti–rabbit antibodies that were conjugated to horseradish peroxidase (Amersham Biosciences, Piscataway, NJ). Proteins were visualized by chemiluminescence.
Nickel pull down
Cells were lysed in 6M guanidine-HCl, 0.1 M Na2HPO4/NaH2PO4, 10 mM imidazole, followed by nickel bead purification and immunoblot analysis.
Lentiviral transduction and generation of stable cell lines
Lentiviruses were produced by transfecting pLKO-1 (for the shRNA-knockdown) or pLV-bc-CMV (for cDNA expression) plasmids together with helper plasmids pCMV-VSVG, pMDLg-RRE (gag/pol), and pRSV-REV into HEK293T cells. Cell supernatants were harvested 48 h after transfection and were either used to infect cells or stored at −80°C. To obtain stable cell lines, cells were infected at low confluence (20%) for 24 h with lentiviral supernatants diluted 1:1 with normal culture medium in the presence of 5 ng/ml of polybrene (Sigma). Forty-eight hours after infection, cells were placed under 5 μg/ml puromycin selection for 1 wk and then passaged before use. To obtain TopFlash-GFP stable cell lines, pCDNA3.1 TopFlash-GFP construct were transfected into C2C12 and HeLa cells, cells were placed under 1 μg/ml neomycin selection for 1 wk, and then passaged before use. His-ubiquitin expressing HEK293T cells were obtained by transfection with His-ubiquitin construct followed by selection in 1 μg/ml puromycin.
ALP staining and mineralization assays
Histochemical examination of ALP activity in cells was performed using naphtol AS-MX phosphate (Sigma) and fast blue RR salt (Sigma) (van Dinther et al., 2010). For mineralization assays cells were washed with phosphate-buffered saline (PBS), fixed with 3.7% formaldehyde, again washed with PBS, and then incubated with 2% alizarin red S solution (pH 4.2) for 2 min and washed with distilled water (van Dinther et al., 2010).
QRT-PCR (quantitative real-time–PCR)
Total RNA was isolated using NucleoSpin RNA II kit (BIOKÉ, Leiden, the Netherlands) reagent. RNA (1 μg) was reverse-transcribed using the RevertAid First Strand cDNA Synthesis Kits (Fermentas, Burlington, Canada). QRT-PCR was accomplished with SYBR Green incorporation (Applied Bioscience, Somerset, NJ) using a StepOne Plus real-time PCR system (Applied Bioscience). Results were normalized to those obtained with GAPDH. Primers used for QRT-PCR were:
mGAPDH forward, 5′-AACTTTGGCATTGTGGAAGG-3′
mGAPDH reverse, 5′-ACACATTGGGGGTAGGAACA-3′
mRunx2 forward, 5′-GAATGCTTCATTCGCCTCAC-3′
mRunx2 reverse, 5′-GTGACCTGCAGAGATTAACC-3′
mOpg (osteoprotegerin) forward, 5′-CGCAAAAGTGTGGAATAGATGTCA-3′
mOpg (osteoprotegerin) reverse, 5′-GGTAGGAACAGCAAACCTGAAGA-3′
mAxin2 forward, 5′-GGTTCCGGCTATGTCTTTGC-3′
mAxin2 reverse, 5′-CAGTGCGTCGCTGGATAACTC-3′
mDKK1 forward, 5′-GCTGCATGAGGCACGCTAT-3′
mDKK1 reverse, 5′-AGAGGGCATGCATATTCCATT-3′
mRANKL forward, 5′-GGGCCACAGCGCTTCTC-3′
mRANKL reverse, 5′-TGGGCCACATCCAACCAT-3′
Preparation of cytosolic and nuclear fractions
Cytosolic and membrane fractions were prepared using the ProteoExtract kit (Calbiochem, San Diego, CA) according to the manufacturer’s standard procedures.
Supplementary Material
Acknowledgments
We are grateful to C. Löwik for KS483 cells, and Xi He of Harvard Medical School and C. Liu of Novartis Institutes for Biomedical Research for constructs. We are grateful for Shengcai Lin for Axin antibody. This work was supported by the Netherlands Organization for Scientific Research (NWO 918.66.066) and Center for Biomedical Genetics.
Abbreviations used:
- ALP
alkaline phosphatase
- APC
adenomatous polyposis coli
- CMV
cytomegalovirus
- Dvl
Disheveled
- FAF1
Fas-associated factor 1
- GSK3β
glycogen synthase kinase 3β
- HA
hemagglutinin
- LEF
lymphoid enhancer factor
- LRP
low-density lipoprotein receptor-related protein
- OPG
osteoprotegerin
- RANKL
receptor activator of nuclear factor κB ligand
- RT
reverse transcription
- shRNA
small hairpin RNA
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E10-12-0985) on March 16, 2011.
REFERENCES
- Adham IM, Khulan J, Held T, Schmidt B, Meyer BI, Meinhardt A, Engel W. Fas-associated factor (FAF1) is required for the early cleavage-stages of mouse embryo. Mol Hum Reprod. 2008;14:207–213. doi: 10.1093/molehr/gan009. [DOI] [PubMed] [Google Scholar]
- Altomare DA, Menges CW, Pei J, Zhang L, Skele-Stump KL, Carbone M, Kane AB, Testa JR. Activated TNF-alpha/NF-kappaB signaling via down-regulation of Fas-associated factor 1 in asbestos-induced mesotheliomas from Arf knockout mice. Proc Natl Acad Sci USA. 2009;106:3420–3425. doi: 10.1073/pnas.0808816106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bea S, et al. Uniparental disomies, homozygous deletions, amplifications, and target genes in mantle cell lymphoma revealed by integrative high-resolution whole-genome profiling. Blood. 2009;113:3059–3069. doi: 10.1182/blood-2008-07-170183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behrens J, von Kries JP, Kuhl M, Bruhn L, Wedlich D, Grosschedl R, Birchmeier W. Functional interaction of beta-catenin with the transcription factor LEF-1. Nature. 1996;382:638–642. doi: 10.1038/382638a0. [DOI] [PubMed] [Google Scholar]
- Bilic J, Huang YL, Davidson G, Zimmermann T, Cruciat CM, Bienz M, Niehrs C. Wnt induces LRP6 signalosomes and promotes dishevelled-dependent LRP6 phosphorylation. Science. 2007;316:1619–1622. doi: 10.1126/science.1137065. [DOI] [PubMed] [Google Scholar]
- Bjorling-Poulsen M, Seitz G, Guerra B, Issinger OG. The pro-apoptotic FAS-associated factor 1 is specifically reduced in human gastric carcinomas. Int J Oncol. 2003;23:1015–1023. [PubMed] [Google Scholar]
- Boyden LM, Mao J, Belsky J, Mitzner L, Farhi A, Mitnick MA, Wu D, Insogna K, Lifton RP. High bone density due to a mutation in LDL-receptor-related protein 5. N Engl J Med. 2002;346:1513–1521. doi: 10.1056/NEJMoa013444. [DOI] [PubMed] [Google Scholar]
- Brunner E, Peter O, Schweizer L, Basler K. pangolin encodes a Lef-1 homologue that acts downstream of Armadillo to transduce the Wingless signal in Drosophila. Nature. 1997;385:829–833. doi: 10.1038/385829a0. [DOI] [PubMed] [Google Scholar]
- Cheyette BN, Waxman JS, Miller JR, Takemaru K, Sheldahl LC, Khlebtsova N, Fox EP, Earnest T, Moon RT. Dapper, a Dishevelled-associated antagonist of beta-catenin and JNK signaling, is required for notochord formation. Dev Cell. 2002;2:449–461. doi: 10.1016/s1534-5807(02)00140-5. [DOI] [PubMed] [Google Scholar]
- Chu K, Niu X, Williams LT. A Fas-associated protein factor, FAF1, potentiates Fas-mediated apoptosis. Proc Natl Acad Sci USA. 1995;92:11894–11898. doi: 10.1073/pnas.92.25.11894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frohlich T, Risau W, Flamme I. Characterization of novel nuclear targeting and apoptosis-inducing domains in FAS associated factor 1. J Cell Sci. 1998;111((Pt 16)):2353–2363. doi: 10.1242/jcs.111.16.2353. [DOI] [PubMed] [Google Scholar]
- Gaur T, et al. Canonical WNT signaling promotes osteogenesis by directly stimulating Runx2 gene expression. J Biol Chem. 2005;280:33132–33140. doi: 10.1074/jbc.M500608200. [DOI] [PubMed] [Google Scholar]
- Glass DA, et al. Canonical Wnt signaling in differentiated osteoblasts controls osteoclast differentiation. Dev Cell. 2005;8:751–764. doi: 10.1016/j.devcel.2005.02.017. [DOI] [PubMed] [Google Scholar]
- Gong Y, et al. LDL receptor-related protein 5 (LRP5) affects bone accrual and eye development. Cell. 2001;107:513–523. doi: 10.1016/s0092-8674(01)00571-2. [DOI] [PubMed] [Google Scholar]
- Hidalgo A, et al. Microarray comparative genomic hybridization detection of chromosomal imbalances in uterine cervix carcinoma. BMC Cancer. 2005;5:77. doi: 10.1186/1471-2407-5-77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmen SL, Zylstra CR, Mukherjee A, Sigler RE, Faugere MC, Bouxsein ML, Deng L, Clemens TL, Williams BO. Essential role of beta-catenin in postnatal bone acquisition. J Biol Chem. 2005;280:21162–21168. doi: 10.1074/jbc.M501900200. [DOI] [PubMed] [Google Scholar]
- Hsu SC, Galceran J, Grosschedl R. Modulation of transcriptional regulation by LEF-1 in response to Wnt-1 signaling and association with beta-catenin. Mol Cell Biol. 1998;18:4807–4818. doi: 10.1128/mcb.18.8.4807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohn AD, Moon RT. Wnt and calcium signaling: beta-catenin-independent pathways. Cell Calcium. 2005;38:439–446. doi: 10.1016/j.ceca.2005.06.022. [DOI] [PubMed] [Google Scholar]
- Korinek V, Barker N, Morin PJ, van Wichen D, de Weger R, Kinzler KW, Vogelstein B, Clevers H. Constitutive transcriptional activation by a beta-catenin-Tcf complex in APC−/− colon carcinoma. Science. 1997;275:1784–1787. doi: 10.1126/science.275.5307.1784. [DOI] [PubMed] [Google Scholar]
- Kramer I, Halleux C, Keller H, Pegurri M, Gooi JH, Weber PB, Feng JQ, Bonewald LF, Kneissel M (2010). Osteocyte Wnt/beta-catenin signaling is required for normal bone homeostasis. Mol Cell Biol. 30:3071–3085. doi: 10.1128/MCB.01428-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Little RD, et al. A mutation in the LDL receptor-related protein 5 gene results in the autosomal dominant high-bone-mass trait. Am J Hum Genet. 2002;70:11–19. doi: 10.1086/338450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu C, Li Y, Semenov M, Han C, Baeg GH, Tan Y, Zhang Z, Lin X, He X. Control of beta-catenin phosphorylation/degradation by a dual-kinase mechanism. Cell. 2002;108:837–847. doi: 10.1016/s0092-8674(02)00685-2. [DOI] [PubMed] [Google Scholar]
- Logan CY, Nusse R. The Wnt signaling pathway in development and disease. Annu Rev Cell Dev Biol. 2004;20:781–810. doi: 10.1146/annurev.cellbio.20.010403.113126. [DOI] [PubMed] [Google Scholar]
- MacDonald BT, Tamai K, He X. Wnt/beta-catenin signaling: components, mechanisms, and diseases. Dev Cell. 2009;17:9–26. doi: 10.1016/j.devcel.2009.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikels AJ, Nusse R. Purified Wnt5a protein activates or inhibits beta-catenin-TCF signaling depending on receptor context. PLoS Biol. 2006;4:e115. doi: 10.1371/journal.pbio.0040115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moon RT, Kohn AD, De Ferrari GV, Kaykas A. WNT and beta-catenin signalling: diseases and therapies. Nat Rev Genet. 2004;5:691–701. doi: 10.1038/nrg1427. [DOI] [PubMed] [Google Scholar]
- Nelson WJ, Nusse R. Convergence of Wnt, beta-catenin, and cadherin pathways. Science. 2004;303:1483–1487. doi: 10.1126/science.1094291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park MY, Jang HD, Lee SY, Lee KJ, Kim E. Fas-associated factor-1 inhibits nuclear factor-kappaB (NF-kappaB) activity by interfering with nuclear translocation of the RelA (p65) subunit of NF-kappaB. J Biol Chem. 2004;279:2544–2549. doi: 10.1074/jbc.M304565200. [DOI] [PubMed] [Google Scholar]
- Park MY, Moon JH, Lee KS, Choi HI, Chung J, Hong HJ, Kim E. FAF1 suppresses IkappaB kinase (IKK) activation by disrupting the IKK complex assembly. J Biol Chem. 2007;282:27572–27577. doi: 10.1074/jbc.C700106200. [DOI] [PubMed] [Google Scholar]
- Reya T, Clevers H. Wnt signalling in stem cells and cancer. Nature. 2005;434:843–850. doi: 10.1038/nature03319. [DOI] [PubMed] [Google Scholar]
- Ryu SW, Chae SK, Lee KJ, Kim E. Identification and characterization of human Fas associated factor 1, hFAF1. Biochem Biophys Res Commun. 1999;262:388–394. doi: 10.1006/bbrc.1999.1217. [DOI] [PubMed] [Google Scholar]
- Ryu SW, Lee SJ, Park MY, Jun JI, Jung YK, Kim E. Fas-associated factor 1, FAF1, is a member of Fas death-inducing signaling complex. J Biol Chem. 2003;278:24003–24010. doi: 10.1074/jbc.M302200200. [DOI] [PubMed] [Google Scholar]
- Song EJ, Yim SH, Kim E, Kim NS, Lee KJ. Human Fas-associated factor 1, interacting with ubiquitinated proteins and valosin-containing protein, is involved in the ubiquitin-proteasome pathway. Mol Cell Biol. 2005;25:2511–2524. doi: 10.1128/MCB.25.6.2511-2524.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spencer GJ, Utting JC, Etheridge SL, Arnett TR, Genever PG. Wnt signalling in osteoblasts regulates expression of the receptor activator of NFkappaB ligand and inhibits osteoclastogenesis in vitro. J Cell Sci. 2006;119:1283–1296. doi: 10.1242/jcs.02883. [DOI] [PubMed] [Google Scholar]
- Takemaru K, Yamaguchi S, Lee YS, Zhang Y, Carthew RW, Moon RT. Chibby, a nuclear beta-catenin-associated antagonist of the Wnt/Wingless pathway. Nature. 2003;422:905–909. doi: 10.1038/nature01570. [DOI] [PubMed] [Google Scholar]
- van Dinther M, Visser N, de Gorter DJ, Doorn J, Goumans MJ, de Boer J, ten Dijke P. ALK2 R206H mutation linked to fibrodysplasia ossificans progressiva confers constitutive activity to the BMP type I receptor and sensitizes mesenchymal cells to BMP-induced osteoblast differentiation and bone formation. J Bone Miner Res. 2010;25:1208–1215. doi: 10.1359/jbmr.091110. [DOI] [PubMed] [Google Scholar]
- Yanagisawa J, et al. The molecular interaction of Fas and FAP-1. A tripeptide blocker of human Fas interaction with FAP-1 promotes Fas-induced apoptosis. J Biol Chem. 1997;272:8539–8545. doi: 10.1074/jbc.272.13.8539. [DOI] [PubMed] [Google Scholar]
- Zeng X, Tamai K, Doble B, Li S, Huang H, Habas R, Okamura H, Woodgett J, He X. A dual-kinase mechanism for Wnt coreceptor phosphorylation and activation. Nature. 2005;438:873–877. doi: 10.1038/nature04185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Gao X, Wen J, Ning Y, Chen YG. Dapper 1 antagonizes Wnt signaling by promoting dishevelled degradation. J Biol Chem. 2006;281:8607–8612. doi: 10.1074/jbc.M600274200. [DOI] [PubMed] [Google Scholar]
- Zhou F, Zhang L, Gong K, Lu G, Sheng B, Wang A, Zhao N, Zhang X, Gong Y. LEF-1 activates the transcription of E2F1. Biochem Biophys Res Commun. 2008a;365:149–153. doi: 10.1016/j.bbrc.2007.10.138. [DOI] [PubMed] [Google Scholar]
- Zhou F, Zhang L, Wang A, Song B, Gong K, Hu M, Zhang X, Zhao N, Gong Y. The association of GSK3 beta with E2F1 facilitates nerve growth factor-induced neural cell differentiation. J Biol Chem. 2008b;283:14506–14515. doi: 10.1074/jbc.M706136200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.