Abstract
Community structure is one of the main structural features of networks, revealing both their internal organization and the similarity of their elementary units. Despite the large variety of methods proposed to detect communities in graphs, there is a big need for multi-purpose techniques, able to handle different types of datasets and the subtleties of community structure. In this paper we present OSLOM (Order Statistics Local Optimization Method), the first method capable to detect clusters in networks accounting for edge directions, edge weights, overlapping communities, hierarchies and community dynamics. It is based on the local optimization of a fitness function expressing the statistical significance of clusters with respect to random fluctuations, which is estimated with tools of Extreme and Order Statistics. OSLOM can be used alone or as a refinement procedure of partitions/covers delivered by other techniques. We have also implemented sequential algorithms combining OSLOM with other fast techniques, so that the community structure of very large networks can be uncovered. Our method has a comparable performance as the best existing algorithms on artificial benchmark graphs. Several applications on real networks are shown as well. OSLOM is implemented in a freely available software (http://www.oslom.org), and we believe it will be a valuable tool in the analysis of networks.
Introduction
The analysis and modeling of networked datasets are probably the hottest research topics within the modern science of complex systems [1]–[7]. The main reason is that, despite its simplicity, the network representation can disclose some relevant features of the system at large, involving its structure, its function, as well as the interplay between structure and function. The elementary units of the system are reduced to simple points, called vertices (or nodes), while their pairwise relationships/interactions are pictured as edges (or links). It is fairly easy to spot the two main ingredients of a graph in many instances. Therefore networks can be found everywhere: in biology (e. g., proteins and their interactions), ecology (e. g., species and their trophic interactions), society (e. g., people and their acquaintanceships). Other noteworthy examples include the Internet (routers/autonomous systems and their physical and/or wireless connections), the World Wide Web (URLs and their hyperlinks), etc.
The structure of most networks, beneath the intrinsic disorder due to the stochastic character of their generation mechanisms, reveals a high degree of organization. In particular, vertices with similar properties or function have a higher chance to be linked to each other than random pairs of vertices and tend to form highly cohesive subgraphs, which are called communities (also modules or clusters). Examples of communities are groups of mutual acquaintances in social networks [8]–[10], subsets of Web pages on the same subject [11], compartments in food webs [12], [13], functional modules in protein interaction networks [14], biochemical pathways in metabolic networks [15], [16], etc.
Detecting communities in graphs may help to identify functional subunits of the system and to uncover similarities among vertices that are not apparent in the absence of detailed (non-topological) information. Vertices belonging to the same community may be classified according to their structural position within the cluster, which may be correlated to their role. Vertices in the core of the cluster may have a function of control and stability within the module, whereas boundary vertices are likely to be mediators between different parts of the graph. The community structure of a network can also be a powerful visual representation of the system: instead of visualizing all the vertices and edges of the network (which is impossible on large systems), one could display its communities and their mutual connections, obtaining a far more compact and understandable description of the graph as a whole. It is thus not surprising that community detection in graphs has been so extensively investigated over the last few years [17]. A huge variety of different methods have been designed by a truly interdisciplinary community of scholars, including physicists, computer scientists, mathematicians, biologists, engineers and social scientists.
However, most algorithms currently available cannot handle important network features. Many methods are designed to find clusters in undirected graphs, and cannot be easily (or not at all) extended to directed graphs. However, there are many datasets for which edge directedness is an essential feature. Citation networks, food webs and the Web graph are but a few examples. Similar problems arise when edges carry weights, indicating the strength of the interaction/affinity between vertices, although extensions are generally easier in this case.
Likewise, the great majority of algorithms are not capable to deal with the peculiar features of community structure. For example, each vertex is typically assigned to a single cluster, while in several instances, like in social networks, vertices are typically shared between two or more clusters. In such cases communities are overlapping (and partitions become covers) and very few methods account for this possibility [18]–[25], which considerably increases the complexity of the problem. Furthermore, community structure is very often hierarchical, i.e. it consists of communities which include (or are included by) other communities. Hierarchies are common in human societies and are crucial for an efficient management of large organizations. Simon pointed out that hierarchy gives robustness and stability to complex systems, yielding an evolutionary advantage on the long run [26]. However, most community finding methods typically look for the “best” partition of a network, disregarding the possible existence of hierarchical structure. Instead, a method should be able to recognize if there is hierarchical structure and, if yes, identify the corresponding levels [27]–[29].
It is also very important for a method to distinguish communities from pseudo-communities. The existence of clusters indicate a preference by some groups of vertices to link to each other. But, if the linking probability is the same for all pairs of vertices, like in random graphs, no communities are expected. In this case, concentrations of edges within groups of vertices are simply the result of random fluctuations, they do not represent potentially non-trivial structures. Many algorithms are not able to see this difference and find clusters in random graphs as well, although they are not meaningful. Scholars have just begun to assess the issue of significance of clusters [30], [31].
Finally, given the recent availability of time-stamped networked datasets, it is now
possible to carry out quantitative studies on the dynamics of community structure,
about which very little is known [32]–[37]. A simple way to treat dynamic datasets is to analyze
snapshots of the system at different times separately, and then map communities of
different snapshots onto each other, such that one can follow the dynamic of each
cluster in time. However, focusing on individual snapshots means disregarding the
information on the system at previous times. Ideally a partition/cover of the system
at time  should be faithful both to its structure at time
should be faithful both to its structure at time
 and to its history [34], [37].
and to its history [34], [37].
In this paper we propose the first method able to meet all requirements listed above, the Order Statistics Local Optimization Method (OSLOM). It is a method that optimizes locally the statistical significance of clusters, defined with respect to a global null model. The concept of statistical significance is inspired by recent work of some of the authors [31], [38]. The paper is structured as follows. After introducing the method, we test its performance on artificial benchmark graphs, comparing it with the performances of the best algorithms currently available. Next, we pass to the analysis of real networks, followed by a final discussion on the work. Some of the tests on artificial and real networks are reported in the Supporting Information S1.
Methods
Statistical significance of clusters
In this section we explain how to estimate the statistical significance of a given cluster. OSLOM will use the significance as a fitness measure in order to evaluate the clusters. Following our previous work [31], we define it as the probability of finding the cluster in a random null model, i. e. in a class of graphs without community structure. We choose the configuration model [39] as our null model. This is a model designed to build random networks with a given distribution of the number of neighbors of a vertex (degree). The networks are generated by joining randomly vertices under the constraint that each vertex has a fixed number of neighbors, taken from the pre-assigned degree distribution. This is basically the same null model adopted by Newman and Girvan to define modularity [40].
We start from a graph  with
with
 vertices and
vertices and 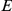 edges. The
framework for the analysis is sketched in Fig. 1. We are given a subgraph
edges. The
framework for the analysis is sketched in Fig. 1. We are given a subgraph
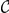 , whose significance is to be assessed, a vertex
, whose significance is to be assessed, a vertex
 and the degree of the vertices of the rest of the graph
and the degree of the vertices of the rest of the graph
 . The degree of subgraph
. The degree of subgraph 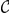 is
is
 ,
,  is the degree of
is the degree of
 , and the rest of vertices have a total degree
, and the rest of vertices have a total degree
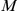 . We can separate the above quantities in the
contributions internal or external to
. We can separate the above quantities in the
contributions internal or external to 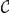 (
( and
and 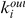 ); the internal
degree of
); the internal
degree of  is
is  (Fig. 1).
(Fig. 1).
Figure 1. A schematic representation of a subgraph
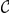 , whose
significance is to be assessed.
, whose
significance is to be assessed.
The subgraph 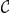 is
embedded within a random graph generated by the configuration model. The
degrees of all vertices of the network are fixed, in the figure we have
highlighted the degrees of
is
embedded within a random graph generated by the configuration model. The
degrees of all vertices of the network are fixed, in the figure we have
highlighted the degrees of  (
(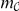 ), of the
vertex
), of the
vertex  at the
center of the analysis (
at the
center of the analysis ( ) and of
the rest of the graph
) and of
the rest of the graph  (
(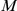 ). These
quantities are expressed as sums of contributions which are internal to
their own set of vertices (as
). These
quantities are expressed as sums of contributions which are internal to
their own set of vertices (as 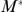 ) or
related to subgraph
) or
related to subgraph 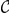 (in or
out). This notation is used in the distribution of Eq. 1.
(in or
out). This notation is used in the distribution of Eq. 1.
Let us suppose that 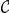 is a subgraph of
graphs generated by the configuration model, where each vertex maintains the
degree it has on the graph
is a subgraph of
graphs generated by the configuration model, where each vertex maintains the
degree it has on the graph  at study. We
assume that the internal degree
at study. We
assume that the internal degree 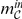 of the subgraph is
fixed. If all the other edges of the network are randomly drawn, the probability
that
of the subgraph is
fixed. If all the other edges of the network are randomly drawn, the probability
that  has
has  neighbors in
neighbors in
 can be written as [38]
can be written as [38]
| (1) |
This equation enumerates the possible
configurations of the network with  connections
between
connections
between  and
and 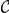 . The factorials of
the formula express the multiplicity of configurations with fixed values of
. The factorials of
the formula express the multiplicity of configurations with fixed values of
 ,
,  ,
,
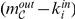 and
and 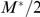 , whereas the power
of
, whereas the power
of  in the numerator stays for the multiplicity coming from
the permutation of the extremes of edges lying between
in the numerator stays for the multiplicity coming from
the permutation of the extremes of edges lying between
 and
and  . Several of the
terms in the expression can actually be written as a function of constants and
. Several of the
terms in the expression can actually be written as a function of constants and
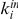 , such as
, such as 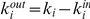 and
and
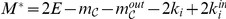 . The normalization factor
. The normalization factor
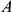 includes terms not depending on
includes terms not depending on
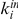 and ensures that
and ensures that
| (2) |
Further details on the numerical implementation of the formula in Eq. 1, as well as on the different approximations taken and their limits, are included in the Supporting Information S1.
The probability of Eq. 1 provides a tool to rank the vertices external to
 according to the likelihood of their topological
relation with the group. If vertex
according to the likelihood of their topological
relation with the group. If vertex  shares many more
edges with the vertices of subgraph
shares many more
edges with the vertices of subgraph 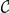 than expected in
the null model, we could consider the inclusion of
than expected in
the null model, we could consider the inclusion of
 in
in  , since the
relationship between
, since the
relationship between  and
and
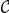 is “unexpectedly” strong. In order to
perform the ranking the cumulative probability
is “unexpectedly” strong. In order to
perform the ranking the cumulative probability  of having a number
of internal connections equal or larger than
of having a number
of internal connections equal or larger than 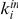 is estimated,
following Ref. [31]. Given that the vertex degree is a discrete variable,
the cumulative distribution has a specific step-wise profile for each value of
is estimated,
following Ref. [31]. Given that the vertex degree is a discrete variable,
the cumulative distribution has a specific step-wise profile for each value of
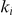 . In order to facilitate the comparison of vertices with
different degrees, we implement a bootstrap strategy by assigning to each vertex
. In order to facilitate the comparison of vertices with
different degrees, we implement a bootstrap strategy by assigning to each vertex
 a value of
a value of  ,
,
 , randomly drawn from the interval
, randomly drawn from the interval
 . This choice is important for a meaningful
estimate of the clusters' significance; other options (e. g., taking
the middle points of the interval) could lead to the identification of
meaningful clusters in random graphs. The bootstrap introduces a
stochastic element in the assessment procedure, which will, in turn, lead to the
use of Monte Carlo techniques.
. This choice is important for a meaningful
estimate of the clusters' significance; other options (e. g., taking
the middle points of the interval) could lead to the identification of
meaningful clusters in random graphs. The bootstrap introduces a
stochastic element in the assessment procedure, which will, in turn, lead to the
use of Monte Carlo techniques.
The variable  bears the information regarding the likelihood of the
topological relation of each vertex with
bears the information regarding the likelihood of the
topological relation of each vertex with 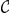 and has an
important feature: it is a uniform random variable distributed between zero and
one for vertices of our null model graphs. Calculating its order statistic
distributions is thus a relatively easy task. The first candidate among the
external vertices to be part of
and has an
important feature: it is a uniform random variable distributed between zero and
one for vertices of our null model graphs. Calculating its order statistic
distributions is thus a relatively easy task. The first candidate among the
external vertices to be part of  is the vertex with
the lowest value of
is the vertex with
the lowest value of  , that we indicate
, that we indicate
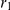 . The cumulative distribution of
. The cumulative distribution of
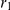 in the null model is then given by
in the null model is then given by
| (3) |
where 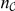 is the number of
vertices in
is the number of
vertices in 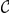 . In general, let
. In general, let  be the value of
variable
be the value of
variable  with rank
with rank 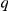 (in increasing
order of the variable
(in increasing
order of the variable  ). Its cumulative
distribution is (Fig.
2):
). Its cumulative
distribution is (Fig.
2):
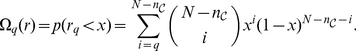 |
(4) |
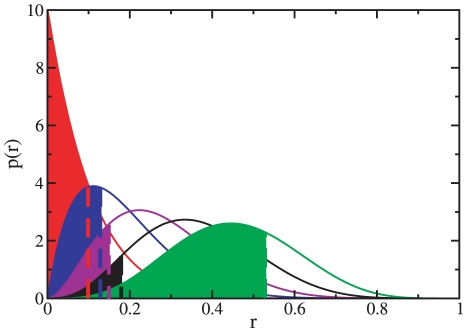
Figure 2. Probability distributions of the scores
 of
vertices external to a given subgraph
of
vertices external to a given subgraph  of the
graph.
of the
graph.
The score 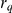 is the
is the
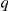 -th
smallest score of the external vertices. In this particular case there
are
-th
smallest score of the external vertices. In this particular case there
are  external vertices. In the figure, we plot
external vertices. In the figure, we plot
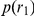 ,
,
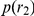 ,
,
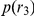 ,
,
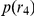 ,
,
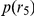 (from left
to right). As an example, the shaded areas show the cumulative
probability
(from left
to right). As an example, the shaded areas show the cumulative
probability  for a few
values of
for a few
values of  that would
correspond to the values estimated in a practical situation. In this
case, the black area,
that would
correspond to the values estimated in a practical situation. In this
case, the black area, 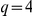 , is the
least extensive and so
, is the
least extensive and so 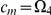 . If
. If
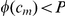 , the
vertices with scores
, the
vertices with scores 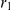 ,
,
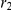 ,
,
 and
and
 will be
added to
will be
added to 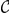 .
.
The reason for the use of order statistics is that we assume that clustering
methods tend to include in each community those vertices which are most strongly
connected to vertices of the community. Due to correlations (the vertices in the
clusters tend to be connected), we cannot calculate the statistics of the
internal connections to the clusters, but we can do it safely for the external
vertices. The values of the different 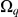 inform us of how
much the external vertices of a group are compatible with the statistics
expected in the null model. To evaluate the full group, we define
inform us of how
much the external vertices of a group are compatible with the statistics
expected in the null model. To evaluate the full group, we define
 among all the neighbors of
among all the neighbors of
 , where
, where 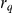 are their
corresponding ranked values for the
are their
corresponding ranked values for the  variable. The
distribution of
variable. The
distribution of 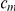 can be easily
tabulated numerically since it only depends on
can be easily
tabulated numerically since it only depends on  . The cumulative
distribution will be denoted as
. The cumulative
distribution will be denoted as 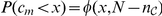 . In the following,
we call
. In the following,
we call 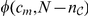 the score of the cluster
the score of the cluster
 .
.
Single cluster analysis
Now that a score to evaluate the statistical significance of the clusters has
been introduced, the next step is to optimize the score across the network by
dividing it into proper clusters. We describe first the optimization of a single
cluster score and will extend later the method to deal with the full network.
First of all one has to give the method a certain tolerance, in the following
referred to as  . This parameter
establishes when a given value of the score is considered significant. Our
procedure consists of two phases: first, we explore the possibility of adding
external vertices to the subgraph
. This parameter
establishes when a given value of the score is considered significant. Our
procedure consists of two phases: first, we explore the possibility of adding
external vertices to the subgraph  ; second,
non-significant vertices in
; second,
non-significant vertices in 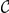 are pruned. They
are described below and illustrated schematically in Fig. 3.
are pruned. They
are described below and illustrated schematically in Fig. 3.
Figure 3. Schematic diagram of the single cluster analysis.
For each vertex
 outside
outside
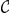 and
connected to it by at least one edge the variable
and
connected to it by at least one edge the variable
 is
computed. Then we calculate
is
computed. Then we calculate 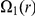 for the
vertex with the smallest
for the
vertex with the smallest  , by using
Eq. 3. If
, by using
Eq. 3. If  , we add
the corresponding vertex to the subgraph, which we now call
, we add
the corresponding vertex to the subgraph, which we now call
 . If
. If
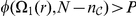 , one
checks the second best vertex, the third best vertex, etc. If there is
finally a vertex, say the
, one
checks the second best vertex, the third best vertex, etc. If there is
finally a vertex, say the  -th best
vertex, for which
-th best
vertex, for which 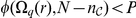 , one
includes all
, one
includes all  best
vertices into subgraph
best
vertices into subgraph  , yielding
subgraph
, yielding
subgraph 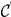 . At this
point, no other vertex outside
. At this
point, no other vertex outside  deserves
to enter the community since all the external vertices are compatible
with the statistics of the random configuration model. It may also
happen that the inequality
deserves
to enter the community since all the external vertices are compatible
with the statistics of the random configuration model. It may also
happen that the inequality 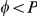 above
holds for no external vertex, in which case we add no vertices to
above
holds for no external vertex, in which case we add no vertices to
 and
and
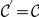 . Either
way, we pass to the second stage with the subgraph
. Either
way, we pass to the second stage with the subgraph
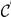 .
.For each vertex
 in
in
 the
variable
the
variable 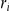 with
respect to the set
with
respect to the set  is
estimated. We pick the “worst” vertex
is
estimated. We pick the “worst” vertex
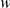 of the
cluster, i. e. the vertex with the highest value of
of the
cluster, i. e. the vertex with the highest value of
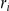 . To check
for its significance we repeat step 1 for the subgraph
. To check
for its significance we repeat step 1 for the subgraph
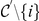 . If
. If
 turns out
to be significant, we keep it inside
turns out
to be significant, we keep it inside 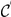 and the
analysis of the cluster is completed. Otherwise,
and the
analysis of the cluster is completed. Otherwise,
 is moved
out of
is moved
out of 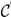 and one
searches for the worst internal vertex of
and one
searches for the worst internal vertex of
 . At some
point we end up with a cluster
. At some
point we end up with a cluster  , whose
internal vertices are all significant and the process stops.
, whose
internal vertices are all significant and the process stops.
The two-steps procedure is a way to “clean up”
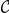 . A cluster is left unchanged only if all the external
vertices are compatible with the null model and all the internal vertices are
not. A few remarks are important here:
. A cluster is left unchanged only if all the external
vertices are compatible with the null model and all the internal vertices are
not. A few remarks are important here:
There can be both good vertices outside
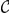 and
bad ones inside. It is important to perform the
complete procedure described above, which guarantees that the final
cluster is significant with respect to the present null model (see also
Ref. [31]).
and
bad ones inside. It is important to perform the
complete procedure described above, which guarantees that the final
cluster is significant with respect to the present null model (see also
Ref. [31]).The procedure is not deterministic, because of the stochastic component in the computation of the cumulative probability
 . So one
shall repeat all the steps several times. The cluster analysis may
deliver a subgraph
. So one
shall repeat all the steps several times. The cluster analysis may
deliver a subgraph  , in
general different from
, in
general different from  , or an
empty subgraph. For each vertex
, or an
empty subgraph. For each vertex  we compute
the participation frequency
we compute
the participation frequency 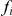 , defined
as the ratio between the number of times
, defined
as the ratio between the number of times  belongs to
any non-empty
belongs to
any non-empty 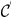 and the
total number of iterations leading to non-empty subgraphs. In general,
we consider the subgraph
and the
total number of iterations leading to non-empty subgraphs. In general,
we consider the subgraph 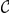 to be a
significant cluster if the single cluster analysis yields a non-empty
subgraph
to be a
significant cluster if the single cluster analysis yields a non-empty
subgraph 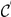 in more
than
in more
than 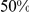 iterations. The final “cleaned”
cluster includes those vertices for which
iterations. The final “cleaned”
cluster includes those vertices for which
 .
.In the worst-case scenario, the complexity of the cluster analysis scales with the number of vertices of
 , times the
number of neighbors of
, times the
number of neighbors of 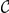 , times the
number of loops needed to have reliable values for the
, times the
number of loops needed to have reliable values for the
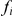 's.
The situation can be considerably improved by keeping track of the order
of the external vertices at each step (using suitable data structures)
and by computing the score only for some reasonably good vertices. For
instance, one could pick just those vertices with
's.
The situation can be considerably improved by keeping track of the order
of the external vertices at each step (using suitable data structures)
and by computing the score only for some reasonably good vertices. For
instance, one could pick just those vertices with
 . We
numerically checked that changing this threshold does not affect the
results, but leads to a faster algorithm.
. We
numerically checked that changing this threshold does not affect the
results, but leads to a faster algorithm.
Network analysis
The previous procedure deals with a single cluster
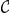 . It finds the external significant vertices and includes
them into
. It finds the external significant vertices and includes
them into  . It also prunes those internal vertices that are not
statistically relevant. Now we extend this procedure by introducing an algorithm
able to analyze the full network. In order to do so, we follow the method
proposed by some of the authors in Ref. [23]. The starting point
is a single vertex, taken at random, in the absence of any information. Let us
suppose that we start from a random vertex
. It also prunes those internal vertices that are not
statistically relevant. Now we extend this procedure by introducing an algorithm
able to analyze the full network. In order to do so, we follow the method
proposed by some of the authors in Ref. [23]. The starting point
is a single vertex, taken at random, in the absence of any information. Let us
suppose that we start from a random vertex  and that our first
group is
and that our first
group is  . The method proceeds as follows:
. The method proceeds as follows:
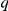 vertices are added to
vertices are added to
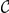 ,
considering the most significant among the neighbors of the cluster. The
number
,
considering the most significant among the neighbors of the cluster. The
number 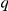 is taken
from a distribution, which in principle can be arbitrary. We choose a
power law with exponent
is taken
from a distribution, which in principle can be arbitrary. We choose a
power law with exponent 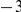 .
.Perform the single cluster analysis.
We repeat the whole procedure starting from several vertices in order to explore
different regions of the network. This yields a final set of clusters that may
overlap. Such type of local optimization was originally implemented in the Local
Fitness Method [23], to handle overlapping communities. The algorithm
stops when it keeps finding similar modules over and over.
Ideally one wishes to encounter the exact same clusters repeatedly. However, the
stochastic element introduced when calculating the vertex score can lead
vertices, whose score is close to the threshold, to change their group
assignments from one realization to another. This can be a problem when we are
trying to decide whether two groups in different instances correspond to the
same cluster. As a practical rule, we say that two groups
 and
and 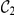 are similar if
are similar if
 , in which case they deserve further attention. Indeed,
it turns out that many of the clusters found are very similar or combinations of
each other. This leads to a very important question: given a set of significant
clusters, which ones should be kept?
, in which case they deserve further attention. Indeed,
it turns out that many of the clusters found are very similar or combinations of
each other. This leads to a very important question: given a set of significant
clusters, which ones should be kept?
Let us consider the problem of choosing between two clusters
 and
and 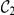 and the union of
the two,
and the union of
the two, 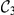 . A solution is to consider the subgraph
. A solution is to consider the subgraph
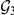 of the vertices in
of the vertices in 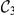 and see if
and see if
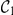 and
and  are significant as
modules of
are significant as
modules of 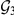 . Strictly speaking we consider
. Strictly speaking we consider
 and
and  which are the
cleaned up clusters within
which are the
cleaned up clusters within  (i.e. with respect
to subgraph
(i.e. with respect
to subgraph  only, neglecting the rest of the network). We discard
only, neglecting the rest of the network). We discard
 if
if 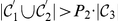 , where we set
, where we set
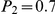 . Otherwise we discard
. Otherwise we discard 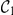 and
and
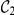 and we keep the union
and we keep the union 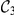 . Instead, if we
have to decide among a set of
. Instead, if we
have to decide among a set of  clusters and their
union, the condition to prefer the submodules is
clusters and their
union, the condition to prefer the submodules is 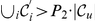 .
.
In general, we check if each cluster has significant submodules, by looking for modules in the subgraph given by the cluster and using the condition above to decide which ones to take. This leads to a set of significant minimal clusters, where minimal means that they have no significant internal cluster structure, according to the condition above. We also need to check whether unions of such minimal clusters do have internal cluster structure, according to our rule, to decide whether the clusters have to be kept separated or merged. After doing this, we still end up with many similar modules. Given a pair of similar modules (in the sense defined above), we first check if their union has significant cluster structure: if it does not, we merge the two clusters, otherwise we systematically prefer the bigger one (if they are equal-sized, we pick the cluster with smaller score).
After the completion of this procedure, the output is a cover of the network. To reduce the stochasticity introduced by the bootstrap, the procedure is repeated in order to obtain several covers. All clusters of the covers are analyzed as described above to select among them the ones which will appear in the final output.
The parameter values may affect the outcome of OSLOM. The value of the
significance level 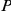 plays an important
role for the determination of the size of the clusters found by OSLOM. In
general, small values of
plays an important
role for the determination of the size of the clusters found by OSLOM. In
general, small values of 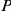 lead to the
identification of large clusters, and large values of
lead to the
identification of large clusters, and large values of
 allow the identification of small clusters. Likewise,
large values of the parameter
allow the identification of small clusters. Likewise,
large values of the parameter 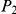 , which controls
the internal structure of modules, generally lead to the identification of large
clusters. The influence of the parameter values is however relevant only when
the community structure of the network is not pronounced. When modules are well
defined, the results of OSLOM do not depend on the particular choice of the
parameter values.
, which controls
the internal structure of modules, generally lead to the identification of large
clusters. The influence of the parameter values is however relevant only when
the community structure of the network is not pronounced. When modules are well
defined, the results of OSLOM do not depend on the particular choice of the
parameter values.
OSLOM
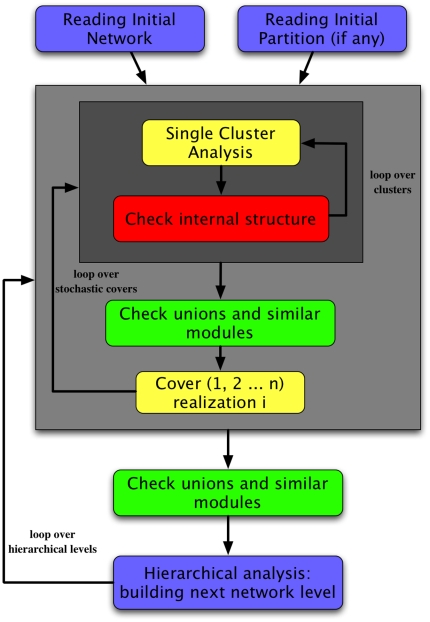
We have described the cleaning of a single cluster and how the full network is analyzed. In the following, all the ingredients are assembled together to form the algorithm that we call OSLOM (Order Statistics Local Optimization Method). A flux diagram summarizing how it works can be seen in Fig. 4. OSLOM consists of three phases:
Figure 4. Flux diagram of OSLOM.
The levels of grey of the squares represent different loop levels. One can provide an initial partition/cover as input, from which the algorithm starts operating, or no input, in which case the algorithm will build the clusters about individual vertices, chosen at random. OSLOM performs first a cleaning procedure of the clusters, followed by a check of their internal structure and by a decision on possible cluster unions. This is repeated with different choices of random numbers in order to obtain better statistics and a more reliable information. The final step is to generate a super-network for the next level of the hierarchical analysis.
First, it looks for significant clusters, until convergence;
Second, it analyzes the resulting set of clusters, trying to detect their internal structure or possible unions thereof;
Third, it detects the hierarchical structure of the clusters.
To speed up the method, one can start from a given partition/cover delivered by another (fast) algorithm or from a priori information. In those cases, the first step will be to clean up the given clusters.
Once the set of minimal significant clusters has been found, the analysis of the
hierarchies consists of the following steps. We construct a new network formed
by clusters, where each cluster is turned into a supervertex and there are edges
between supervertices if the representative clusters are linked to each other.
The resulting superedges are weighted by the number of edges between the initial
clusters. There is the problem of properly assigning edges between clusters, if
the edges are incident on overlapping vertices. Suppose to have an edge whose
endvertices  and
and 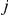 belong to
belong to
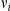 and
and 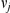 clusters,
respectively. This edge lies simultaneously between any pair of clusters
clusters,
respectively. This edge lies simultaneously between any pair of clusters
 and
and 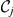 , with
, with
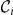 including
including  and
and
 including
including  . The contribution
of the edge to the superedge between
. The contribution
of the edge to the superedge between  and
and
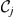 equals
equals  . The resulting
non-integer weights may lead to non-integer values for the weight of superedges,
whereas we need integer values in order to use Eq. 1. For this reason, the
weight of each superedge is rounded to the nearest integer value. We stress that
the weight we deal with here indicates just how to “split” edges, it
is not related to the weight that edges may carry. If the original network is
weighted, the rescaled weight of an edge is
. The resulting
non-integer weights may lead to non-integer values for the weight of superedges,
whereas we need integer values in order to use Eq. 1. For this reason, the
weight of each superedge is rounded to the nearest integer value. We stress that
the weight we deal with here indicates just how to “split” edges, it
is not related to the weight that edges may carry. If the original network is
weighted, the rescaled weight of an edge is  ,
,
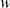 being the weight of the edge in the network. Once the
supernetwork has been built, one applies the method again, obtaining the second
hierarchical level. The latter is turned again into a supernetwork, as we
explained above, and so on, until the method produces no clusters. In this way
OSLOM recovers the hierarchical community structure of the original graph.
being the weight of the edge in the network. Once the
supernetwork has been built, one applies the method again, obtaining the second
hierarchical level. The latter is turned again into a supernetwork, as we
explained above, and so on, until the method produces no clusters. In this way
OSLOM recovers the hierarchical community structure of the original graph.
We will describe next the main features of OSLOM, and what it adds to the state of the art in community detection.
Significant clusters
The main characteristic of OSLOM is that it is based on a fitness measure,
the score, that is tightly related to the significance of the clusters in
the configuration model. In fact, the single cluster analysis is designed to
optimize the cluster significance as defined in Ref. [31]. Therefore the
output of OSLOM consists of clusters that are unlikely to be found in an
equivalent random graph with the same degree sequence. The tolerance
 , fixed initially, determines whether such clusters
are “unexpectedly unlikely”, and therefore significant, or not.
So, if the method is fed with a random graph, the output will include very
few clusters or even none at all.
, fixed initially, determines whether such clusters
are “unexpectedly unlikely”, and therefore significant, or not.
So, if the method is fed with a random graph, the output will include very
few clusters or even none at all.
Homeless vertices
The vertices in a random network will be deemed as homeless. Homeless vertices are those that are not assigned to any cluster. This is a very important feature that OSLOM includes. The presence of random noise or non-significant vertices is an issue that may occur in many real systems. However, very few clustering techniques take into account this possibility. In OSLOM, it comes as a natural output. We will quantitatively analyze this feature when we test the method on benchmark graphs.
Overlapping communities
A natural output of OSLOM is the possibility for clusters to overlap. Since each cluster is “cleaned” independently of the others, a fraction of its vertices may belong also to other clusters, eventually. We will show the efficiency of OSLOM in unveiling overlapping vertices in suitably designed benchmarks.
Cluster hierarchy
Another relevant feature of OSLOM is the analysis of the hierarchical structure of the clusters. As mentioned above, the third phase of our method includes a procedure to take care of this issue. The results are very good on hierarchical benchmarks.
OSLOM generally finds different depths in different hierarchical branches. In fact, when the algorithm is applied not all vertices are grouped, as some of them are homeless. The coexistence of homeless vertices with proper clusters yields a hierarchical structure with branches of different depths.
Weighted networks
OSLOM can be generalized to weighted graphs as well. We assume that the
contributions to the probability of having a connection between two vertices
 and
and 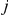 with a certain
weight
with a certain
weight 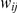 , given the vertex degrees
, given the vertex degrees
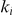 and
and  and their
strengths,
and their
strengths,  and
and
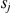 , is separable in two different terms in the
configuration model: one for the topology and another for the weight [38]. The
strength of a vertex is defined as the sum of the weights of all the edges
incident on it. We approximate the weight contribution
by
, is separable in two different terms in the
configuration model: one for the topology and another for the weight [38]. The
strength of a vertex is defined as the sum of the weights of all the edges
incident on it. We approximate the weight contribution
by
| (5) |
where
 is the harmonic mean of the average weights of
vertices
is the harmonic mean of the average weights of
vertices  and
and  , defined as
, defined as
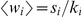 and
and  , respectively.
The idea behind this expression is that the weight of an edge of the null
model should be proportional to the average weight of its endvertices. We
proposed the harmonic average because it is more sensitive to the small
values of
, respectively.
The idea behind this expression is that the weight of an edge of the null
model should be proportional to the average weight of its endvertices. We
proposed the harmonic average because it is more sensitive to the small
values of 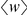 .
.
We use this distribution to define a new variable
 , accounting for the probability of having a certain
weight on a given edge with the strengths of the vertices and the general
weight distribution known. We combine this variable
, accounting for the probability of having a certain
weight on a given edge with the strengths of the vertices and the general
weight distribution known. We combine this variable
 with its topological counterpart,
with its topological counterpart,
 , obtaining a new variable
, obtaining a new variable
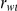 . This is a non-trivial task since both probabilities
are defined on a different set of elements (see the Supporting
Information S1). For
. This is a non-trivial task since both probabilities
are defined on a different set of elements (see the Supporting
Information S1). For  we can
estimate, as before, the order statistic distributions and we proceed just
as we do for unweighted graphs.
we can
estimate, as before, the order statistic distributions and we proceed just
as we do for unweighted graphs.
Directed graphs
OSLOM can be easily generalized to handle directed graphs. For that, we need
to define two uniformly distributed random variables
 and
and 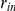 . The former is
based on the probability that vertex
. The former is
based on the probability that vertex  has outgoing
edges ending on vertices of the given subgraph
has outgoing
edges ending on vertices of the given subgraph
 , the latter is based on the probability that
, the latter is based on the probability that
 has incoming edges originating from vertices of
has incoming edges originating from vertices of
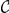 . These two probabilities are computed through
analogous formulas as in Eq. 1 or numerical approximations to it. The final
score of vertex
. These two probabilities are computed through
analogous formulas as in Eq. 1 or numerical approximations to it. The final
score of vertex  is given by
the product
is given by
the product  . We are able
to calculate the distribution of this product and therefore to estimate its
order statistics (just as for the weighted case, see Section 1.1. of Supporting
Information S1). The rest of the clustering method proceeds as
explained above. If graphs have edges with both directions and weights, we
have four variables for each vertex:
. We are able
to calculate the distribution of this product and therefore to estimate its
order statistics (just as for the weighted case, see Section 1.1. of Supporting
Information S1). The rest of the clustering method proceeds as
explained above. If graphs have edges with both directions and weights, we
have four variables for each vertex:  ,
,
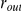 and the corresponding versions for the weights. The
final score is given again by the product of these four variables.
and the corresponding versions for the weights. The
final score is given again by the product of these four variables.
Dynamical networks
Time-stamped networked datasets are usually divided into snapshots,
condensing the relational information between vertices within different time
windows. Snapshots are typically analyzed separately, whereas it would be
more informative to combine the information from different time slices. For
instance, consider two snapshots  and
and
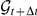 at times
at times  and
and
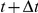 , respectively. A simple idea is to find the
partition/cover of the network at time
, respectively. A simple idea is to find the
partition/cover of the network at time  , by applying
the method to the corresponding snapshot, and to use the result as an input
for the application of the method to the network at time
, by applying
the method to the corresponding snapshot, and to use the result as an input
for the application of the method to the network at time
 . In this way one can see how the community structure
at time
. In this way one can see how the community structure
at time  “evolves” to that at time
“evolves” to that at time
 . This is a rather general approach, it can be
adopted for other algorithms for community detection, like greedy
optimization techniques. OSLOM has the useful property that it can start
from any initial partition/cover, which can be given as input. In this way
the clusters found in
. This is a rather general approach, it can be
adopted for other algorithms for community detection, like greedy
optimization techniques. OSLOM has the useful property that it can start
from any initial partition/cover, which can be given as input. In this way
the clusters found in 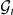 can be used as
initial condition for the analysis of
can be used as
initial condition for the analysis of 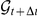 . With this
approach, the new partition/cover is closer to that in
. With this
approach, the new partition/cover is closer to that in
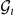 and we are able to track the groups' evolution.
Naturally, if the two snapshots are very different from each other (because
they refer to times between which the system has changed considerably, for
instance), OSLOM produces a partition/cover in
and we are able to track the groups' evolution.
Naturally, if the two snapshots are very different from each other (because
they refer to times between which the system has changed considerably, for
instance), OSLOM produces a partition/cover in
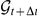 that is uncorrelated with that of
that is uncorrelated with that of
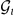 .
.
Complexity
The complexity of OSLOM cannot be estimated exactly, as it depends on the
specific features of the community structure at study. Therefore we carried
out a numerical study of the complexity, whose results are shown in Fig. 5. We apply the
method on the LFR benchmark [41], that we have
used extensively to test the performance of OSLOM. We have used both the
standard version of the algorithm and a fast implementation, in which the
algorithm acts on the partition delivered by a quick method. For each
version we have considered undirected and unweighted LFR benchmark graphs
with two different levels of mixtures between the clusters
( and
and 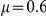 , corresponding
to well separated and well mixed clusters). The other parameters needed to
build the LFR benchmark graphs are the same as for the graphs used in Fig. 6. The diagram of
Fig. 5 shows the
execution time (in seconds) as a function of the number
, corresponding
to well separated and well mixed clusters). The other parameters needed to
build the LFR benchmark graphs are the same as for the graphs used in Fig. 6. The diagram of
Fig. 5 shows the
execution time (in seconds) as a function of the number
 of vertices of the graphs. The processes were run on
a workstation HP Z800. The time scales as a power law of
of vertices of the graphs. The processes were run on
a workstation HP Z800. The time scales as a power law of
 with good approximation, if the graphs are not too
small. The behavior seems to depend neither on how mixed communities are,
nor on the particular implementation of the algorithm (there seems to be
just a factor between the corresponding curves). Power law fits of the
large-N portion of the curves yield an exponent
with good approximation, if the graphs are not too
small. The behavior seems to depend neither on how mixed communities are,
nor on the particular implementation of the algorithm (there seems to be
just a factor between the corresponding curves). Power law fits of the
large-N portion of the curves yield an exponent
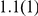 , which implies that the complexity is essentially
linear in this case.
, which implies that the complexity is essentially
linear in this case.
Figure 5. Complexity of OSLOM.
The diagram shows how the execution time of two different implementations of the algorithm scales with the network size (expressed by the number of vertices), for LFR benchmark graphs.
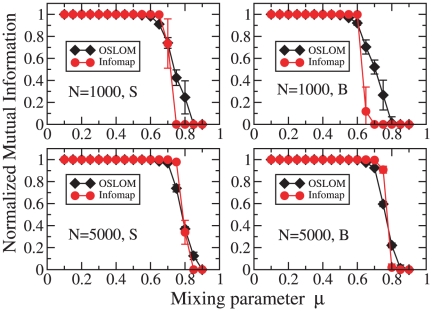
Figure 6. Tests on undirected and unweighted LFR benchmark graphs without overlapping communities.
The parameters of the graphs are: average degree
 ,
maximum degree
,
maximum degree  ,
exponents of the power law distributions are
,
exponents of the power law distributions are
 for
degree and
for
degree and 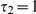 for
community size, S and B mean that community sizes are in the range
for
community size, S and B mean that community sizes are in the range
 (“small”) and
(“small”) and 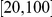 (“big”), respectively. We considered two network sizes:
(“big”), respectively. We considered two network sizes:
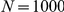 (top)
and
(top)
and 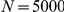 (bottom). The two curves refer to OSLOM (diamonds) and Infomap
(circles).
(bottom). The two curves refer to OSLOM (diamonds) and Infomap
(circles).
Results
Artificial networks
In this section we test OSLOM against artificial benchmarks, comparing its performance with those of the best algorithms currently available. We mostly adopted the LFR benchmark [41], [42], a class of graphs with planted community structure and heterogeneous distributions of vertex degree and community size. Tests on the well known Girvan-Newman (GN) benchmark [8] are shown in the Supporting Information S1. In this section we present tests on undirected and unweighted networks, with and without hierarchical structure and overlapping communities. We also show how OSLOM handles the presence of randomness in the graph structure. Tests on weighted networks and on directed networks can be found in the Supporting Information S1.
In the following sections, for each network, we compose the results of 10 iterations for the network analysis for the first hierarchical level and the results of 50 iterations for higher levels, if any. The single cluster analysis was repeated 100 times for each cluster.
LFR benchmark
The LFR benchmark [41], [42], like the GN
benchmark, is a particular case of the planted
 -partition model
[43],
which is the simplest possible model of networks with communities. The
planted
-partition model
[43],
which is the simplest possible model of networks with communities. The
planted  -partition model is a class of graphs whose vertices
are divided into
-partition model is a class of graphs whose vertices
are divided into 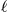 equal-sized
groups, such that the probability that two vertices of the same group are
linked is
equal-sized
groups, such that the probability that two vertices of the same group are
linked is 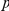 , while the probability that two vertices of
different groups are linked is
, while the probability that two vertices of
different groups are linked is 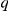 , with
, with
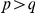 . The planted
. The planted 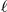 -partition
model is too simple to describe real networks. Vertices have essentially the
same degree and communities have the same size, at odds with empirical
analysis showing that both features typically are broadly distributed [19], [44]–[48]. Therefore we
have recently proposed a generalization of the model, the LFR benchmark, by
introducing power-law distributions for the vertex degree and the community
size, with exponents
-partition
model is too simple to describe real networks. Vertices have essentially the
same degree and communities have the same size, at odds with empirical
analysis showing that both features typically are broadly distributed [19], [44]–[48]. Therefore we
have recently proposed a generalization of the model, the LFR benchmark, by
introducing power-law distributions for the vertex degree and the community
size, with exponents 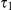 and
and
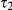 , respectively [41]. The LFR
benchmark poses a far harder challenge to algorithms than the benchmark by
Girvan and Newman, which is regularly used in the literature, and is more
suitable to spot their limits. We are of course aware that the communities
of the model are still too simple to match the communities of real networks.
Other features should be introduced, to tailor the model graphs onto the
real graphs. This is certainly doable, and could be specialized to the
particular domain of applicability one is interested in. Still, the clusters
of the LFR benchmark are a much better proxy of real communities than the
clusters of other benchmark graphs.
, respectively [41]. The LFR
benchmark poses a far harder challenge to algorithms than the benchmark by
Girvan and Newman, which is regularly used in the literature, and is more
suitable to spot their limits. We are of course aware that the communities
of the model are still too simple to match the communities of real networks.
Other features should be introduced, to tailor the model graphs onto the
real graphs. This is certainly doable, and could be specialized to the
particular domain of applicability one is interested in. Still, the clusters
of the LFR benchmark are a much better proxy of real communities than the
clusters of other benchmark graphs.
Vertices of the LFR benchmark have a fixed degree (in this case taken from
the given power law distribution), so the two parameters
 and
and 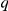 of the planted
of the planted
 -partition model are not independent and we choose as
independent variable the mixing parameter
-partition model are not independent and we choose as
independent variable the mixing parameter
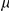 , which is the ratio of the number of external
neighbors of a vertex by the total degree of the vertex. Small values of
, which is the ratio of the number of external
neighbors of a vertex by the total degree of the vertex. Small values of
 indicate well separated clusters, whereas for higher
and higher values communities become more and more mixed to each other.
indicate well separated clusters, whereas for higher
and higher values communities become more and more mixed to each other.
As a term of comparison we used Infomap [49], which has proved to
be very accurate on artificial benchmark graphs [50]. Fig. 6 shows the
comparative performance of OSLOM and Infomap on the LFR benchmark, with
undirected and unweighted edges and non-overlapping clusters. As a measure
of similarity between the planted partition and that recovered by the
algorithm we adopted the Normalized Mutual Information (NMI) [51], in the
extended version proposed in Ref. [23], which enables
one to compare both partitions and covers. We used this definition also for
hard planted partitions, since modules found by OSLOM may be overlapping. In
all tests on artificial graphs each point is always an average over
 realizations.
realizations.
The plots correspond to two network sizes,  and
and
 , and two ranges of community size,
, and two ranges of community size,
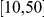 (“small”) and
(“small”) and
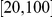 (“big”), that we indicate with the
letters S and B, respectively. In this way we can check how much the
performance of the algorithm is affected by the network size and the average
size of the communities. The other network parameters are given in the
caption. From the plots we conclude that OSLOM and Infomap have a basically
equivalent performance.
(“big”), that we indicate with the
letters S and B, respectively. In this way we can check how much the
performance of the algorithm is affected by the network size and the average
size of the communities. The other network parameters are given in the
caption. From the plots we conclude that OSLOM and Infomap have a basically
equivalent performance.
It is important to test the performance of the algorithms on large graphs as
well, given the increasing availability of large networked datasets. The
question is if and how their performance is affected by the network size.
Fig. 7 shows that
both OSLOM and Infomap are effective at finding communities on large LFR
graphs. We remark that the inferior accuracy of OSLOM when communities are
better defined comes from the fact that the method occasionally finds
homeless vertices, i.e. vertices that are not significantly linked to any
cluster. These are vertices that happen not to have a significant excess of
neighbors within their community with respect to the number of neighbors in
the other communities, despite the fact that the average number of internal
neighbors is high. This happens because of fluctuations, and the method
judges such vertices as not belonging to any group, which makes sense. This
issue of the homeless vertices is a general feature of OSLOM. One should not
judge it negatively, though. If a vertex  happens to
have a number of external neighbors which is appreciably higher than the
expected external degree of the vertex
happens to
have a number of external neighbors which is appreciably higher than the
expected external degree of the vertex 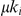 , the condition
, the condition
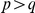 of the planted
of the planted  -partition
model does not hold, so in principle the vertex should not be put in its
original community. The confusion derives from the fact that the condition
-partition
model does not hold, so in principle the vertex should not be put in its
original community. The confusion derives from the fact that the condition
 holds on average.
holds on average.
Figure 7. Tests on large undirected and unweighted LFR benchmark graphs without overlapping communities.
The network sizes are 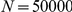 (left)
and
(left)
and  (right), the maximum degree
(right), the maximum degree 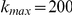 and
the community size ranges from
and
the community size ranges from  to
to
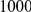 . The
other parameters are the same as those used for the graphs of Fig. 6. The two
curves refer to OSLOM (diamonds) and Infomap (circles).
. The
other parameters are the same as those used for the graphs of Fig. 6. The two
curves refer to OSLOM (diamonds) and Infomap (circles).
LFR benchmark with overlapping communities
The LFR benchmark also accounts for overlapping communities, by assigning to each vertex an equal number of neighbors in different clusters [42]. To simplify things, we assume that each vertex belongs to the same number of communities. We cannot use Infomap for the comparison, as it delivers “hard” partitions, without overlaps between clusters. So we used two recent methods, that have a good performance on LFR graphs with overlapping communities: COPRA [52], based on label propagation [53], and MOSES [54], based on stochastic block modeling [55]. COPRA and MOSES are more efficient to detect overlapping communities in LFR benchmark graphs than the popular Clique Percolation Method (CPM) [19], which is the reason why we do not use the CPM here. In Fig. 8 we show how the performance of each method decays with the fraction of overlapping vertices, for different choices of the mixing parameter and for the small (S) and big (B) communities defined above. Since in social networks there may be many vertices belonging to several groups, we also considered the extreme situation of graphs consisting entirely of overlapping vertices. In this case, by increasing the number of memberships of the vertices communities become more fuzzy and it gets harder and harder for any method to correctly identify the modules. From Fig. 8 we deduce that OSLOM significantly outperforms COPRA in both tests and MOSES in the test with overlapping and non-overlapping vertices, while the performances of OSLOM and MOSES are quite close when all vertices are overlapping.
Figure 8. Test on undirected and unweighted LFR benchmark with overlapping communities.
The parameters are: 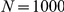 ,
,
 ,
,
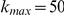 ,
,
 ,
,
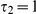 . S and
B indicate the usual ranges of community sizes we use:
. S and
B indicate the usual ranges of community sizes we use:
 and
and
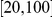 ,
respectively. We tested OSLOM against two recent methods to find
covers in graphs: COPRA [52] and MOSES
[54]. The left panel displays the normalized
mutual information (NMI) between the planted cover and the one
recovered by the algorithm, as a function of the fraction of
overlapping vertices. Each overlapping vertex is shared between two
clusters. The four curves correspond to different values of the
mixing parameter
,
respectively. We tested OSLOM against two recent methods to find
covers in graphs: COPRA [52] and MOSES
[54]. The left panel displays the normalized
mutual information (NMI) between the planted cover and the one
recovered by the algorithm, as a function of the fraction of
overlapping vertices. Each overlapping vertex is shared between two
clusters. The four curves correspond to different values of the
mixing parameter  (
(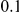 and
and
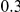 ) and
to the community size ranges S and B. The right panel shows a test
on graphs whose vertices are all shared between clusters. Each
vertex is member of the same number of clusters. The plot shows the
NMI as a function of the number of memberships of the vertices. Each
curve corresponds to a given value of the average degree
) and
to the community size ranges S and B. The right panel shows a test
on graphs whose vertices are all shared between clusters. Each
vertex is member of the same number of clusters. The plot shows the
NMI as a function of the number of memberships of the vertices. Each
curve corresponds to a given value of the average degree
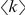 . The
graph parameters are
. The
graph parameters are 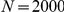 ,
,
 ,
,
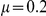 ,
,
 ,
,
 .
Community sizes are in the range
.
Community sizes are in the range 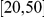 .
.
Hierarchical LFR benchmark
OSLOM is capable to handle hierarchical community structure as well. To test
its performance we have designed an algorithm that produces a version of the
LFR benchmark with hierarchy. To keep things simple, we consider a two-level
hierarchical structure (Fig.
9). The idea is to use the wiring procedure of the original
algorithm twice, first for the micro-communities and then for the
macro-communities. In order to do so, we need two mixing parameters:
 , the fraction of neighbors of each vertex belonging
to different macro-communities;
, the fraction of neighbors of each vertex belonging
to different macro-communities;  , the fraction
of neighbors of each vertex belonging to the same macro-community but to
different micro-communities.
, the fraction
of neighbors of each vertex belonging to the same macro-community but to
different micro-communities.
Figure 9. A realization of the hierarchical LFR benchmark with two levels.
Stars indicate overlapping vertices.
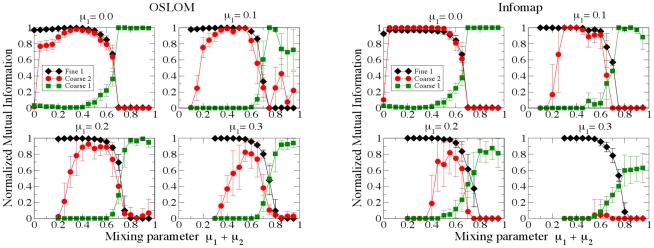
The question is whether the algorithm is able to recover both planted
partitions of the benchmark, which we call Fine
(micro-communities) and Coarse (macro-communities). The
partitions found by the algorithm can be one, two or more, we call them
partition 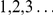 . In the test, whose results are illustrated in Fig. 10, we compare the
Fine partition with partition 1 (Fine 1), the Coarse partition with
partition 2 (Coarse 2), and the Coarse partition with partition 1 (Coarse
1). We compare OSLOM with a recent extension of Infomap to networks with
hierarchical community structure [56]. In the plots we show
how the similarity of the three pairs of partitions mentioned above varies
by increasing
. In the test, whose results are illustrated in Fig. 10, we compare the
Fine partition with partition 1 (Fine 1), the Coarse partition with
partition 2 (Coarse 2), and the Coarse partition with partition 1 (Coarse
1). We compare OSLOM with a recent extension of Infomap to networks with
hierarchical community structure [56]. In the plots we show
how the similarity of the three pairs of partitions mentioned above varies
by increasing 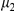 but keeping
but keeping
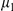 constant (we picked the values
constant (we picked the values
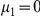 ,
, 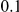 ,
,
 ,
,  ). For a better
comparison of the panels we put on the x-axis the sum
). For a better
comparison of the panels we put on the x-axis the sum
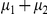 , representing the fraction of neighbors of a vertex
not belonging to its micro-community. We find that, when
, representing the fraction of neighbors of a vertex
not belonging to its micro-community. We find that, when
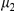 increases, the Fine partition becomes difficult to
resolve and, for
increases, the Fine partition becomes difficult to
resolve and, for 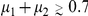 , it cannot be
found anymore and both algorithms can only find the Coarse partition.
Instead, for smaller value of
, it cannot be
found anymore and both algorithms can only find the Coarse partition.
Instead, for smaller value of  , the
algorithms can recover both levels. OSLOM performs better than Infomap if
, the
algorithms can recover both levels. OSLOM performs better than Infomap if
 is not too small.
is not too small.
Figure 10. Test on hierarchical LFR benchmark graphs (unweighted, undirected and without overlapping clusters).
We compare three pairs of partitions: the lowest hierarchical
partition found by the algorithm (indicated by
 ) with
the set of micro-communities of the benchmark (Fine); the lowest
hierarchical partition found by the algorithm with the set of
macro-communities of the benchmark (Coarse); the second lowest
hierarchical partition found by the algorithm (indicated by
) with
the set of micro-communities of the benchmark (Fine); the lowest
hierarchical partition found by the algorithm with the set of
macro-communities of the benchmark (Coarse); the second lowest
hierarchical partition found by the algorithm (indicated by
 ) with
the set of macro-communities of the benchmark. The corresponding
similarities are plotted as a function of
) with
the set of macro-communities of the benchmark. The corresponding
similarities are plotted as a function of
 , for
fixed
, for
fixed  . There
are
. There
are 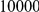 vertices, the average degree
vertices, the average degree  , the
maximum degree
, the
maximum degree 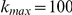 , the
size of the macro-communities lies between
, the
size of the macro-communities lies between
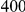 and
and
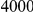 vertices, the size of the micro-communities lies between
vertices, the size of the micro-communities lies between
 and
and
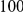 vertices. The exponents of the degree and community size
distributions are
vertices. The exponents of the degree and community size
distributions are 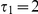 and
and
 .
.
Random graphs and noise
We check whether OSLOM is also able to recognize the absence, and not simply the presence, of community structure. In random graphs vertices are connected to each other at random, modulo some basic constraints like, e. g., keeping some prescribed degree distribution or sequence. In this way, there are by definition no groups of vertices that preferentially link to each other, so there are no communities. There may be subgraphs with an internal edge density higher than the average edge density of the whole network, but they originate from stochastic fluctuations (noise). A good community finding algorithm should be able to recognize that such subgraphs are false positives, and discard them. Here we want to see if OSLOM distinguishes “order” from “noise”. For this purpose, we carried out two tests.
In Fig. 11 we applied
OSLOM and Infomap to Erdös-Rényi random graphs [57] and
scale-free networks [58]. The goal is to see whether the algorithms
recognize that there are no actual communities. Good answers are the
partition with as many communities as vertices, or the partition with all
vertices in the same community. Let us call  the partition
found by the algorithm at hand. Clusters in
the partition
found by the algorithm at hand. Clusters in 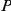 containing at
least two vertices and smaller than the whole network indicate that the
method has been fooled. The fraction of graph vertices belonging to those
clusters is a measure of reliability: the lower this number, the better the
algorithm. In Fig. 11 we
show this variable as a function of the average degree
containing at
least two vertices and smaller than the whole network indicate that the
method has been fooled. The fraction of graph vertices belonging to those
clusters is a measure of reliability: the lower this number, the better the
algorithm. In Fig. 11 we
show this variable as a function of the average degree
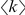 of the random graphs we considered. For OSLOM it
remains very low for all values of
of the random graphs we considered. For OSLOM it
remains very low for all values of 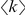 . This is not
surprising, since OSLOM estimates the statistical significance of clusters,
and is therefore ideal to detect stochastic fluctuations. Infomap instead
finds many non-trivial clusters when
. This is not
surprising, since OSLOM estimates the statistical significance of clusters,
and is therefore ideal to detect stochastic fluctuations. Infomap instead
finds many non-trivial clusters when  is low,
whereas it correctly recognizes the absence of community structure if
is low,
whereas it correctly recognizes the absence of community structure if
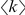 increases.
increases.
Figure 11. Test on random graphs.
We plot the fraction of vertices belonging to non-trivial clusters
(i.e. to clusters with more than one and less than
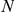 vertices, where
vertices, where  is as
usual the size of the graph), as a function of the average degree of
the graph. The curves correspond to Erdös-Rényi graphs
(diamonds) and scale-free networks (circles). All graphs have
is as
usual the size of the graph), as a function of the average degree of
the graph. The curves correspond to Erdös-Rényi graphs
(diamonds) and scale-free networks (circles). All graphs have
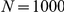 vertices. The only parameter needed to build Erdös-Rényi
graphs is the probability that a pair of vertices is connected,
which is determined by the average degree
vertices. The only parameter needed to build Erdös-Rényi
graphs is the probability that a pair of vertices is connected,
which is determined by the average degree
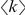 . The
scale-free networks were built with the configuration model [39], starting from a fixed degree sequence for
the vertices obeying the predefinite power law distribution. The
parameters of the distribution are: degree exponent
. The
scale-free networks were built with the configuration model [39], starting from a fixed degree sequence for
the vertices obeying the predefinite power law distribution. The
parameters of the distribution are: degree exponent
 ,
maximum degree
,
maximum degree  .
.
The second test deals with graphs consisting of an ordered
part, with well-defined clusters, and a noisy part,
consisting of vertices randomly attached to the rest of the network. The
ordered part is an LFR benchmark graph with  vertices and
represents the starting configuration of our system. The noisy vertices (up
to
vertices and
represents the starting configuration of our system. The noisy vertices (up
to 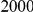 in number) are successively added in sequence, and a
newly added vertex is linked to the other ones via preferential attachment
[58].
The initial degree of the noisy vertices is drawn from a power law
distribution with
in number) are successively added in sequence, and a
newly added vertex is linked to the other ones via preferential attachment
[58].
The initial degree of the noisy vertices is drawn from a power law
distribution with 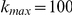 and exponent
and exponent
 . We measure two things, as a function of the number
of noisy vertices: the similarity between the set of noisy vertices and the
set of homeless vertices found by OSLOM, which is expressed by the Jaccard
Index [59]
(Fig. 12, left); the
similarity between the planted partition of the ordered part of the graph
and the subset of the partition found by OSLOM including (only) the vertices
of the ordered part, which is expressed by the normalized mutual information
(Fig. 12, right). We
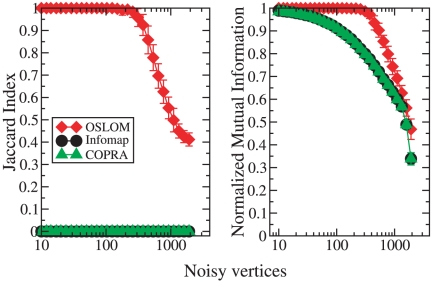
compare OSLOM with Infomap and COPRA [52]. We find that OSLOM
correctly separates the clusters and the noise up to a number of about
. We measure two things, as a function of the number
of noisy vertices: the similarity between the set of noisy vertices and the
set of homeless vertices found by OSLOM, which is expressed by the Jaccard
Index [59]
(Fig. 12, left); the
similarity between the planted partition of the ordered part of the graph
and the subset of the partition found by OSLOM including (only) the vertices
of the ordered part, which is expressed by the normalized mutual information
(Fig. 12, right). We
compare OSLOM with Infomap and COPRA [52]. We find that OSLOM
correctly separates the clusters and the noise up to a number of about
 noisy vertices, which represent almost a third of
the whole network. Infomap and COPRA, instead, do not recognize the noisy
vertices, no matter how small their number is. Also, they tend to mix noisy
vertices with the clusters of the planted partition of the ordered part, as
shown by the fact that the partition they recover never exactly match the
planted partition, not even when just a few noisy vertices are present.
These results are actually understandable in the case of Infomap, which is
based on the minimization of the code length required to describe random
walks taking place on the graph: singletons (clusters consisting of single
vertices) are generally not admitted because they increase the amount of
information required to map the process, due to the high number of
transitions of the walker from the singletons to the rest of the graph and
back.
noisy vertices, which represent almost a third of
the whole network. Infomap and COPRA, instead, do not recognize the noisy
vertices, no matter how small their number is. Also, they tend to mix noisy
vertices with the clusters of the planted partition of the ordered part, as
shown by the fact that the partition they recover never exactly match the
planted partition, not even when just a few noisy vertices are present.
These results are actually understandable in the case of Infomap, which is
based on the minimization of the code length required to describe random
walks taking place on the graph: singletons (clusters consisting of single
vertices) are generally not admitted because they increase the amount of
information required to map the process, due to the high number of
transitions of the walker from the singletons to the rest of the graph and
back.
Figure 12. Test on graphs including communities and noise.
The communities are those of an LFR benchmark graph (undirected,
unweighted and without overlapping clusters), with
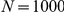 ,
,
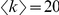 ,
,
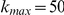 ,
,
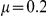 . The
cluster size ranges from
. The
cluster size ranges from  to
to
 vertices. The noise comes by adding vertices which are randomly
linked to the existing vertices, via preferential attachment. The
test consists in checking whether the community finding algorithm at
study (here OSLOM, Infomap and COPRA) is able to find the
communities of the planted partition of the LFR benchmark and to
recognize as homeless the other vertices.
vertices. The noise comes by adding vertices which are randomly
linked to the existing vertices, via preferential attachment. The
test consists in checking whether the community finding algorithm at
study (here OSLOM, Infomap and COPRA) is able to find the
communities of the planted partition of the LFR benchmark and to
recognize as homeless the other vertices.
Real networks
In this section we discuss the application of OSLOM to networks from the real world. In Table 1 we list the networks considered in our analysis, along with some basic statistics obtained from the detection of their community structure with OSLOM.
Table 1. Basic statistics of the real networks we analyzed, including the main features of their community structure, detected by OSLOM.
| Network | N | E |

|
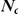
|

|

|

|
| Zachary's club | 34 | 78 | 4.59 | 2 | 17.0 | 1.03 | 0.0294 |
| Dolphins | 62 | 159 | 5.13 | 2 | 32.5 | 1.08 | 0.0322 |
| Football | 115 | 613 | 10.7 | 11 | 10.0 | 1.00 | 0.0434 |
| UK commuting |
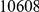
|
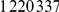
|
230.07 | 248 | 45.43 | 1.06 | 0.00386 |
| C. elegans | 453 |
|
8.94 | 25 | 17.04 | 1.22 | 0.229 |
| Word association |

|

|
8.82 | 261 | 22.48 | 1.35 | 0.395 |
| Live Journal |
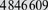
|

|
17.6 |
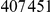
|
10.01 | 1.19 | 0.294 |
| www.uk |

|

|
15.81 |
|
28.08 | 1.02 | 0.125 |
| US airports 2009 (jan) | 448 |

|
34.19 | 11 | 33.81 | 1.28 | 0.352 |
| US airports 2009 (mar) | 456 |
|
37.24 | 6 | 67.83 | 1.22 | 0.272 |
| US airports 2009 (jun) | 453 |
|
37.42 | 9 | 45.33 | 1.28 | 0.315 |
| US airports 2009 (sep) | 452 |

|
34.81 | 9 | 41.55 | 1.26 | 0.347 |
From left to right, we list the number of vertices
 and
edges
and
edges  , the
average degree
, the
average degree 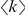 , the
number of clusters
, the
number of clusters 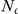 , the
average cluster size
, the
average cluster size  , the
average number of memberships per vertex
, the
average number of memberships per vertex
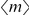 and
the fraction
and
the fraction  of
vertices not assigned to any cluster (homeless vertices). The values
related to the community structure refer to the lowest hierarchical
level.
of
vertices not assigned to any cluster (homeless vertices). The values
related to the community structure refer to the lowest hierarchical
level.
We analyzed different types of systems: social, information, biological and infrastructural networks. Here we discuss only some of them, the rest of the analysis can be found in the Supporting Information S1.
The word association network
This network is built on the University of South Florida Free Association
Norms [60]. Here the presence of an edge between words
 and
and 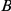 indicates that
some people associate
indicates that
some people associate 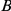 to the word
to the word
 . This network is considered a paradigmatic example
of graph with overlapping communities [19], since several words may
have various meanings and belong to different groups of words. In Fig. 13 we see a few
subgraphs of the word association network, revolving around four keywords:
bright, knowledge,
music and play. We see that the
keywords are shared among several clusters, which are semantically highly
homogeneous. For instance, bright belongs to three groups,
centered on the words color, shine and
smart, respectively, which makes sense. In the same
subgraph, the words sun and dark are also
overlapping vertices, belonging to the groups of color and
shine, as one might expect. In the subgraph centered on
knowledge, one distinguishes the groups referring to
the words mind, intelligent,
expert and college/university. Here
there are many overlapping vertices, like the word
intelligence, shared between the groups of
mind and intelligent, and a bunch of
terms indicating (mostly) professional status within schools and/or
universities, like student, professor,
teacher, etc., which lie between the groups of
expert and college/university. In the
third subgraph, the word music is shared by the groups of
instrument, song/dance and
noise/sound: other overlapping vertices are the words
sing and voice, lying between
song/dance and noise/sound, and the
words bass and saxophone, belonging to the
groups of song/dance and instrument.
Finally, the word play sits between the communities of
sport, music and
youth/kid; other overlapping vertices in this subgraph
include game, children,
toy, etc.
. This network is considered a paradigmatic example
of graph with overlapping communities [19], since several words may
have various meanings and belong to different groups of words. In Fig. 13 we see a few
subgraphs of the word association network, revolving around four keywords:
bright, knowledge,
music and play. We see that the
keywords are shared among several clusters, which are semantically highly
homogeneous. For instance, bright belongs to three groups,
centered on the words color, shine and
smart, respectively, which makes sense. In the same
subgraph, the words sun and dark are also
overlapping vertices, belonging to the groups of color and
shine, as one might expect. In the subgraph centered on
knowledge, one distinguishes the groups referring to
the words mind, intelligent,
expert and college/university. Here
there are many overlapping vertices, like the word
intelligence, shared between the groups of
mind and intelligent, and a bunch of
terms indicating (mostly) professional status within schools and/or
universities, like student, professor,
teacher, etc., which lie between the groups of
expert and college/university. In the
third subgraph, the word music is shared by the groups of
instrument, song/dance and
noise/sound: other overlapping vertices are the words
sing and voice, lying between
song/dance and noise/sound, and the
words bass and saxophone, belonging to the
groups of song/dance and instrument.
Finally, the word play sits between the communities of
sport, music and
youth/kid; other overlapping vertices in this subgraph
include game, children,
toy, etc.
Figure 13. Application of OSLOM to real networks: the word association network.
Stars indicate overlapping vertices.
UK commuting
This is the network of flows of commuters between areas of the United
Kingdom, and therefore it has a clearly geographic character. It is composed
of 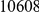 vertices, each representing a ward, i. e. a
geographical division used in the UK census for statistical purposes. The
whole territory of the United Kingdom is divided into wards. Each edge
corresponds to a flow of commuters between the ward of origin and that of
destination, with a weight accounting for the number of commuters per day.
The data were collected during the
vertices, each representing a ward, i. e. a
geographical division used in the UK census for statistical purposes. The
whole territory of the United Kingdom is divided into wards. Each edge
corresponds to a flow of commuters between the ward of origin and that of
destination, with a weight accounting for the number of commuters per day.
The data were collected during the 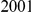 UK census,
when the ward of residence and the ward of work/study was registered for a
sizeable part of the British population. The database can be accessed online
at the site of the Office for National Statistics http://www.ons.gov.uk/census. OSLOM finds three hierarchical
levels (Fig. 14). The
clusters of the second level delimit geographical areas typically centered
about one major town. In the highest level the areas of England, Wales,
Scotland and Northern Ireland are clearly recognizable. Interestingly,
Northern Ireland and Scotland are parts of the same community, due to the
large flow of commuters between the two regions, despite the geographical
separation. Black points represent overlapping vertices.
UK census,
when the ward of residence and the ward of work/study was registered for a
sizeable part of the British population. The database can be accessed online
at the site of the Office for National Statistics http://www.ons.gov.uk/census. OSLOM finds three hierarchical
levels (Fig. 14). The
clusters of the second level delimit geographical areas typically centered
about one major town. In the highest level the areas of England, Wales,
Scotland and Northern Ireland are clearly recognizable. Interestingly,
Northern Ireland and Scotland are parts of the same community, due to the
large flow of commuters between the two regions, despite the geographical
separation. Black points represent overlapping vertices.
Figure 14. Application of OSLOM to real networks: flows of commuters in the UK.
Black points indicate overlapping vertices.
LiveJournal and UK Web
We also applied OSLOM to two large networks. The first is a network of
friendship relationships between users of the on-line community
LiveJournal (www.livejournal.com),
and was downloaded from the Stanford Large Network Dataset Collection
(http://snap.stanford.edu/data/). The second is a crawl of
the Web graph carried out by the Stanford WebBase Project (http://dbpubs.stanford.edu:8091/~testbed/doc2/WebBase/),
within the UK domain (.uk). We remind that the Web graph is a directed graph
whose vertices are Web pages, while the edges are the hyperlinks that enable
one to surf from one page to another. These two systems are too large for
OSLOM, due to the huge variety of possible cluster sizes to explore.
Therefore we applied a two-step method: in the first step, we derived an
initial partition 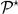 with the
Louvain method [61], which is able to handle large networked
datasets; in the second step, we apply OSLOM to refine the clusters of
with the
Louvain method [61], which is able to handle large networked
datasets; in the second step, we apply OSLOM to refine the clusters of
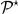 . In principle, this procedure should yield the same
partitions/covers as applying OSLOM directly, if one repeated OSLOM's
cluster search many times. But this would make the calculations too lengthy,
so, in order to complete the analysis within a reasonable time, it is
necessary to keep the number of iterations low. In this way there is the big
advantage of drastically reducing the computational complexity, which makes
large systems tractable, even if results would be more accurate if one could
apply OSLOM from scratch. Clearly, since different iterations are
independent processes, one could sensibly increase the statistics by
distributing the iterations among different processors, if available.
. In principle, this procedure should yield the same
partitions/covers as applying OSLOM directly, if one repeated OSLOM's
cluster search many times. But this would make the calculations too lengthy,
so, in order to complete the analysis within a reasonable time, it is
necessary to keep the number of iterations low. In this way there is the big
advantage of drastically reducing the computational complexity, which makes
large systems tractable, even if results would be more accurate if one could
apply OSLOM from scratch. Clearly, since different iterations are
independent processes, one could sensibly increase the statistics by
distributing the iterations among different processors, if available.
In Fig. 15 we present the
distribution of cluster sizes of the first two hierarchical levels found by
OSLOM. The results are obtained by performing a single iteration on a
workstation HP Z800. For the Web graph, which is the larger system, with
nearly  million vertices and
million vertices and  million edges
(see Table 1), the
analysis was completed in about
million edges
(see Table 1), the
analysis was completed in about  hours. For the
social network of LiveJournal we can compare the results
with the corresponding distributions found by Infomap and the Label
Propagation Method (LPM) proposed by Leung et al. [62], which were computed in
a recent analysis [48]. In that work the original Infomap was used, so
neither Infomap nor the LPM could detect hierarchical community structure
and there is just one cluster size distribution, corresponding to the single
partition recovered. The distributions are broad and quite similar across
different methods. Interestingly, the two hierarchical levels of
LiveJournal (OSLOM 1 and OSLOM 2) are not too
different, indicating a sort of self-similarity of the community structure.
For the Web the two levels are more dissimilar and the distributions have a
clear power law decay (with different exponents) up to a cutoff, which is
approximately the same for both curves (
hours. For the
social network of LiveJournal we can compare the results
with the corresponding distributions found by Infomap and the Label
Propagation Method (LPM) proposed by Leung et al. [62], which were computed in
a recent analysis [48]. In that work the original Infomap was used, so
neither Infomap nor the LPM could detect hierarchical community structure
and there is just one cluster size distribution, corresponding to the single
partition recovered. The distributions are broad and quite similar across
different methods. Interestingly, the two hierarchical levels of
LiveJournal (OSLOM 1 and OSLOM 2) are not too
different, indicating a sort of self-similarity of the community structure.
For the Web the two levels are more dissimilar and the distributions have a
clear power law decay (with different exponents) up to a cutoff, which is
approximately the same for both curves ( vertices).
vertices).
Figure 15. Application of OSLOM to real networks: friendships of LiveJournal users (left) and sample of the .uk domain of the Web graph (right).
We show the distribution of cluster sizes obtained by OSLOM for the first two hierarchical levels (OSLOM 1 and OSLOM 2). For LiveJournal we can compare the distributions with those found with Infomap [49] and the Label Propagation Method (LPM) by Leung et al. [62].
Dynamic datasets: the US air transportation network
For the last application, we used a time-stamped dataset, the US air
transportation network. The data can be downloaded from the Bureau of
Transportation Statistics (US government) (http://www.bts.gov). Vertices
are airports in the USA and edges are weighted by the number of passengers
transported along the corresponding routes. In Fig. 16 we show the geographical location
of the airports and their communities, indicated by the symbols, for three
snapshots, corresponding to the traffic in March, June and September 2009,
respectively. We remind that for dynamical datasets we usually take the
partition/cover 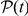 of the system
at time
of the system
at time  , and we use it as initial partition/cover for the
topology of the system at time
, and we use it as initial partition/cover for the
topology of the system at time 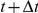 , which is then
refined by OSLOM, in order to “adapt”
, which is then
refined by OSLOM, in order to “adapt”
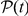 to the current structure. This is done to exploit
the information of more snapshots at the same time. Since the three maps of
Fig. 16 are mostly
illustrative, communities were derived by applying directly OSLOM to the
corresponding snapshots, for simplicity. The diagram indicates the
similarity between networks and their corresponding partitions/covers in
different snapshots. Each snapshot represents the whole traffic of one
trimester, which corresponds to a season, while
to the current structure. This is done to exploit
the information of more snapshots at the same time. Since the three maps of
Fig. 16 are mostly
illustrative, communities were derived by applying directly OSLOM to the
corresponding snapshots, for simplicity. The diagram indicates the
similarity between networks and their corresponding partitions/covers in
different snapshots. Each snapshot represents the whole traffic of one
trimester, which corresponds to a season, while
 year, as we want to measure the variation of the
network structure in consecutive seasons. The similarity between
partitions/covers is computed with the normalized mutual information, as
usual. The similarity of two weighted networks like the ones at study is
measured in the following way. First, one computes the distance
year, as we want to measure the variation of the
network structure in consecutive seasons. The similarity between
partitions/covers is computed with the normalized mutual information, as
usual. The similarity of two weighted networks like the ones at study is
measured in the following way. First, one computes the distance
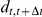 between the matrices
between the matrices  and
and
 :
:  . The matrix
. The matrix
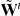 is derived from the standard weight matrix
is derived from the standard weight matrix
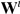 by dividing each edge weight by the sum of all edge
weights. This is done because the traffic flows tend to increase steadily in
time, so comparing the original weight matrices is not appropriate. The
quantity
by dividing each edge weight by the sum of all edge
weights. This is done because the traffic flows tend to increase steadily in
time, so comparing the original weight matrices is not appropriate. The
quantity  is a dissimilarity measure. We turn it to a
similarity index by changing its sign, adding a constant and rescaling the
resulting values. Since we wish to compare the trend of the network
similarity with that of the partition/cover similarity, the additional
constant and the rescaling factor are chosen such to reproduce the average
and the variance of the curve of the normalized mutual information. After
this operation, the two trends are finally comparable. The diagram shows
that both measures follow a yearly periodicity, with peaks corresponding to
the winter season, which is then more stable than the others.
is a dissimilarity measure. We turn it to a
similarity index by changing its sign, adding a constant and rescaling the
resulting values. Since we wish to compare the trend of the network
similarity with that of the partition/cover similarity, the additional
constant and the rescaling factor are chosen such to reproduce the average
and the variance of the curve of the normalized mutual information. After
this operation, the two trends are finally comparable. The diagram shows
that both measures follow a yearly periodicity, with peaks corresponding to
the winter season, which is then more stable than the others.
Figure 16. Application of OSLOM to real networks: US airport network.
The maps show the position of the airports, which are represented by
symbols, indicating the communities found by applying OSLOM directly
to the corresponding network, without exploiting the information of
previous snapshots. The diagram shows the “seasonality”
of air traffic. The normalized mutual information (diamonds) was
computed comparing the cover of the system at time
 adjusted by OSLOM on the network at time
adjusted by OSLOM on the network at time
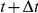 , and
the cover obtained by applying OSLOM directly to the system at time
, and
the cover obtained by applying OSLOM directly to the system at time
 . The
circles are estimates of the similarity of the network matrices of
snapshots separated by
. The
circles are estimates of the similarity of the network matrices of
snapshots separated by  (one
year). For each year we took four snapshots, by cumulating the
traffic of each trimester. The most stable networks are typically in
winter (vertical lines).
(one
year). For each year we took four snapshots, by cumulating the
traffic of each trimester. The most stable networks are typically in
winter (vertical lines).
Discussion
We have introduced OSLOM, the first method that finds clusters in networks based on their statistical significance. It is a multi-purpose technique, capable to handle various types of graphs, accounting for edge direction, edge weights, overlapping communities, hierarchy and network dynamics. Therefore, it can be used for a wide variety of datasets and applications.
We have thoroughly tested OSLOM against the best algorithms currently available on
various types of artificial benchmark graphs, with excellent results. In particular,
OSLOM is superior on directed graphs and in the detection of strongly overlapping
clusters. Moreover, it is an ideal method to recognize the absence of community
structure and/or the presence of randomness in graphs. In some cases OSLOM returns
slightly less accurate results than other methods, because it finds several homeless
vertices when communities are fuzzy. This is due to the fact that, in the
realizations of benchmark graphs, it may happen that some vertices end up having the
same number of neighbors (or even more) in other communities than in their own, due
to fluctuations, even if on average this does not happen. So, the classification of
those vertices, imposed by the planted
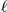 -partition model, is not justified topologically. This is an
important general issue that needs to be assessed in the future, to avoid systematic
errors in the testing procedure.
-partition model, is not justified topologically. This is an
important general issue that needs to be assessed in the future, to avoid systematic
errors in the testing procedure.
OSLOM is a local algorithm, so it respects the nature of community structure, which
is a local feature of networks, the more so the larger the systems at study.
However, the null model adopted to estimate the statistical significance of clusters
is the configuration model, which is global. This is the same null model adopted in
modularity optimization [63], and is responsible for the serious problems of this
technique, like its well known resolution limit [64]. Therefore we perform an
iterative cluster search within the clusters found after the first application of
the method, by considering each cluster as a network on its own. In this way we
progressively limit the horizon of the part of the network under exploration, and we
are able to find the smallest significant clusters, which are the natural building
blocks of the network and the basis of its hierarchical community structure. So the
null model, originally global, gets confined to smaller and smaller portions of the
graph. The actual resolution of the method is thus not due to the null model, but to
the choice of the threshold  . In this paper we have
set
. In this paper we have
set 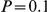 , which is often used in various contexts and delivers an
excellent performance on the benchmark graphs we have adopted. Nevertheless, how
much a real graph deviates from a random graph depends on the
specific system at hand, and it would be more appropriate to estimate the threshold
, which is often used in various contexts and delivers an
excellent performance on the benchmark graphs we have adopted. Nevertheless, how
much a real graph deviates from a random graph depends on the
specific system at hand, and it would be more appropriate to estimate the threshold
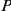 case by case. This is an issue to consider for future work.
We remark that also for modularity optimization one could in principle iteratively
restrict the null model to the clusters found by the method. However, modularity is
based on the expected value of variables estimated on the null
model, neglecting random fluctuations, which is why modularity can attain large
values on specific partitions of random graphs [65]–[67]. OSLOM instead accounts for
those fluctuations, so it is far more reliable, in this respect. Furthermore OSLOM
is a local method, so it does not suffer from the severe problems coming from
modularity's global optimization [68].
case by case. This is an issue to consider for future work.
We remark that also for modularity optimization one could in principle iteratively
restrict the null model to the clusters found by the method. However, modularity is
based on the expected value of variables estimated on the null
model, neglecting random fluctuations, which is why modularity can attain large
values on specific partitions of random graphs [65]–[67]. OSLOM instead accounts for
those fluctuations, so it is far more reliable, in this respect. Furthermore OSLOM
is a local method, so it does not suffer from the severe problems coming from
modularity's global optimization [68].
Another important aspect to emphasize is the need to perform many iterations, to get more accurate results. This is not a specific feature of OSLOM, but it should be done for all community detection techniques with a stochastic character, like methods based on optimization (e. g., modularity optimization). In the literature there is the general attitude to perform a single iteration, and to reduce the complexity of an algorithm to the time required to carry out one iteration. But this is not appropriate, especially on large networks. For instance, by performing a single iteration, vertices lying on the border between clusters may be assigned to a specific cluster, while in many cases they are overlapping. By combining the results of several iterations, instead, it is more likely to distinguish overlapping vertices from the others. Furthermore, one can compute the strength of the membership of vertices in different clusters, from the frequency with which they were classified in each cluster. One can also disambiguate stable from unstable clusters, which could be recovered from specific iterations. So, it is crucial to collect and combine the results of many iterations. Of course, the complexity of the method grows with the number of iterations, but it can be considerably reduced by distributing runs among many different processors, if large computer clusters are available.
The running time of OSLOM is dominated by the exhaustive search of significant vertices, inside and outside the clusters. This search could be carried out with greedy approaches, with a huge computational advantage, and this is an improvement we plan to implement in the near future. On the other hand, if one wishes to attack very large graphs, OSLOM could be used at a second stage, as a refinement technique, to clean the results of an initial partition delivered by a fast algorithm. In this case, since the initial clusters are usually cores or parts of the significant clusters we are looking for, OSLOM converges far more rapidly than its direct application without inputs. We have seen in the previous section that, by combining OSLOM with the Louvain method by Blondel et al., we were able to handle systems with millions of vertices.
We have proposed a recipe to deal with the increasingly more important issue of detecting communities in dynamic networks. The idea is to take advantage of the information of different snapshots at the same time, by “adapting” the partition/cover of the earlier snapshot to the topology of the other one. In this way it is possible to uncover the correlation between the structures of the system at different time stamps.
We have shown the versatility of OSLOM by applying it to various networked datasets. OSLOM provides the first comprehensive toolbox for the analysis of community structure in graphs and is an ideal complement of existing tools for network analysis. The algorithm, with all its variants (including a fast two-step procedure for the analysis of very large networks) is implemented in a freely downloadable and documented software (http://www.oslom.org).
Supporting Information
(PDF)
Acknowledgments
We thank Paolo Bajardi, Steve Gregory and Martin Rosvall for useful suggestions.
Footnotes
Competing Interests: The authors have declared that no competing interests exist.
Funding: A.L. and S.F. gratefully acknowledge ICTeCollective. The project ICTeCollective acknowledges the financial support of the Future and Emerging Technologies (FET) programme within the Seventh Framework Programme for Research of the European Commission, under FET-Open grant number 238597. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Albert R, Barabási AL. Statistical mechanics of complex networks. Rev Mod Phys. 2002;74:47–97. [Google Scholar]
- 2.Dorogovtsev SN, Mendes JFF. Evolution of networks. Adv Phys. 2002;51:1079–1187. [Google Scholar]
- 3.Newman MEJ. The structure and function of complex networks. SIAM Rev. 2003;45:167–256. [Google Scholar]
- 4.Pastor-Satorras R, Vespignani A. Evolution and Structure of the Internet: A Statistical Physics Approach. New York, , NY, USA: Cambridge University Press; 2004. [Google Scholar]
- 5.Boccaletti S, Latora V, Moreno Y, Chavez M, Hwang DU. Complex networks: Structure and dynamics. Phys Rep. 2006;424:175–308. [Google Scholar]
- 6.Barrat A, Barthélemy M, Vespignani A. Dynamical processes on complex networks. Cambridge, UK: Cambridge University Press; 2008. [Google Scholar]
- 7.Caldarelli G. Scale-free networks. Oxford, UK: Oxford University Press; 2007. [Google Scholar]
- 8.Girvan M, Newman ME. Community structure in social and biological networks. Proc Natl Acad Sci USA. 2002;99:7821–7826. doi: 10.1073/pnas.122653799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lusseau D, Newman MEJ. Identifying the role that animals play in their social networks. Proc Royal Soc London B. 2004;271:S477–S481. doi: 10.1098/rsbl.2004.0225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Adamic LA, Glance N. LinkKDD '05: Proceedings of the 3rd international workshop on Link discovery. New York, , NY, USA: ACM Press; 2005. The political blogosphere and the 2004 u.s. election: divided they blog. pp. 36–43. [Google Scholar]
- 11.Flake GW, Lawrence S, Lee Giles C, Coetzee FM. Self-organization and identification of web communities. IEEE Computer. 2002;35:66–71. [Google Scholar]
- 12.Pimm SL. The structure of food webs. Theor Popul Biol. 1979;16:144–158. doi: 10.1016/0040-5809(79)90010-8. [DOI] [PubMed] [Google Scholar]
- 13.Krause AE, Frank KA, Mason DM, Ulanowicz RE, Taylor WW. Compartments revealed in food-web structure. Nature. 2003;426:282–285. doi: 10.1038/nature02115. [DOI] [PubMed] [Google Scholar]
- 14.Jonsson PF, Cavanna T, Zicha D, Bates PA. Cluster analysis of networks generated through homology: automatic identification of important protein communities involved in cancer metastasis. BMC Bioinf. 2006;7:2. doi: 10.1186/1471-2105-7-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Holme P, Huss M, Jeong H. Subnetwork hierarchies of biochemical pathways. Bioinformatics. 2003;19:532–538. doi: 10.1093/bioinformatics/btg033. [DOI] [PubMed] [Google Scholar]
- 16.Guimerà R, Amaral LAN. Functional cartography of complex metabolic networks. Nature. 2005;433:895–900. doi: 10.1038/nature03288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fortunato S. Community detection in graphs. Physics Reports. 2010;486:75–174. [Google Scholar]
- 18.Baumes J, Goldberg MK, Krishnamoorthy MS, Ismail MM, Preston N. Finding communities by clustering a graph into overlapping subgraphs. In: Guimaraes N, Isaias PT, editors. IADIS AC. IADIS; 2005. pp. 97–104. [Google Scholar]
- 19.Palla G, Derényi I, Farkas I, Vicsek T. Uncovering the overlapping community structure of complex networks in nature and society. Nature. 2005;435:814–818. doi: 10.1038/nature03607. [DOI] [PubMed] [Google Scholar]
- 20.Zhang S, Wang RS, Zhang XS. Identification of overlapping community structure in complex networks using fuzzy c-means clustering. Physica A. 2007;374:483–490. [Google Scholar]
- 21.Gregory S. Proceedings of the 11th European Conference on Principles and Practice of Knowledge Discovery in Databases (PKDD 2007) Berlin, Germany: Springer-Verlag; 2007. An algorithm to find overlapping community structure in networks. pp. 91–102. [Google Scholar]
- 22.Nepusz T, Petróczi A, Négyessy L, Bazsó F. Fuzzy communities and the concept of bridgeness in complex networks. Phys Rev E. 2008;77:016107. doi: 10.1103/PhysRevE.77.016107. [DOI] [PubMed] [Google Scholar]
- 23.Lancichinetti A, Fortunato S, Kertész J. Detecting the overlapping and hierarchical community structure in complex networks. New J Phys. 2009;11:033015. [Google Scholar]
- 24.Evans TS, Lambiotte R. Line graphs, link partitions, and overlapping communities. Phys Rev E. 2009;80:016105. doi: 10.1103/PhysRevE.80.016105. [DOI] [PubMed] [Google Scholar]
- 25.Kovács IA, Palotai R, Szalay MS, Csermely P. Community landscapes: An integrative approach to determine overlapping network module hierarchy, identify key nodes and predict network dynamics. PLoS ONE. 2010;5:e12528. doi: 10.1371/journal.pone.0012528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Simon H. The architecture of complexity. Proc Am Phil Soc. 1962;106:467–482. [Google Scholar]
- 27.Sales-Pardo M, Guimerà R, Moreira AA, Amaral LAN. Extracting the hierarchical organization of complex systems. Proc Natl Acad Sci USA. 2007;104:15224–15229. doi: 10.1073/pnas.0703740104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Clauset A, Moore C, Newman MEJ. Airoldi EM, Blei DM, Fienberg SE, Goldenberg A, Xing EP, et al., editors. Structural Inference of Hierarchies in Networks. Statistical Network Analysis: Models, Issues, and New Directions. 2007. pp. 1–13. Springer, Berlin, Germany, volume 4503 of Lect. Notes Comp. Sci.
- 29.Clauset A, Moore C, Newman MEJ. Hierarchical structure and the prediction of missing links in networks. Nature. 2008;453:98–101. doi: 10.1038/nature06830. [DOI] [PubMed] [Google Scholar]
- 30.Bianconi G, Pin P, Marsili M. Assessing the relevance of node features for network structure. Proc Natl Acad Sci USA. 2009;106:11433–11438. doi: 10.1073/pnas.0811511106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lancichinetti A, Radicchi F, Ramasco JJ. Statistical significance of communities in networks. Phys Rev E. 2010;81:046110. doi: 10.1103/PhysRevE.81.046110. [DOI] [PubMed] [Google Scholar]
- 32.Hopcroft J, Khan O, Kulis B, Selman B. Tracking evolving communities in large linked networks. Proc Natl Acad Sci USA. 2004;101:5249–5253. doi: 10.1073/pnas.0307750100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Backstrom L, Huttenlocher D, Kleinberg J, Lan X. KDD '06: Proceedings of the 12th ACM SIGKDD international conference on Knowledge discovery and data mining. New York, NY, USA: ACM; 2006. Group formation in large social networks: membership, growth, and evolution. pp. 44–54. [Google Scholar]
- 34.Chakrabarti D, Kumar R, Tomkins A. KDD '06: Proceedings of the 12th ACM SIGKDD international conference on Knowledge discovery and data mining. New York, NY, USA: ACM; 2006. Evolutionary clustering. pp. 554–560. [Google Scholar]
- 35.Palla G, Barabási AL, Vicsek T. Quantifying social group evolution. Nature. 2007;446:664–667. doi: 10.1038/nature05670. [DOI] [PubMed] [Google Scholar]
- 36.Asur S, Parthasarathy S, Ucar D. KDD '07: Proceedings of the 13th ACM SIGKDD international conference on Knowledge discovery and data mining. New York, NY, USA: ACM; 2007. An event-based framework for characterizing the evolutionary behavior of interaction graphs. pp. 913–921. [Google Scholar]
- 37.Mucha PJ, Richardson T, Macon K, Porter MA, Onnela J. Community Structure in Time-Dependent, Multiscale, and Multiplex Networks. Science. 2010;328:876. doi: 10.1126/science.1184819. [DOI] [PubMed] [Google Scholar]
- 38.Radicchi F, Lancichinetti A, Ramasco JJ. Combinatorial approach to modularity. Phys Rev E. 2010;82:026102. doi: 10.1103/PhysRevE.82.026102. [DOI] [PubMed] [Google Scholar]
- 39.Molloy M, Reed B. A critical point for random graphs with a given degree sequence. Random Struct Algor. 1995;6:161–179. [Google Scholar]
- 40.Newman MEJ, Girvan M. Finding and evaluating community structure in networks. Phys Rev E. 2004;69:026113. doi: 10.1103/PhysRevE.69.026113. [DOI] [PubMed] [Google Scholar]
- 41.Lancichinetti A, Fortunato S, Radicchi F. Benchmark graphs for testing community detection algorithms. Phys Rev E. 2008;78:046110. doi: 10.1103/PhysRevE.78.046110. [DOI] [PubMed] [Google Scholar]
- 42.Lancichinetti A, Fortunato S. Benchmarks for testing community detection algorithms on directed and weighted graphs with overlapping communities. Phys Rev E. 2009;80:016118. doi: 10.1103/PhysRevE.80.016118. [DOI] [PubMed] [Google Scholar]
- 43.Condon A, Karp RM. Algorithms for graph partitioning on the planted partition model. Random Struct Algor. 2001;18:116–140. [Google Scholar]
- 44.Albert R, Jeong H, Barabási AL. Error and attack tolerance of complex networks. Nature. 2000;406:378–382. doi: 10.1038/35019019. [DOI] [PubMed] [Google Scholar]
- 45.Newman MEJ. Detecting community structure in networks. Eur Phys J B. 2004;38:321–330. doi: 10.1103/PhysRevE.69.066133. [DOI] [PubMed] [Google Scholar]
- 46.Radicchi F, Castellano C, Cecconi F, Loreto V, Parisi D. Defining and identifying communities in networks. Proc Natl Acad Sci USA. 2004;101:2658–2663. doi: 10.1073/pnas.0400054101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Clauset A, Newman MEJ, Moore C. Finding community structure in very large networks. Phys Rev E. 2004;70:066111. doi: 10.1103/PhysRevE.70.066111. [DOI] [PubMed] [Google Scholar]
- 48.Lancichinetti A, Kivelä M, Saramäki J, Fortunato S. Characterizing the community structure of complex networks. PLoS ONE. 2010;5:e11976. doi: 10.1371/journal.pone.0011976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rosvall M, Bergstrom CT. Maps of random walks on complex networks reveal community structure. Proc Natl Acad Sci USA. 2008;105:1118–1123. doi: 10.1073/pnas.0706851105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lancichinetti A, Fortunato S. Community detection algorithms: A comparative analysis. Phys Rev E. 2009;80:056117. doi: 10.1103/PhysRevE.80.056117. [DOI] [PubMed] [Google Scholar]
- 51.Danon L, Daz-Guilera A, Duch J, Arenas A. Comparing community structure identification. J Stat Mech. 2005;P09008 [Google Scholar]
- 52.Gregory S. Finding overlapping communities in networks by label propagation. New Journal of Physics. 2010;12:103018. [Google Scholar]
- 53.Raghavan UN, Albert R, Kumara S. Near linear time algorithm to detect community structures in large-scale networks. Phys Rev E. 2007;76:036106. doi: 10.1103/PhysRevE.76.036106. [DOI] [PubMed] [Google Scholar]
- 54.McDaid A, Hurley NJ. Detecting highly overlapping communities with model-based overlapping seed expansion. ASONAM 2010. 2010.
- 55.Nowicki K, Snijders TAB. Estimation and Prediction for Stochastic Blockstructures. J Am Stat Assoc. 2001;96 [Google Scholar]
- 56.Rosvall M, Bergstrom CT. Multilevel compression of random walks on networks reveals hierarchical organization in large integrated systems. Eprint. 2010;arXiv:10100431. doi: 10.1371/journal.pone.0018209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Erdös P, Rényi A. On random graphs. I. Publ Math Debrecen. 1959;6:290–297. [Google Scholar]
- 58.Barabási AL, Albert R. Emergence of scaling in random networks. Science. 1999;286:509–512. doi: 10.1126/science.286.5439.509. [DOI] [PubMed] [Google Scholar]
- 59.Tan PN, Steinbach M, Kumar V. Introduction to Data Mining. New York, USA: Addison Wesley, 1 edition; 2005. [Google Scholar]
- 60.Nelson DL, McEvoy CL, Schreiber TA. The university of south florida word association, rhyme, and word fragment norms. 1998 doi: 10.3758/bf03195588. [DOI] [PubMed] [Google Scholar]
- 61.Blondel VD, Guillaume JL, Lambiotte R, Lefebvre E. Fast unfolding of communities in large networks. J Stat Mech. 2008;P10008 [Google Scholar]
- 62.Leung IXY, Hui P, Liò P, Crowcroft J. Towards real-time community detection in large networks. Phys Rev E. 2009;79:066107. doi: 10.1103/PhysRevE.79.066107. [DOI] [PubMed] [Google Scholar]
- 63.Newman MEJ. From the Cover: Modularity and community structure in networks. Proc Natl Acad Sci USA. 2006;103:8577–8582. doi: 10.1073/pnas.0601602103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Fortunato S, Barthélemy M. Resolution limit in community detection. Proc Natl Acad Sci USA. 2007;104:36–41. doi: 10.1073/pnas.0605965104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Guimerà R, Sales-Pardo M, Amaral LA. Modularity from fluctuations in random graphs and complex networks. Phys Rev E. 2004;70:025101 (R). doi: 10.1103/PhysRevE.70.025101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Reichardt J, Bornholdt S. When are networks truly modular? Physica D. 2006;224:20–26. [Google Scholar]
- 67.Reichardt J, Bornholdt S. Partitioning and modularity of graphs with arbitrary degree distribution. Phys Rev E. 2007;76:015102 (R). doi: 10.1103/PhysRevE.76.015102. [DOI] [PubMed] [Google Scholar]
- 68.Good BH, de Montjoye YA, Clauset A. Performance of modularity maximization in practical contexts. Phys Rev E. 2010;81:046106. doi: 10.1103/PhysRevE.81.046106. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(PDF)