Abstract
Yersinia pestis is the causative agent of the plague. Y. pestis KIM 10+ strain was passaged and selected for loss of the 102 kb pgm locus, resulting in an attenuated strain, KIM D27. In this study, whole genome sequencing was performed on KIM D27 in order to identify any additional differences. Initial assemblies of 454 data were highly fragmented, and various bioinformatic tools detected between 15 and 465 SNPs and INDELs when comparing both strains, the vast majority associated with A or T homopolymer sequences. Consequently, Illumina sequencing was performed to improve the quality of the assembly. Hybrid sequence assemblies were performed and a total of 56 validated SNP/INDELs and 5 repeat differences were identified in the D27 strain relative to published KIM 10+ sequence. However, further analysis showed that 55 of these SNP/INDELs and 3 repeats were errors in the KIM 10+ reference sequence. We conclude that both 454 and Illumina sequencing were required to obtain the most accurate and rapid sequence results for Y. pestis KIMD27. SNP and INDELS calls were most accurate when both Newbler and CLC Genomics Workbench were employed. For purposes of obtaining high quality genome sequence differences between strains, any identified differences should be verified in both the new and reference genomes.
Introduction
Whole genome sequencing (WGS) has broadened the field of microbial forensics by allowing the identification of bacteria not only to the subspecies level, but also to specific sequence types [1]. With the advent of next generation sequencing (NGS) technologies, the application of whole genome sequencing data promises to be even more applicable since it is much more affordable and rapid than traditional Sanger sequencing. The data generated by NGS can be used to fully understand the lineage of naturally or maliciously constructed disease isolates, and could even yield information about past propagation and culturing conditions due to mutations that occur during passage [2], [3]. For forensic purposes, it is necessary to use sequencing methods that yield very accurate results, as well as bioinformatics tools that reveal the similarities and differences with a reference genome such as single nucleotide polymorphisms (SNPs), insertion and deletion events (INDELs), and differences in tandem repeats (VNTRs). To date, genome resequencing efforts with NGS have been used to study the evolutionary lineage of different strains within a single species [2], [4], [5], or to study the types of adaptations that occur during growth in specific conditions [3]. Furthermore, Y. pestis has also been subject of NGS sequencing studies to characterize the phylogeographic diversity within the species [6]. The study found that Y. pestis strains can be grouped into 4 specific lineages (1 root and 3 branches), that can be characterized further by lineage-specific SNPs [6]. Although the conclusions from these studies depend on the accuracy of the data, any errors inherent in the technology used can be tolerated in some applications. In contrast, for forensic applications, NGS data must be practically error-free since the data could potentially be used to identify and prosecute perpetrators of criminal or terrorist acts.
Y. pestis KIM D27, the attenuated laboratory strain, is an isogenic derivative of KIM 10+ (biovar Mediaevalis) that was passaged until the pigmentation phenotype was lost due to loss of the pgm locus [7]. The virulent Y. pestis KIM 10+ was originally isolated from a human pneumonic plague infection in Iran in 1961 [8]. The 102 kb pgm locus is composed of a pathogenicity island and genes involved in pigment synthesis, and its loss results in the attenuated phenotype [7]. The pathogenicity island contains genes for biosynthesis of yersiniabactin [9] and hemin storage (hms) genes [10], that are used for iron scavenging [11], biofilm formation [12], and prevention of damage due to reactive oxygen species [13]. Since loss of the pgm locus renders a strain avirulent [14], [15], it is exempt from the US Department of Health and Human Services (HHS) Select Agent rules [16], and is commonly used in the lab as a model for Yersinia pestis studies. Prior to this report, the genome sequence of Y. pestis KIM D27 has not been reported nor has any comprehensive analysis of the details of its genome alterations relative to the fully virulent KIM 10+ parent been considered as they pertain to avirulent phenotype of KIM D27. This issue has assumed some relevance due to the death of a researcher possibly due to an accidental infection by such an attenuated strain of Y. pestis [17].
We took advantage of the fact that the genome sequence of Y. pestis KIM 10+ was available since its publication in 2002 [18], to test our NGS platforms for forensic purposes by sequencing KIM D27 genomic DNA. Whole genome sequence of KIM D27 confirms the loss of the pgm locus and allowed a detailed comparison with the published KIM 10+ sequence. Initial comparisons indicated that KIM D27 contained numerous SNPs and INDELs, but upon further sequencing and examination, these were shown to be errors in the reference KIM 10+ sequence. However, KIM D27 did possess 2 authentic SNPs and 1 repeat expansion relative to the KIM 10+ parent. The data were used to correct the genome sequence of KIM 10+. It was also concluded that the most accurate sequence for genome comparison purposes results from hybrid 454-Illumina sequencing coupled with the use of multiple bioinformatics tools for identifying differences between genome sequences from closely related strains.
Materials and Methods
Genome sequencing and assembly
Y. pestis KIM D27 genomic DNA was obtained from Olaf Schneewind at the University of Chicago. A 454 Titanium 8 kb paired-end library was constructed and used for sequencing on a full plate of 454 XLR Titanium platform following manufacturer's instructions (Roche, Branford, CT). The overall paired end ratio was 53.9% and the average coverage of the chromosome was 29x. An Illumina paired-end library was constructed and 100 bp sequencing was conducted on Illumina Genome Analyzer IIx following manufacturer's instructions (Illumina, San Diego, CA). Raw sequences from 454 only, 454 and Illumina, or 454, Illumina and Sanger from PCR verification products, were assembled de novo using the 454 Newbler assembler [19] and the Celera Assembler 6.1 [20], with default parameters. In addition, mapping assemblies using the published KIM 10+ (accession no. NC_004088) were performed with 454 Newbler, CLC Genomics Workbench (CLC bio, Cambridge, MA), or Amoscmp -shortReads [21] using default parameters. Mate pair information was used to predict the order and orientation of the contigs, confirming that no genomic rearrangements were present relative to KIM 10+.
Annotation
The KIM D27 genome was annotated through JCVI's auto-annotation pipeline. Whenever possible, annotation followed the nomenclature of KIM 10+. All ORFs ≥ 30 amino acids in length and annotated in other Yersinia genomes were added to the KIM D27 annotation.
SNP, INDELS, and VNTR analysis
The resulting hybrid 454, Illumina, and Sanger assembly was used to align against the Y. pestis KIM 10+ nucleotide sequence in CLC Genomics Workbench or Newbler assembler to detect single base-pair polymorphisms (SNP) or insertion/deletion (INDELS) using a 4 read minimum and 2000 read maximum coverage with a frequency of at least 35%, allowing for at most 3 different variants at any site. The results were filtered to exclude any SNPs or INDELs that had low confidence after assembly. All remaining changes were verified by PCR amplification and Sanger sequencing using primers listed in Table S1. SNP or INDELS were corrected to match Sanger read if it did not match the consensus sequence. Repeats were detected with REputer [22] using default parameters and variations in tandem repeat numbers were characterized with Tandem Repeat Finder [23] and CLC genomics workbench. Errors in the Y. pestis KIM 10+ genome sequence were identified by PCR amplification and Sanger sequencing. KIM 10+ genomic DNA was purchased from Biodefense and Emerging Infections Research Resources Repository (Cat No. NR-2645; Manassas, VA).
Nucleotide sequence accession numbers
The Y. pestis KIM D27 genome was deposited in GenBank under accession number ADDC00000000.
Results
Summary of sequencing and assembly results for Y. pestis KIM D27
The genomic DNA of Y. pestis KIM D27 was purified and sequenced on a full-plate of 454 Life Sciences Genome Sequencer FLX machine (Roche, Branford, CT). The 454 sequencing from 8 kb paired end libraries yielded 677,705 reads that passed filter wells with a 53.9% paired end ratio and a total chromosomal coverage of 29X. 454 sequencing data resulted in highly fragmented assemblies using either Celera Assembler 6.1 [20] or Newbler assembler [19], with slightly better results for AMOScmp (12 contigs) [21] (Table 1). The fragmentation was due in part to the complex repeat structure of Yersinia pestis genome [24]. The effect of repeats on assembly is evident because 123,000 bases of the KIM 10+ were not covered by a uniquely matching read during reference assemblies, most of these associated with repeat regions or multi-copy sequences like rRNA or transposable elements (not including the known pgm deletion discussed below [9]; data not shown). The assemblies showed that the KIM 10+ and KIM D27 genomes had the same overall structure. However, a close inspection of the assembly of 454 sequences resulted in a high number of discrepancies from the KIM 10+ reference genome, including SNPs and INDELs (Figure 1). A majority of these (82%) involved either an A or T residue and most (72%) were primarily associated with poly(A) or poly(T) sequences.
Table 1. Summary of sequencing results for Y. pestis KIM D27 and comparison with KIM10.
| Sequencing technology | Type of assembly | Total number of contigs | |
| de novo | Reference | ||
| 454 | CA | 135 | - |
| Newbler | 306 | 126 | |
| AMOScmp | − | 12 | |
| CLC | − | − | |
| Illumina | Newbler | − | − |
| CLC | 378 | 4 | |
| 454 + Illumina Hybrid | Newbler | − | 120 |
| AMOScmp | − | 1 | |
| CLC Reference | − | 4 | |
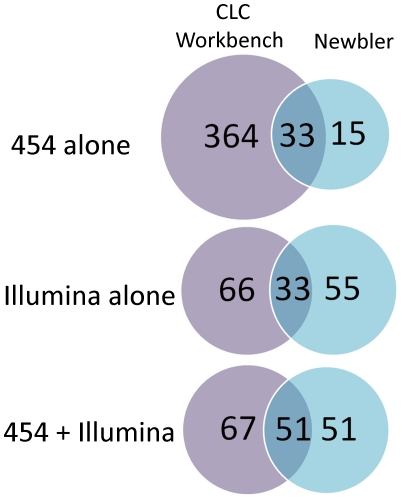
Figure 1. Comparison of SNP and INDELs detection tools based on different sequence assemblies.
Assemblies of KIM D27 sequences from either 454 only, Illumina only, or hybrid assemblies (454 + Illumina) were analyzed for SNPs and INDELs as described in Materials and Methods. The Venn diagram represents the number of identified differences in either CLC Genomics Workbench or Newbler either singly (purple or blue, respectively) or the overlap between both programs (darkened area).
Because of the large number of SNPs and INDELs identified, additional sequencing was performed with a second platform to eliminate technology-specific errors, such as those associated with homopolymer tracts with the 454 platform. One lane of 100 bp Illumina sequencing was done on a paired end library, yielding over 18 million reads and 371X chromosomal coverage. Hybrid mapping assemblies were generated using Newbler reference assembler (120 contigs) [19], or CLC Genomics Workbench (3 contigs; CLC bio, Cambridge, MA) reference assembler and are summarized in Table 1. In our experience, the consensus built by AMOScmp was faulty due to its inability to efficiently handle homopolymeric bases. This assembly, while structurally matching the KIM 10+ reference, contained hundreds of consensus discrepancies with KIM 10+, and thus was not further analyzed.
The sequence of KIM D27 showed that the chromosomal region from 2580534 to 2679386 of KIM 10+ that contains the pgm locus and genes involved in iron acquisition was entirely missing as reported previously [9]. The genetic loss was a consequence of genetic rearrangement across insertion (IS) elements IS100 that border the region. Other organisms, such as Burkholderia mallei, have also lost virulence due to rearrangements across IS element while cultured in the laboratory [25]–[26]. From the coverage data, the ratio between the plasmids was determined to be 1∶2∶4 for pMT-1, pCD1, and pPCP1, compared to the chromosome, respectively.
SNP, INDELs, and repeat analysis and confirmation
A major goal for forensic sequencing and other applications requiring high sequence accuracy is to identify both large and small changes in the genome that could explain the source of a strain or have implications for gene function, for example. Therefore, a detailed analysis of SNPs, INDELs, changes in numbers of repeats and gene-scale gains or losses was conducted. SNPs and INDELs were identified for the chromosome and all three plasmids using CLC Genomics Workbench or Newbler and are summarized in Figure 1. Analysis of 454 data alone produced the most SNP and INDELs (364 by CLC alone, 15 by Newbler alone, 33 by both). The Illumina data alone gave fewer SNPs (59 for CLC alone, 55 by Newbler alone, 48 by both). Hybrid assemblies including both types of data resulted in slightly higher SNPs than Illumina data alone (67 by CLC, 51 by Newbler, 51 by both). For all assemblies, alignment to the KIM 10+ reference sequence identified 3–5 additional SNPs and INDELs, leading to a total of 18 unambiguous SNPs and 38 INDELs. All of these SNPs and INDELs were amplified by PCR and confirmed by Sanger sequencing from KIM D27 source DNA.
In addition to SNP/INDELs, Nucmer alignment [27] of KIM D27 de novo contigs from the CA6.1 assembly and KIM 10+ identified multiple breaks in regions of suspected differences in numbers of repeat units. REputer [22] analysis showed that 21 of these regions contained repeat elements (e.g., VNTR). After PCR and Sanger sequencing, 16 of these sequences matched the published KIM 10+ sequence. However, 4 of the repeats in KIM D27 appeared to have expanded while 1 repeat had contracted relative to the KIM 10+ published genome (Table 2). Since KIM D27 is a laboratory derived isolate of KIM 10+, the 56 SNP/INDELs and 5 repeat differences seemed suspiciously high, and it was possible that the reference KIM 10+ sequence had some errors. Therefore, Y. pestis KIM 10+ DNA was purchased from the Biodefense and Emerging Infections Research Resources Repository (Cat No. NR-2645; Manassas, VA) and used as template for PCR and Sanger sequencing. 55 of 56 SNP/INDELs and 2 repeat expansions and 1 contraction were found to be errors in the KIM 10+ sequence, since the PCR products had the same sequence as KIM D27. Because the trace reads from the KIM 10+ sequence were no longer available we are unable to determine the source of the error. One true SNP between KIM D27 and KIM 10+ was confirmed at coordinate 1505634 on KIM 10+, which results in an Alanine to Valine change in hisS (Table 2); one true additional 17 bp tandem repeat expansion was found at coordinate 3346611 upstream of the modE gene (Table 3). Lastly, one large expansion (∼3 kb) had occurred in KIM D27 within a gene for a putative outer membrane surface protein (invasin homolog, y3884) that is 9 kb in KIM 10+ and 12 kb in KIM D27 (Table 3 and Figure 2). The difference in size was due to multiple copies of a 289 bp repeat as identified by Tandem Repeat Finder [23]. Interestingly, the KIM 10+ PCR reactions appeared mixed (Figure 2), suggesting that either the genomic DNA was derived from a mixed population of repeats resulting in gene lengths from 7–13 kb or that the repeat is unstable, resulting from copy number variation on replication.
Table 2. Repeat expansion or reduction in Y. pestis KIM D27.
| Type | KIM 10+ coordinates | Size in KIM 10+ (bp) | KIM D27 coordinates | Size in KIM D27 (bp) | Coding | Comment |
| VNTR | 125066–125542 | 476† | 125066–125429 | 363 | N | Tandem repeat. Five copies in KIM 10+; four in KIM D27. |
| VNTR | 704203–704423 | 220† | 704091–704433 | 342 | N | Repeat area expanded in D27. |
| Expansion* | 3346556–3346654 | 98 | 3245775–3245890 | 115 | N | 17 bp insertion (TTTCTATCTATGTTGTTA) extends N-term of modE(y3040) |
| IS Element | 4232115–4232834 | 719† | 4131351–4132781 | 1430 | Y | Tandem repeat of IS154. One copy in reference, two copies in PCR. |
| VNTR* | 4328042–4329222 | 1180 | 4227881–4233335 | 5454 | Y | Multiple copies of a 289 bp motif; PCR results in ladder effect in y3884 |
Based on published sequence. PCR reactions showed these sites were the same length in KIM 10+ DNA as was observed for KIM D27.
These sequences were different between KIM 10+ and KIM D27.
Table 3. SNP/INDEL confirmed in Y. pestis KIM10+.
| Type of substitution | Coordinate | Ref | Var | Nearest gene | Annotation | Coding | Result | Homopolymer or repetivite region |
| SNP (pCD1) | 65595 | A | G | y0087.1N | putative transposase | N | repetitive region | |
| SNP | 121683 | A | G | y0111 | transposase | Y | No amino acid change | |
| SNP | 142073 | C | G | y0129 | gltD; glutamate synthase beta subunit | Y | P to A at residue 223 | 4 C |
| INDEL | 276834 | − | G | y0258 | hypothetical protein | Y | Extends N-term of ORF | 3 G |
| INDEL | 327388 | G | − | y0305 | transposase | N | Possibly extends ORF | |
| INDEL | 655265 | − | C | y0579 | tyrB; tyrosine biosynthesis | N | ||
| INDEL | 887688 | − | G | y0795 | mrcB; murein sacculus | N | 3 G | |
| INDEL | 897819 | A | − | y0800 | chloride channel protein | Y | Moves stop 2 aa up | 4 A |
| SNP | 988057 | G | T | y0879 | putative sugar transport | N | ||
| SNP | 1086724 | T | C | y0962 | hypothetical protein | Y | No amino acid change | |
| INDEL | 1176674 | G | − | y1043 | tes; acyl-CoA thioesterase II | N | 5 G | |
| SNP | 1184200 | A | G | y1049 | acrB; multidrug efflux pump | Y | No amino acid change | 6 A |
| INDEL | 1375636 | − | C | y1224 | NrdE truncation | N | Extends N-term of ORF | 7 C |
| INDEL/SNP | 1415467 | A | − | y1265 | kdpA; potassium transporter | Y | Changes balance out | repetitive region |
| INDEL/SNP | 1415469 | − | G | y1265 | kdpA; potassium transporter | Y | Changes balance out | repetitive region |
| SNP* | 1505634 | C | T | y1354 | hisS | Y | A to V at residue 88 | |
| INDEL | 1544038 | − | A | y1389 | Transcriptional regulator | N | 3 A | |
| INDEL | 1616527 | − | G | y1457 | hypothetical protein, putative peroxidase | N | ||
| SNP | 1758361 | G | T | y1590 | fabB | N | May change promoter | |
| INDEL | 1830493 | C | − | y1655 | hypothetical protein | N | ||
| INDEL | 2006614 | − | C | y1821 | mgtA; magnesium transporter | N | repetitive region | |
| INDEL | 2006622 | A | − | y1821 | mgtA; magnesium transporter | N | repetitive region | |
| INDEL | 2006743 | − | T | y1821 | mgtA; magnesium transporter | N | repetitive region | |
| INDEL | 2006767 | − | T | y1821 | mgtA; magnesium transporter | N | repetitive region | |
| INDEL | 2006775 | − | T | y1821 | mgtA; magnesium transporter | N | repetitive region | |
| INDEL | 2006778 | − | T | y1821 | mgtA; magnesium transporter | N | repetitive region | |
| INDEL | 2006781 | − | T | y1821 | mgtA; magnesium transporter | N | repetitive region | |
| INDEL | 2006785 | − | T | y1821 | mgtA; magnesium transporter | N | repetitive region | |
| INDEL | 2006791 | − | T | y1821 | mgtA; magnesium transporter | N | repetitive region | |
| INDEL | 2006801 | − | T | y1821 | mgtA; magnesium transporter | N | repetitive region | |
| SNP | 2006807 | C | A | y1821 | mgtA; magnesium transporter | N | repetitive region | |
| INDEL | 2018833 | A | − | y1834 | hypothetical proteins | Y | Unites 2 ORFs | 3 A |
| SNP | 2021584 | C | T | y1834 | hypothetical protein | Y | No amino acid change | |
| SNP | 2078371 | C | T | y1880 | hypothetical protein | Y | K to N at residue 33 | |
| INDEL | 2253705 | G | − | y2047 | Tryptophan synthase alpha subunit | Y | Shorter C-term of trpA | |
| INDEL | 2377088 | A | − | y2150 | purU | N | Extends N-term of ORF | 3 A |
| INDEL | 2377158 | T | − | y2150 | purU | Y | 2nd aa, restores reading frame due to other purU change | 4 T |
| SNP | 2470103 | A | C | y2242 | chaperone | Y | No amino acid change | |
| INDEL | 2563999 | − | A | y2328 | hypothetical protein | Y | Another stop available | 5 A |
| INDEL | 2564024 | − | G | y2328 | hypothetical protein | Y | Another stop available | 3 G |
| SNP | 2786829 | G | A | y2524 | ftn | Y | No amino acid change | repetitive region |
| SNP | 2786831 | G | A | y2524 | ftn | Y | S to F at residue 11 | repetitive region |
| SNP | 2786834 | G | A | y2524 | ftn | Y | A to G at residue 10 | repetitive region |
| INDEL | 2959407 | G | − | y2681 | hypothetical protein | Y | Joins both ORFs | 3 G |
| SNP | 2978605 | A | G | y2697 | hypothetical protein | Y | V to G at residue 289 | |
| SNP | 2981487 | C | T | y2698 | hypothetical protein | Y | No amino acid change | |
| SNP | 2981565 | C | G | y2698 | hypothetical protein | Y | No amino acid change | |
| INDEL | 3231270 | − | G | y2925 | hypothetical protein | N | ||
| SNP | 3533369 | A | G | y3211 | hypothetical protein | Y | N to S at residue 188 | |
| INDEL | 3546848 | − | A | y3221 | PTS permease | Y | Extends ORFs at C-term | |
| INDEL | 3782193 | − | C | y3410 | Non-ribosomal peptide synthase | Y | Changes balance out | 3 C |
| INDEL | 3782201 | − | G | y3410 | Non-ribosomal peptide synthase | Y | Changes balance out | 4 G |
| INDEL | 3782212 | A | − | y3410 | Non-ribosomal peptide synthase | Y | Changes balance out | |
| INDEL | 3824911 | C | − | y3437 | hypothetical protein | Y | Joins both ORFs | |
| INDEL | 4154719 | G | − | y3736 | NadR disrupted | Y | Functional NadR | 3 G |
| INDEL | 4363489 | G | − | y3907 | hypothetical protein | Y | Joins both ORFs | 5 G |
| INDEL | 4470283 | T | − | y4034 | phosphoethanolamine transferase | Y | No amino acid change |
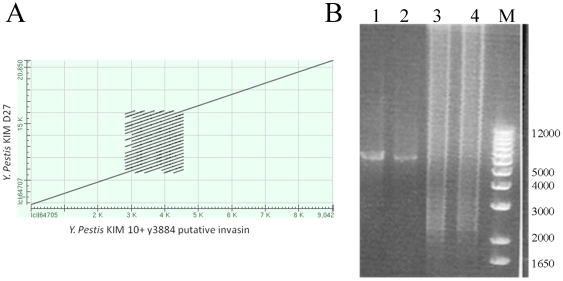
Figure 2. A. Nucmer alignement of putative invasin gene from KIM 10+ and KIM D27.
Regions of homology are depicted as diagonal lines. The 289 bp repeat aligns with itself numerous times and results in the square pattern. B. PCR amplification of the invasin gene from KIM 10+ and KIM D27. The chromosomal region 4228610 – 4234900 was amplified from genomic DNA from either strain. The predicted 6,290 bp product was observed as the major product from KIM D27 (lanes 1 and 2), but a ladder effect was observed when KIM 10+ DNA was used (lanes 3 and 4).
Tables 2 and 3 show the effects of the corrections to the KIM 10+ genome annotation as a result of the identified errors. 23 of the 55 sequence corrections occurred in non-coding regions of the chromosome, while 9 others were silent substitutions within coding regions. 7 of the corrections result in amino acid substitutions. Three of the corrections introduce stop codons, although two of these occur within 2 amino acids of the C-terminus of the protein, and thus are predicted to have little effect on the structure or function of the peptide. In every case where the ORF was extended or truncated, the corrected sequence matched the published sequence of other Yersinia genomes (data not shown).
Determination of the source of the errors in the 454-only assembly
In order to ascertain the potential universality of the problems with the 454-only assembly, an analysis of candidate INDELs conflicts from other 454-only genomes sequenced at the JCVI was performed (Table 4). For each species, the closest finished genome available was chosen as the reference. SNPs were not analyzed because the reference genome did not come from the same strain. The genome sequences of query strains were compared with completed reference sequences published in GenBank, and all INDELs that could result from 454 sequencing errors were identified (Table 4). Y. pestis had among the highest average coverage (i.e. the number of reads that represent any given nucleotide) and number of INDELs, but all genomes contained potential 454-introduced errors. There was no relation between the number of homopolymeric-INDELs and % GC content of the genome. As expected, in general the higher the sequencing coverage, the lower the number of INDELs (P<0.05), although this was not true for Y. pestis.
Table 4. Search for conflicting INDELs in 454-only genomes.
| Organism | Technology | % Genome Covered | %GC | Average Coverage | Conflicting INDELs | Short Sequence Repeats |
| E. coli strain HMP | paired end | 99% | 51% | 29x | 12 | 57 |
| P. gingivalis W83 | fragment | 100% | 48% | 18x | 21 | 58 |
| E. coli strain HMP | fragment | 99% | 51% | 19x | 95 | 64 |
| M. tuberculosis strain | fragment | 99% | 66% | 16x | 118 | 78 |
| M. tuberculosis strain | fragment | 98% | 66% | 13x | 189 | 97 |
| M. tuberculosis strain | fragment | 98% | 66% | 13x | 206 | 94 |
| Y. pestis KIM D27 | paired end | 98% | 48% | 29x | 411 | 246 |
Since the errors were mostly present at poly(A) and poly(T) sequences it is possible that the large number of errors in the KIM D27 454 sequence were due to the strain having an aberrantly high amount of these homopolymer sequences in its genome. The genomes of several species were analyzed for incidence of 4–9 base A or T sequences in CLC Genomics Workbench. Y. pestis had an A or T homopolymer frequency of 11.00 per kb, close to the average (13.23) (Figure 3) and an overall number of A or T homopolymers of 49,536 compared to the average of 52,200. This indicates that the number of 454 errors was not simply an effect of having a high frequency or occurrence of A and T homopolymers. However, the number of simple sequence repeats (SSR; i.e., motifs between 2 and 25 nucleotides that are repeated in tandem) had a positive correlation with the number of INDELs (P<0.005), suggesting that genomes with highly repetitive sequences pose significant difficulties for sequencing and assembly (Table 4). However, increasing sequencing coverage and incorporating Illumina data yielded a better genome assembly than either the Sanger only [18], or 454 only data.
Figure 3. Instances of poly(A) and poly(T) in sequenced genomes.
All 4–9 bp poly(A) and poly(T) in published genomes were counted and plotted per kilobase in each genome.
Discussion
In this study we tested our next generation sequencing (NGS) platforms for the purposes of rapidly and accurately sequencing and characterizing disease causing microorganisms. Using the 454 sequencing platform we obtained the genomic sequence of Y. pestis KIM D27 within days of obtaining the genomic DNA. After various attempts at assembling the genome, we discovered that 454-only sequencing yielded large numbers of SNP, INDELS, and VNTR differences when compared to the parental genome, most within poly(A) or poly(T) homopolymer tracts. Consequently, we complemented the 454 data with Illumina sequencing data to improve the consensus sequence. We found that optimal sequencing of Y.pestis KIM D27 required hybrid assemblies of 454 paired end data and Illumina reads. The hybrid assembly also resulted in highly effective SNP identification, with neither CLC Genomics workbench nor Newbler making false positive calls. The inclusion of multiple SNP calling programs resulted in no false positives called by more than one tool, providing the most confidence in the SNPs that were called. In our experience, the primary strengths of 454 sequencing were that the long reads allow de novo assembly and detection of structural changes in a genome, and rapid sequencing and assembly (less than 1 week in the case of Y. pestis KIM D27). However, the consensus quality is not optimal due to artifacts in homopolymer tracts, especially in genomes with high numbers of short repeats. In turn, the coverage depth acquired by Illumina sequencing provides very accurate consensus sequence generation and analysis, including mapping assemblies, SNP and INDEL calling. However, Illumina sequencing requires more time to generate data, and in general, de novo assemblies are more fragmented than 454-sequence assemblies. We found that the Celera Assembler was able to handle both 454 and hybrid assemblies better and resulted in more robust, accurate assemblies than any of the other programs used. However, mapping assemblies were best performed by CLC Genomics Workbench. We also found that SNP and INDELs were best identified by using several bioinformatic tools, comparing their results, and curating the list of candidate modifications manually.
Analysis of other 454-only genomes from JCVI indicated that, while the severity of the A and T homopolymer error rate was excessive for Y. pestis, the problem is likely to be present in all of the genomes. While the problem can be partially obviated by increased 454 sequencing depth, we found that the error rate was not correlated with the level of coverage, but rather with the number of simple sequence repeats present in the genome. Indeed, a recent study using 454 only sequencing of B. anthracis found that the number of potential SNP/INDELs variants among 8 derivatives from either the Ames or Sterne lineages could be as high as 97 differences [28], and was not related to the level of coverage. The authors were able to verify roughly one half of those differences as true differences using PCR and Sanger sequencing. However, there still remain one half of those differences that can be attributed to sequencing errors. Thus, it might be more cost effective to add Illumina sequencing instead of additional 454 sequencing.
The Y. pestis KIM D27 genome sequence did not reveal any additional genome alterations that would contribute to its attenuated phenotype beyond those already discussed [9], [11], [14], [15]. This analysis, however, revealed that KIM 10+ and to a lesser extent KIM D27, encode a highly variable region within a putative invasin gene (y3040; Figure 2), which may confer adaptation to different hosts, or immune reactions. An additional variable region was detected in the molybdate transport transciriptional regulator, modE, which extended the N-terminus of the peptide by 17 residues. A similar extension is observed in Y. pestis Antigua and Y. pestsis Angola. Interestingly, the additional residues add a secretion signal as predicted by SignalP [29], suggesting there might be a different response to molybdates in KIM D27 than KIM 10+. Lastly, the modification to the hisS gene at residue A88V is not predicted to have a major effect on the protein, because valine is not an uncommon residue at that location among members of the HisS family. However, a modification in HisS could result in differences in the histidine biosynthesis pathway regulation between the two strains that could results through adaptation to laboratory growth.
Importantly, the fact that out of the 56 SNPs and INDELs and 5 repeat size changes initially validated in KIM D27, all but 1 SNPs and 2 repeat expansions (Tables 2 and 3) were found to be sequence errors in the reference genome has significant implications for forensic microbiology and applications involving comparisons with published reference genomes. Specifically, newly observed alterations must be analyzed in both the new isolate as well as the reference genome to discard the possibility of errors in published sequences. Our predicted error levels in the recently 454-only sequenced genomes (see Table 3) combined with those found in older Sanger-only genome sequences would make it virtually impossible to achieve high confidence SNP determinations using solely 454-sequence data, eliminating the possibility of definitive conclusions about the source of disease-causing strains in an investigation such as the Anthrax attacks. Additionally, the limitations of observing large chromosomal rearrangements using only Illumina (or other short read technology), obligates the use of a combined sequencing approach for forensic investigations.
Supporting Information
List of primers used to validate SNPs and INDELs in Yersinia pestis KIM10 and KIM D27.
(XLS)
Footnotes
Competing Interests: The authors have declared that no competing interests exist.
Funding: This project has been funded in whole with federal funds from the National Institute of Allergy and Infectious Diseases, National Institutes of Health, Department of Health and Human Services under contract number HHSN272200900007C. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Read TD, Salzberg SL, Pop M, Shumway M, Umayam L, et al. Comparative genome sequencing for discovery of novel polymorphisms in Bacillus anthracis. Science. 2002;296:2028–2033. doi: 10.1126/science.1071837. [DOI] [PubMed] [Google Scholar]
- 2.Sjodin A, Svensson K, Lindgren M, Forsman M, Larsson P. Whole-genome sequencing reveals distinct mutational patterns in closely related laboratory and naturally propagated Francisella tularensis strains. PLoS One. 2010;5:e11556. doi: 10.1371/journal.pone.0011556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Conrad TM, Joyce AR, Applebee MK, Barrett CL, Xie B, et al. Whole-genome resequencing of Escherichia coli K-12 MG1655 undergoing short-term laboratory evolution in lactate minimal media reveals flexible selection of adaptive mutations. Genome Biol. 2009;10:R118. doi: 10.1186/gb-2009-10-10-r118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jarvik T, Smillie C, Groisman EA, Ochman H. Short-term signatures of evolutionary change in the Salmonella enterica serovar typhimurium 14028 genome. J Bacteriol. 2010;192:560–567. doi: 10.1128/JB.01233-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kuroda M, Serizawa M, Okutani A, Sekizuka T, Banno S, et al. Genome-wide single nucleotide polymorphism typing method for identification of Bacillus anthracis species and strains among B. cereus group species. J Clin Microbiol. 2010;48:2821–2829. doi: 10.1128/JCM.00137-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Morelli G, Song Y, Mazzoni CJ, Eppinger M, Roumagnac P, et al. Yersinia pestis genome sequencing identifies patterns of global phylogenetic diversity. Nat Genet. 2010;42:1140–1143. doi: 10.1038/ng.705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lahteenmaki K, Virkola R, Saren A, Emody L, Korhonen TK. Expression of plasminogen activator pla of Yersinia pestis enhances bacterial attachment to the mammalian extracellular matrix. Infect Immun. 1998;66:5755–5762. doi: 10.1128/iai.66.12.5755-5762.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Finegold MJ, Petery JJ, Berendt RF, Adams HR. Studies on the pathogenesis of plague. Blood coagulation and tissue responses of Macaca mulatta following exposure to aerosols of Pasteurella pestis. Am J Pathol. 1968;53:99–114. [PMC free article] [PubMed] [Google Scholar]
- 9.Buchrieser C, Rusniok C, Frangeul L, Couve E, Billault A, et al. The 102-kilobase pgm locus of Yersinia pestis: sequence analysis and comparison of selected regions among different Yersinia pestis and Yersinia pseudotuberculosis strains. Infect Immun. 1999;67:4851–4861. doi: 10.1128/iai.67.9.4851-4861.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lillard JW, Jr, Fetherston JD, Pedersen L, Pendrak ML, Perry RD. Sequence and genetic analysis of the hemin storage (hms) system of Yersinia pestis. Gene. 1997;193:13–21. doi: 10.1016/s0378-1119(97)00071-1. [DOI] [PubMed] [Google Scholar]
- 11.Carniel E. The Yersinia high-pathogenicity island: an iron-uptake island. Microbes Infect. 2001;3:561–569. doi: 10.1016/s1286-4579(01)01412-5. [DOI] [PubMed] [Google Scholar]
- 12.Forman S, Bobrov AG, Kirillina O, Craig SK, Abney J, et al. Identification of critical amino acid residues in the plague biofilm Hms proteins. Microbiology. 2006;152:3399–3410. doi: 10.1099/mic.0.29224-0. [DOI] [PubMed] [Google Scholar]
- 13.Paauw A, Leverstein-van Hall MA, van Kessel KP, Verhoef J, Fluit AC. Yersiniabactin reduces the respiratory oxidative stress response of innate immune cells. PLoS One. 2009;4:e8240. doi: 10.1371/journal.pone.0008240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Brubaker RR. Mutation rate to nonpigmentation in Pasteurella pestis. J Bacteriol. 1969;98:1404–1406. doi: 10.1128/jb.98.3.1404-1406.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Iteman I, Guiyoule A, de Almeida AM, Guilvout I, Baranton G, et al. Relationship between loss of pigmentation and deletion of the chromosomal iron-regulated irp2 gene in Yersinia pestis: evidence for separate but related events. Infect Immun. 1993;61:2717–2722. doi: 10.1128/iai.61.6.2717-2722.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Perry RD, Bearden SW. Isolation and confirmation of Yersinia pestis mutants exempt from select agent regulations. Curr Protoc Microbiol Chapter 5: Unit 5B. 2008;2 doi: 10.1002/9780471729259.mc05b02s11. [DOI] [PubMed] [Google Scholar]
- 17.Easton J. Chicago: University of Chicago News Release; 2009. Rare infection may have caused death of Chicago scientist. [Google Scholar]
- 18.Deng W, Burland V, Plunkett G, 3rd, Boutin A, Mayhew GF, et al. Genome sequence of Yersinia pestis KIM. J Bacteriol. 2002;184:4601–4611. doi: 10.1128/JB.184.16.4601-4611.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Margulies M, Egholm M, Altman WE, Attiya S, Bader JS, et al. Genome sequencing in microfabricated high-density picolitre reactors. Nature. 2005;437:376–380. doi: 10.1038/nature03959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Miller JR, Delcher AL, Koren S, Venter E, Walenz BP, et al. Aggressive assembly of pyrosequencing reads with mates. Bioinformatics. 2008;24:2818–2824. doi: 10.1093/bioinformatics/btn548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pop M, Phillippy A, Delcher AL, Salzberg SL. Comparative genome assembly. Briefings in bioinformatics. 2004;5:237. doi: 10.1093/bib/5.3.237. [DOI] [PubMed] [Google Scholar]
- 22.Kurtz S, Schleiermacher C. REPuter: fast computation of maximal repeats in complete genomes. Bioinformatics. 1999;15:426–427. doi: 10.1093/bioinformatics/15.5.426. [DOI] [PubMed] [Google Scholar]
- 23.Benson G. Tandem repeats finder: a program to analyze DNA sequences. Nucleic Acids Res. 1999;27:573–580. doi: 10.1093/nar/27.2.573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Darling AE, Miklos I, Ragan MA. Dynamics of genome rearrangement in bacterial populations. PLoS Genet. 2008;4:e1000128. doi: 10.1371/journal.pgen.1000128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schutzer SE, Schlater LR, Ronning CM, DeShazer D, Luft BJ, et al. Characterization of clinically-attenuated Burkholderia mallei by whole genome sequencing: candidate strain for exclusion from Select Agent lists. PLoS One. 2008;3:e2058. doi: 10.1371/journal.pone.0002058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Losada L, Ronning CM, DeShazer D, Woods D, Fedorova N, et al. Continuing Evolution of Burkholderia mallei Through Genome Reduction and Large-Scale Rearrangements. Genome Biol Evol 2010. 2010:102–116. doi: 10.1093/gbe/evq003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kurtz S, Ohlebusch E, Schleiermacher C, Stoye J, Giegerich R. Computation and visualization of degenerate repeats in complete genomes. Proc Int Conf Intell Syst Mol Biol. 2000;8:228–238. [PubMed] [Google Scholar]
- 28.Chen PE, Willner KM, Butani A, Dorsey S, George M, et al. Rapid Identification of Genetic Modifications in Bacillus anthracis Using Whole Genome Draft Sequences Generated by 454 Pyrosequencing. PLoS ONE. 2010;5:e12397. doi: 10.1371/journal.pone.0012397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dyrløv Bendtsen J, Nielsen H, von Heijne G, Brunak S. Improved Prediction of Signal Peptides: SignalP 3.0. Journal of Molecular Biology. 2004;340:783–795. doi: 10.1016/j.jmb.2004.05.028. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
List of primers used to validate SNPs and INDELs in Yersinia pestis KIM10 and KIM D27.
(XLS)