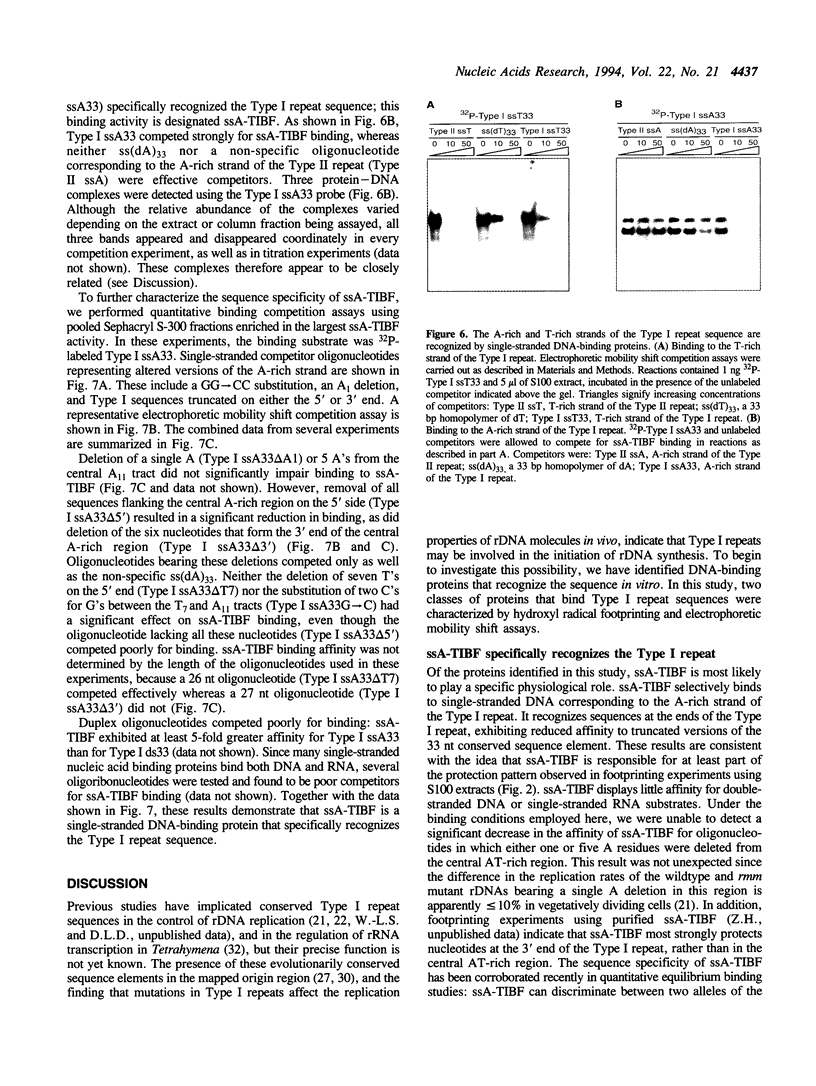
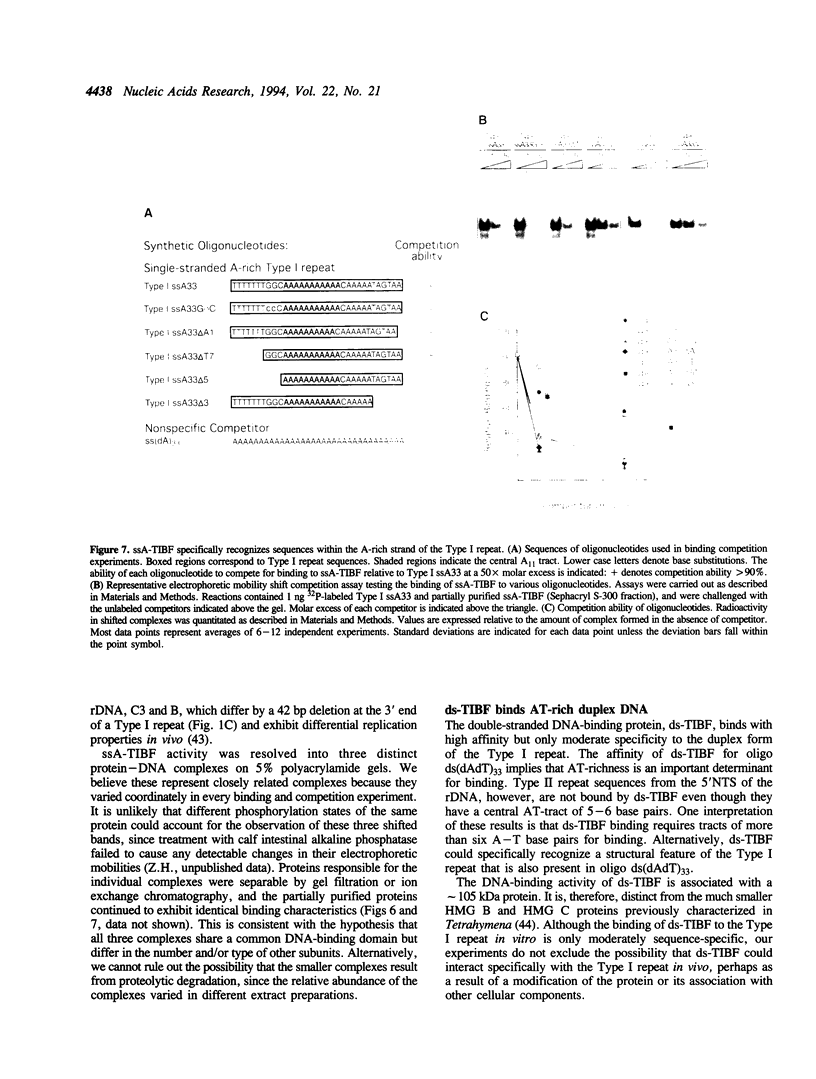
Abstract
An origin of DNA replication has been mapped within the 5' non-transcribed spacer region of the amplified macronuclear rRNA genes (rDNA) of Tetrahymena thermophila. Mutations in 33 nt conserved AT-rich Type I repeat sequences located in the origin region cause defects in the replication and/or maintenance of amplified rDNA in vivo. Fe(II)EDTA cleavage footprinting of restriction fragments containing the Type I repeat showed that most of the conserved nucleotides were protected by proteins in extracts of Tetrahymena cells. Two classes of proteins that bound the Type I repeat were identified and characterized using synthetic oligonucleotides in electrophoretic mobility shift assays. One of these, ds-TIBF, bound preferentially to duplex DNA and exhibited only moderate specificity for Type I repeat sequences. In contrast, a single-stranded DNA-binding protein, ssA-TIBF, specifically recognized the A-rich strand of the Type I repeat sequence. Deletion of the 5' or 3' borders of the conserved sequence significantly reduced binding of ssA-TIBF. The binding properties of ssA-TIBF, coupled with genetic evidence that Type I sequences function as cis-acting rDNA replication control elements in vivo, suggest a possible role for ssA-TIBF in rDNA replication in Tetrahymena.
Full text
PDF





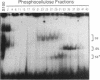
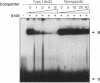
Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bell S. P., Kobayashi R., Stillman B. Yeast origin recognition complex functions in transcription silencing and DNA replication. Science. 1993 Dec 17;262(5141):1844–1849. doi: 10.1126/science.8266072. [DOI] [PubMed] [Google Scholar]
- Bell S. P., Stillman B. ATP-dependent recognition of eukaryotic origins of DNA replication by a multiprotein complex. Nature. 1992 May 14;357(6374):128–134. doi: 10.1038/357128a0. [DOI] [PubMed] [Google Scholar]
- Benbow R. M., Zhao J., Larson D. D. On the nature of origins of DNA replication in eukaryotes. Bioessays. 1992 Oct;14(10):661–670. doi: 10.1002/bies.950141004. [DOI] [PubMed] [Google Scholar]
- Bergemann A. D., Johnson E. M. The HeLa Pur factor binds single-stranded DNA at a specific element conserved in gene flanking regions and origins of DNA replication. Mol Cell Biol. 1992 Mar;12(3):1257–1265. doi: 10.1128/mcb.12.3.1257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borowiec J. A., Dean F. B., Bullock P. A., Hurwitz J. Binding and unwinding--how T antigen engages the SV40 origin of DNA replication. Cell. 1990 Jan 26;60(2):181–184. doi: 10.1016/0092-8674(90)90730-3. [DOI] [PubMed] [Google Scholar]
- Bradford M. M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976 May 7;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- Bramhill D., Kornberg A. A model for initiation at origins of DNA replication. Cell. 1988 Sep 23;54(7):915–918. doi: 10.1016/0092-8674(88)90102-x. [DOI] [PubMed] [Google Scholar]
- Caddle M. S., Dailey L., Heintz N. H. RIP60, a mammalian origin-binding protein, enhances DNA bending near the dihydrofolate reductase origin of replication. Mol Cell Biol. 1990 Dec;10(12):6236–6243. doi: 10.1128/mcb.10.12.6236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cech T. R., Brehm S. L. Replication of the extrachromosomal ribosomal RNA genes of Tetrahymena thermophilia. Nucleic Acids Res. 1981 Jul 24;9(14):3531–3543. doi: 10.1093/nar/9.14.3531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Challberg M. D., Kelly T. J. Animal virus DNA replication. Annu Rev Biochem. 1989;58:671–717. doi: 10.1146/annurev.bi.58.070189.003323. [DOI] [PubMed] [Google Scholar]
- Challoner P. B., Amin A. A., Pearlman R. E., Blackburn E. H. Conserved arrangements of repeated DNA sequences in nontranscribed spacers of ciliate ribosomal RNA genes: evidence for molecular coevolution. Nucleic Acids Res. 1985 Apr 11;13(7):2661–2680. doi: 10.1093/nar/13.7.2661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cockell M., Frutiger S., Hughes G. J., Gasser S. M. The yeast protein encoded by PUB1 binds T-rich single stranded DNA. Nucleic Acids Res. 1994 Jan 11;22(1):32–40. doi: 10.1093/nar/22.1.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DePamphili M. L. How transcription factors regulate origins of DNA replication in eukaryotic cells. Trends Cell Biol. 1993 May;3(5):161–167. doi: 10.1016/0962-8924(93)90137-p. [DOI] [PubMed] [Google Scholar]
- DePamphilis M. L. Eukaryotic DNA replication: anatomy of an origin. Annu Rev Biochem. 1993;62:29–63. doi: 10.1146/annurev.bi.62.070193.000333. [DOI] [PubMed] [Google Scholar]
- Diffley J. F., Cocker J. H. Protein-DNA interactions at a yeast replication origin. Nature. 1992 May 14;357(6374):169–172. doi: 10.1038/357169a0. [DOI] [PubMed] [Google Scholar]
- Diffley J. F. Global regulators of chromosome function in yeast. Antonie Van Leeuwenhoek. 1992 Aug;62(1-2):25–33. doi: 10.1007/BF00584460. [DOI] [PubMed] [Google Scholar]
- Dobbs D. L., Shaiu W. L., Benbow R. M. Modular sequence elements associated with origin regions in eukaryotic chromosomal DNA. Nucleic Acids Res. 1994 Jul 11;22(13):2479–2489. doi: 10.1093/nar/22.13.2479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Echols H. Nucleoprotein structures initiating DNA replication, transcription, and site-specific recombination. J Biol Chem. 1990 Sep 5;265(25):14697–14700. [PubMed] [Google Scholar]
- Estes H. G., Robinson B. S., Eisenberg S. At least three distinct proteins are necessary for the reconstitution of a specific multiprotein complex at a eukaryotic chromosomal origin of replication. Proc Natl Acad Sci U S A. 1992 Dec 1;89(23):11156–11160. doi: 10.1073/pnas.89.23.11156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fairman M. P., Stillman B. Cellular factors required for multiple stages of SV40 DNA replication in vitro. EMBO J. 1988 Apr;7(4):1211–1218. doi: 10.1002/j.1460-2075.1988.tb02933.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fangman W. L., Brewer B. J. Activation of replication origins within yeast chromosomes. Annu Rev Cell Biol. 1991;7:375–402. doi: 10.1146/annurev.cb.07.110191.002111. [DOI] [PubMed] [Google Scholar]
- Foss M., McNally F. J., Laurenson P., Rine J. Origin recognition complex (ORC) in transcriptional silencing and DNA replication in S. cerevisiae. Science. 1993 Dec 17;262(5141):1838–1844. doi: 10.1126/science.8266071. [DOI] [PubMed] [Google Scholar]
- Fried M., Crothers D. M. Equilibria and kinetics of lac repressor-operator interactions by polyacrylamide gel electrophoresis. Nucleic Acids Res. 1981 Dec 11;9(23):6505–6525. doi: 10.1093/nar/9.23.6505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galli I., Iguchi-Ariga S. M., Ariga H. The AT-rich tract of the SV40 ori core: negative synergism and specific recognition by single stranded and duplex DNA binding proteins. Nucleic Acids Res. 1992 Jul 11;20(13):3333–3339. doi: 10.1093/nar/20.13.3333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garner M. M., Revzin A. A gel electrophoresis method for quantifying the binding of proteins to specific DNA regions: application to components of the Escherichia coli lactose operon regulatory system. Nucleic Acids Res. 1981 Jul 10;9(13):3047–3060. doi: 10.1093/nar/9.13.3047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greider C. W., Blackburn E. H. Identification of a specific telomere terminal transferase activity in Tetrahymena extracts. Cell. 1985 Dec;43(2 Pt 1):405–413. doi: 10.1016/0092-8674(85)90170-9. [DOI] [PubMed] [Google Scholar]
- Heintz N. H., Dailey L., Held P., Heintz N. Eukaryotic replication origins as promoters of bidirectional DNA synthesis. Trends Genet. 1992 Nov;8(11):376–381. doi: 10.1016/0168-9525(92)90298-i. [DOI] [PubMed] [Google Scholar]
- Hofmann J. F., Gasser S. M. Identification and purification of a protein that binds the yeast ARS consensus sequence. Cell. 1991 Mar 8;64(5):951–960. doi: 10.1016/0092-8674(91)90319-t. [DOI] [PubMed] [Google Scholar]
- Kapler G. M., Blackburn E. H. A weak germ-line excision mutation blocks developmentally controlled amplification of the rDNA minichromosome of Tetrahymena thermophila. Genes Dev. 1994 Jan;8(1):84–95. doi: 10.1101/gad.8.1.84. [DOI] [PubMed] [Google Scholar]
- Kapler G. M. Developmentally regulated processing and replication of the Tetrahymena rDNA minichromosome. Curr Opin Genet Dev. 1993 Oct;3(5):730–735. doi: 10.1016/s0959-437x(05)80091-7. [DOI] [PubMed] [Google Scholar]
- Kapler G. M., Orias E., Blackburn E. H. Tetrahymena thermophila mutants defective in the developmentally programmed maturation and maintenance of the rDNA minichromosome. Genetics. 1994 Jun;137(2):455–466. doi: 10.1093/genetics/137.2.455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuno K., Kuno S., Matsushima K., Murakami S. Evidence for binding of at least two factors, including T-rich strand-binding factor(s) to the single-stranded ARS1 sequence in Saccharomyces cerevisiae. Mol Gen Genet. 1991 Nov;230(1-2):45–48. doi: 10.1007/BF00290649. [DOI] [PubMed] [Google Scholar]
- Larson D. D., Blackburn E. H., Yaeger P. C., Orias E. Control of rDNA replication in Tetrahymena involves a cis-acting upstream repeat of a promoter element. Cell. 1986 Oct 24;47(2):229–240. doi: 10.1016/0092-8674(86)90445-9. [DOI] [PubMed] [Google Scholar]
- Li J. J., Herskowitz I. Isolation of ORC6, a component of the yeast origin recognition complex by a one-hybrid system. Science. 1993 Dec 17;262(5141):1870–1874. doi: 10.1126/science.8266075. [DOI] [PubMed] [Google Scholar]
- Malkas L. H., Baril E. F. Sequence recognition protein for the 17-base-pair A + T-rich tract in the replication origin of simian virus 40 DNA. Proc Natl Acad Sci U S A. 1989 Jan;86(1):70–74. doi: 10.1073/pnas.86.1.70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marahrens Y., Stillman B. A yeast chromosomal origin of DNA replication defined by multiple functional elements. Science. 1992 Feb 14;255(5046):817–823. doi: 10.1126/science.1536007. [DOI] [PubMed] [Google Scholar]
- Micklem G., Rowley A., Harwood J., Nasmyth K., Diffley J. F. Yeast origin recognition complex is involved in DNA replication and transcriptional silencing. Nature. 1993 Nov 4;366(6450):87–89. doi: 10.1038/366087a0. [DOI] [PubMed] [Google Scholar]
- Miyahara K., Hashimoto N., Higashinakagawa T., Pearlman R. E. Common sequence elements are important for transcription and replication of the extrachromosomal rRNA-encoding genes of Tetrahymena. Gene. 1993 May 30;127(2):209–213. doi: 10.1016/0378-1119(93)90721-e. [DOI] [PubMed] [Google Scholar]
- Natale D. A., Umek R. M., Kowalski D. Ease of DNA unwinding is a conserved property of yeast replication origins. Nucleic Acids Res. 1993 Feb 11;21(3):555–560. doi: 10.1093/nar/21.3.555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newlon C. S., Theis J. F. The structure and function of yeast ARS elements. Curr Opin Genet Dev. 1993 Oct;3(5):752–758. doi: 10.1016/s0959-437x(05)80094-2. [DOI] [PubMed] [Google Scholar]
- Niles E. G., Sutiphong J., Haque S. Structure of the Tetrahymena pyriformis rRNA gene. Nucleotide sequence of the transcription initiation region. J Biol Chem. 1981 Dec 25;256(24):12849–12856. [PubMed] [Google Scholar]
- Palen T. E., Cech T. R. Chromatin structure at the replication origins and transcription-initiation regions of the ribosomal RNA genes of Tetrahymena. Cell. 1984 Apr;36(4):933–942. doi: 10.1016/0092-8674(84)90043-6. [DOI] [PubMed] [Google Scholar]
- Pan W. C., Orias E., Flacks M., Blackburn E. H. Allele-specific, selective amplification of a ribosomal RNA gene in Tetrahymena thermophila. Cell. 1982 Mar;28(3):595–604. doi: 10.1016/0092-8674(82)90214-8. [DOI] [PubMed] [Google Scholar]
- Schmidt A. M., Herterich S. U., Krauss G. A single-stranded DNA binding protein from S. cerevisiae specifically recognizes the T-rich strand of the core sequence of ARS elements and discriminates against mutant sequences. EMBO J. 1991 Apr;10(4):981–985. doi: 10.1002/j.1460-2075.1991.tb08032.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulman I. G., Cook R. G., Richman R., Allis C. D. Tetrahymena contain two distinct and unusual high mobility group (HMG)-like proteins. J Cell Biol. 1987 Jun;104(6):1485–1494. doi: 10.1083/jcb.104.6.1485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stillman B. Initiation of chromosomal DNA replication in eukaryotes. Lessons from lambda. J Biol Chem. 1994 Mar 11;269(10):7047–7050. [PubMed] [Google Scholar]
- Traut W., Fanning E. Sequence-specific interactions between a cellular DNA-binding protein and the simian virus 40 origin of DNA replication. Mol Cell Biol. 1988 Feb;8(2):903–911. doi: 10.1128/mcb.8.2.903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tullius T. D., Dombroski B. A. Hydroxyl radical "footprinting": high-resolution information about DNA-protein contacts and application to lambda repressor and Cro protein. Proc Natl Acad Sci U S A. 1986 Aug;83(15):5469–5473. doi: 10.1073/pnas.83.15.5469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vieira J., Messing J. Production of single-stranded plasmid DNA. Methods Enzymol. 1987;153:3–11. doi: 10.1016/0076-6879(87)53044-0. [DOI] [PubMed] [Google Scholar]
- Wobbe C. R., Weissbach L., Borowiec J. A., Dean F. B., Murakami Y., Bullock P., Hurwitz J. Replication of simian virus 40 origin-containing DNA in vitro with purified proteins. Proc Natl Acad Sci U S A. 1987 Apr;84(7):1834–1838. doi: 10.1073/pnas.84.7.1834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wold M. S., Kelly T. Purification and characterization of replication protein A, a cellular protein required for in vitro replication of simian virus 40 DNA. Proc Natl Acad Sci U S A. 1988 Apr;85(8):2523–2527. doi: 10.1073/pnas.85.8.2523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yaeger P. C., Orias E., Shaiu W. L., Larson D. D., Blackburn E. H. The replication advantage of a free linear rRNA gene is restored by somatic recombination in Tetrahymena thermophila. Mol Cell Biol. 1989 Feb;9(2):452–460. doi: 10.1128/mcb.9.2.452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu G. L., Blackburn E. H. Amplification of tandemly repeated origin control sequences confers a replication advantage on rDNA replicons in Tetrahymena thermophila. Mol Cell Biol. 1990 May;10(5):2070–2080. doi: 10.1128/mcb.10.5.2070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu G. L., Blackburn E. H. Transformation of Tetrahymena thermophila with a mutated circular ribosomal DNA plasmid vector. Proc Natl Acad Sci U S A. 1989 Nov;86(21):8487–8491. doi: 10.1073/pnas.86.21.8487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeidler R., Hobert O., Johannes L., Faulhammer H., Krauss G. Characterization of two novel single-stranded DNA-specific autonomously replicating sequence-binding proteins from Saccharomyces cerevisiae, one of which is adenylosuccinate synthetase. J Biol Chem. 1993 Sep 25;268(27):20191–20197. [PubMed] [Google Scholar]