Abstract
Infections caused by the fungus Aspergillus are a major cause of morbidity and mortality in immunocompromised populations. To identify genes required for virulence that could be used as targets for novel treatments, we mapped quantitative trait loci (QTL) affecting virulence in the progeny of a cross between two strains of A. nidulans (FGSC strains A4 and A91). We genotyped 61 progeny at 739 single nucleotide polymorphisms (SNP) spread throughout the genome, and constructed a linkage map that was largely consistent with the genomic sequence, with the exception of one potential inversion of ∼527 kb on Chromosome V. The estimated genome size was 3705 cM and the average intermarker spacing was 5.0 cM. The average ratio of physical distance to genetic distance was 8.1 kb/cM, which is similar to previous estimates, and variation in recombination rate was significantly positively correlated with GC content, a pattern seen in other taxa. To map QTL affecting virulence, we measured the ability of each progeny strain to kill model hosts, larvae of the wax moth Galleria mellonella. We detected three QTL affecting in vivo virulence that were distinct from QTL affecting in vitro growth, and mapped the virulence QTL to regions containing 7–24 genes, excluding genes with no sequence variation between the parental strains and genes with only synonymous SNPs. None of the genes in our QTL target regions have been previously associated with virulence in Aspergillus, and almost half of these genes are currently annotated as “hypothetical”. This study is the first to map QTL affecting the virulence of a fungal pathogen in an animal host, and our results illustrate the power of this approach to identify a short list of unknown genes for further investigation.
Introduction
Aspergillus is a genus of ubiquitous fungi that typically grow on decaying organic matter [1] but can also cause life-threatening infections in immunocompromised patients. For example, Aspergillus infections are responsible for approximately 9–17% of deaths in the first year following transplantation among liver, heart and lung transplant recipients [2], [3]. Even with treatment, systemic infections are associated with mortality rates between 30–90%, depending on the patient group [4], [5], underlining the need for new antifungal agents. Currently, most antimicrobial drugs block basic functions of pathogens rather than targeting specific virulence traits [6]; therefore, understanding the genes that contribute to virulence could facilitate the identification and targeting of pathogen-specific pathways.
The virulence of Aspergillus species is determined by multiple factors that include the ability to acquire iron, grow at mammalian body temperature, and adhere to the host respiratory epithelium, as well as the ability to produce mycotoxins, conidial pigments and melanin [4], [5], [7], [8], [9], [10], [11]. Given this trait complexity, and natural variation in virulence-related traits within Aspergillus species [12], [13], [14], [15], it should be possible to identify virulence-related genes using quantitative trait locus (QTL) mapping. QTL are genomic regions that contribute to variation in complex traits such as virulence and are identified through association between genetic markers and phenotype. Because QTL mapping uses molecular markers, this approach is unbiased and can identify genes and/or regulatory regions that are either unknown or not expected to contribute to a given phenotype. Despite an enormous number of QTL studies of animals and plants, there has been relatively little QTL work with fungi. The few studies that have mapped QTL affecting virulence-related traits in fungi have been extremely successful in terms of gene identification. For example, genes contributing to variation in the ability to grow at elevated temperatures among Saccharomyces cerevisiae isolates from human patients have been mapped [16], as has a gene affecting virulence traits in the pathogenic yeast, Cryptococcus neoformans [17].
Typically, the first step to mapping QTL is to cross two different wild-type strains. Most systemic Aspergillus infections are caused by A. fumigatus [1] but when we initiated this work a sexual cycle had not yet been observed in this species (it has been demonstrated since [18]). We therefore examined A. nidulans, a species that is also responsible for some infections [1], and readily undergoes sexual reproduction. Although responsible for fewer infections than A. fumigatus, A. nidulans is more resistant to certain antifungal drugs [19], [20], more resistant to human phagocytic defenses in vitro [21] and more virulent in patients with chronic granulomatous disease [22] than A. fumigatus. In addition to its clinical relevance, for decades, A. nidulans has been a model organism for the study of a variety of cellular processes [23].
The objective of the present study was to map QTL affecting virulence in A. nidulans. Because Aspergillus virulence is determined by multiple factors, we took an unbiased approach to detect variation in virulence regardless of the underlying causes. Specifically, we mapped QTL affecting the ability of A. nidulans to kill an animal host. Although infection of immunosuppressed rodents is the model that most closely approximates human disease, QTL mapping requires testing many progeny in replicate, and therefore we used a well-characterized insect host: larvae of the wax moth, Galleria mellonella. Although there are major differences between mammalian and insect immune systems, pathogenic fungi often require the same traits for virulence in mammalian and non-vertebrate hosts [24]. Furthermore, some signaling pathways involved in the innate immune response are conserved among insects and mammals, and there are also parallels between phagocytosis by insect hemocytes and by human neutrophils [24]. Correlation between virulence in insect and mammalian models has been observed in A. fumigatus [25], [26], [27], Candida albicans [28] and Yersinia pseudotuberculosis [29]. In addition to our in vivo measure of virulence, we also measured growth on solid medium so that we could distinguish between QTL with specific effects on virulence from QTL affecting growth both in vitro and in vivo. Because iron acquisition and tolerance of low iron conditions are thought to be important virulence factors [30], [31], we measured growth on both low iron and iron-supplemented media.
Mapping QTL also requires a linkage map that describes the distance between loci in terms of how frequently recombination occurs (genetic distance), rather than the number of base pairs (physical distance). Although there is already a linkage map for A. nidulans, the existing map is based largely on phenotypic markers [32] and so would not be suitable for mapping in progeny from two wild-type strains. We therefore created a single nucleotide polymorphism (SNP)-based linkage map by genotyping a panel of progeny at SNP throughout the genome.
Results
Linkage map
Of the 768 SNPs genotyped, 29 were excluded because they were not polymorphic, or had heterozygous or missing genotypes for many samples, including parent strains (Fungal Genetics Stock Centre A4 and A91), leaving a total of 739 markers. Several progeny had near-identical genotypes (>700 genotypes in common): 7 were identical to A4, 7 were identical to A91, while there were 12 groups of identical progeny genotypes ranging in size from 2–5 strains. There were 61 unique progeny genotypes (not including parental genotypes), and only these genotypes were included in linkage and QTL mapping.
Building linkage groups using all markers and requiring LOD scores of 6 or more and a maximum recombination frequency of 0.2 to establish linkage yielded 30 linkage groups and 3 unlinked loci (cntg-29-52692; cntg-43-189692; cntg-84-580184). Linkage groups and the three unlinked markers were combined to correspond to A. nidulans chromosomes on the basis of markers located on separate linkage groups but known to be located on the same contig, and/or contigs located on separate linkage groups known to be located on the same chromosome [33], [34]. In almost all cases where linkage groups were combined in this way, the recombination frequency between adjacent markers was 0.23 or lower, and the support for linkage was a LOD score of 3.7 or higher. However, the recombination frequency between markers cntg-55-265971 and cntg-55-175295 was 0.29, for which the LOD score was 2.22.
Combining linkage groups in this way and using MapDisto to order the markers, we obtained marker orders that were largely consistent with the genome sequence. Where there were discrepancies between the order calculated by MapDisto and that based on the genome sequence, we calculated map length based on the marker order from the genome sequence. In some cases, the genome sequence yielded a shorter map length than the MapDisto distance, and in 9 other cases the genome sequence yielded a map length within 20% of the MapDisto order (considering only the contentious markers and not the entire chromosome), and we adopted the genome order for further analyses. However, in one case the map length based on the genome sequence was substantially longer than that using the MapDisto order. On Chromosome V, the MapDisto order between markers cntg-157-107424 and cntg-98-505170 (Table S1) yielded a map length of 21.3 cM for this region, whereas the marker order from the genome sequence yielded a length of 58.5 cM. This discrepancy was due to an inversion of all of the markers from contigs 88 and 89, and no other markers. It is therefore not clear whether this is an error in the genome assembly in which the order of these two contigs was reversed. This region has been suggested to contain the centromere and has been difficult to map previously [35]. Four markers were on contigs not placed on chromosomes in the current genome sequence: we mapped cntg-185-3866 to Chromosome VII, cntg-202-4715 to Chromosome I, and cntg-221-4469 and cntg-243-2551 to Chromosome II.
We present the linkage map used in subsequent analyses in Table S1, rather than as a figure because of the large number and density of markers. Table 1 summarizes the results of linkage mapping. Chromosomes ranged in size from 331.3 cM (Chromosome IV) to 577.2 cM (Chromosome VII), with an estimated genome size of 3705 cM. The average intermarker spacing per chromosome ranged from 3.8 cM (Chromosome I) to 6.3 cM (Chromosome V).
Table 1. Summary of linkage mapping in cross between A. nidulans strains A4 and A91.
| Chromosome | No. of markers | Average marker spacing (cM) | Genetic length covered by markers (cM) | Genetic length including chromosome ends (cM) | Physical length covered by markers (kb) | Ratio of physical distance to genetic distance (kb/cM) |
| 1 | 101 | 3.8 | 383.2 | 390.9 | 3664 | 9.6 |
| 2 | 106 | 4.2 | 439.7 | 448.1 | 3986 | 9.1 |
| 3 | 82 | 5.3 | 425.5 | 436.0 | 3357 | 7.9 |
| 4 | 74 | 4.4 | 322.5 | 331.3 | 2732 | 8.5 |
| 5 | 77 | 6.3 | 480.3 | 492.9 | 3071 | 6.4 |
| 6 | 77 | 6.1 | 460.4 | 472.5 | 3313 | 7.2 |
| 7 | 110 | 5.2 | 566.8 | 577.2 | 4464 | 7.9 |
| 8 | 112 | 4.9 | 546.3 | 556.2 | 4825 | 8.8 |
| Total | 739 | 5.0 | 3624.7 | 3705.0 | 29412 | Average 8.1 |
Variation in recombination rate is correlated with GC content
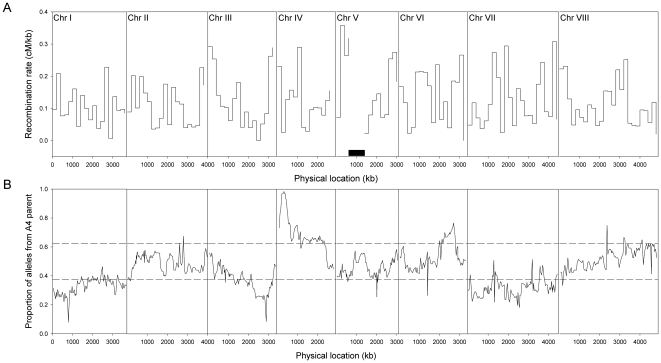
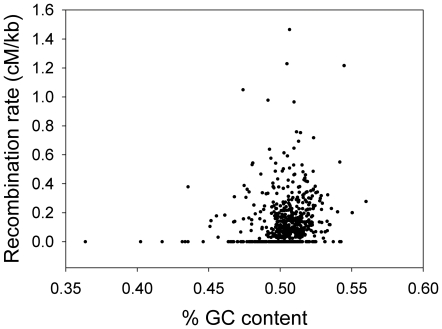
Our linkage map provided the genetic positions of the markers and the A. nidulans genomic sequence [34], [36] provided their physical positions (included in Table S1). The average ratio of physical distance to genetic distance per chromosome varied from 6.4 kb/cM (Chromosome V) to 9.6 kb/cM (Chromosome I), with further variation within chromosomes shown in Fig. 1A. Variation in recombination rate (cM/kb) between intervals was significantly positively correlated with GC content (Spearman rank r = 0.19; N = 647; P<0.0001; Fig. 2); this relationship remained significant when intervals with extreme GC content (<45% or >55%) were removed (Spearman rank r = 0.17; N = 637; P<0.0001). We analyzed the correlation between recombination rate and GC content over different scales by averaging these parameters within non-overlapping windows of various sizes. The relationship remained significant up to a window size of 450 kb (Spearman rank r = 0.33; N = 68; P<0.007), but was not significant at larger scales. We did not include intervals located within the Chromosome V inversion in these analyses in case the inversion affected recombination rates.
Figure 1. Recombination rate and segregation ratios throughout the genome.
(A) Variation in recombination rate averaged over non-overlapping 200 kb windows across Chromosomes I–VIII. Recombination rate is not displayed for the potential inversion on Chromosome V, which is denoted by the black rectangle. (B) Marker segregation ratios across Chromosomes I–VIII. The 95% confidence interval for a 1∶1 ratio is indicated by horizontal dashed lines at 0.375 and 0.625.
Figure 2. Correlation between the recombination rate (cM/kb) and GC content of 647 intermarker intervals.
The number of intermarker intervals is smaller than the number of markers because intervals within the potential inversion on Chromosome V are not included, and four markers are not placed in the current genome assembly.
Many markers show skewed segregation ratios
We observed skewed segregation ratios throughout much of the genome: Markers on most of Chromosomes I and VII and approximately half of Chromosome III were significantly skewed towards A91 alleles (not accounting for multiple tests), whereas markers on most of Chromosome IV and part of Chromosome VI were significantly skewed towards A4 alleles (Fig. 1B). Segregation data for all markers are available in Table S1.
Growth is a quantitative trait
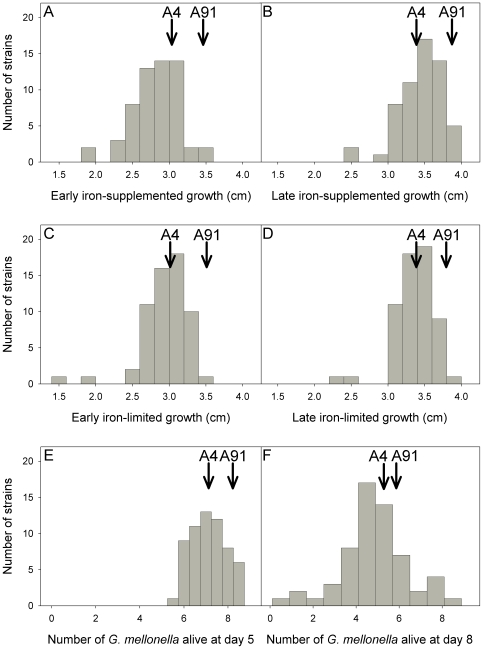
There was significant variation among strains in early growth (colony diameter at day 3) and late growth (the difference in colony diameter between days 3 and 6) on both iron-limited and iron-supplemented medium (P<0.0001 in all cases). Differences between the parental strains were also significant, with A91 showing more growth than A4 (P<0.0001 in all cases). The distributions of all traits are approximately normal, with only two strains showing very poor growth in vitro (Fig. 3). Furthermore, the progeny are distributed asymmetrically around the parental strains (Fig. 3), with few progeny showing higher growth than strain A91.
Figure 3. Phenotypic distributions of traits measured in 61 unique progeny genotypes.
(A) early iron-supplemented growth, (B) late iron-supplemented growth, (C) early iron-limited growth, (D) late iron-limited growth, and number of G.mellonella alive at (E) 5-days post injection and at (F) 8-days post injection.
Surprisingly, early growth was significantly higher on iron-limited (2.97±0.03 cm standard error) compared to iron-supplemented medium (2.85±0.03 cm; paired t92 = −5.04; P<0.0001), whereas late growth showed the expected pattern (iron supplemented: 3.48±0.03 cm; iron-limited: 3.38±0.03 cm; paired t92 = 4.02; P<0.0001). There was a significant correlation between growth on iron-limited and iron-supplemented medium (early growth: Pearson r92 = 0.73, P<0.0001; late growth: Pearson r92 = 0.62, P<0.0001).
In vivo virulence is a quantitative trait
None of the G. mellonella larvae from any of the negative controls died. There was little mortality in the first 3 days after injection with A. nidulans, therefore we limited analysis to G. mellonella survival on days 4 through 8. Variation among strains in the number of G. mellonella surviving was significant on days 7 and 8 post-injection (day 7: F94,270 = 1.77; P = 0.0002; day 8: F94,270 = 1.98; P<0.0001), but not on the other days. However, the difference between the two parental strains was significant on days 4 and 5 (day 4: F1,270 = 6.47; P = 0.01; day 5: F1,269 = 8.32; P = 0.004). Injection with strain A4 resulted in lower G. mellonella survival, i.e., A4 had higher virulence compared to A91. We therefore analyzed the results of survival on days 5 and 8 in further analyses; the distributions of these traits are shown in Fig. 3.
QTL mapping reveals loci affecting virulence are distinct from those affecting growth
We performed composite interval mapping with various window sizes (5 cM, 10 cM and 20 cM) and regression models (forward, backward, or forward and backward), which yielded similar results. However, increasing the number of background markers increased the statistical support for QTL, and therefore resulted in the identification of a greater number of significant QTL. Because the number of significant QTL was sensitive to the number of background markers, we report results from the forward and backward regression model (probability into, 0.05; probability out, 0.1), in which the number of background markers was determined automatically rather than by user input. Significance thresholds were calculated by permutation and did not make any assumptions about trait distribution.
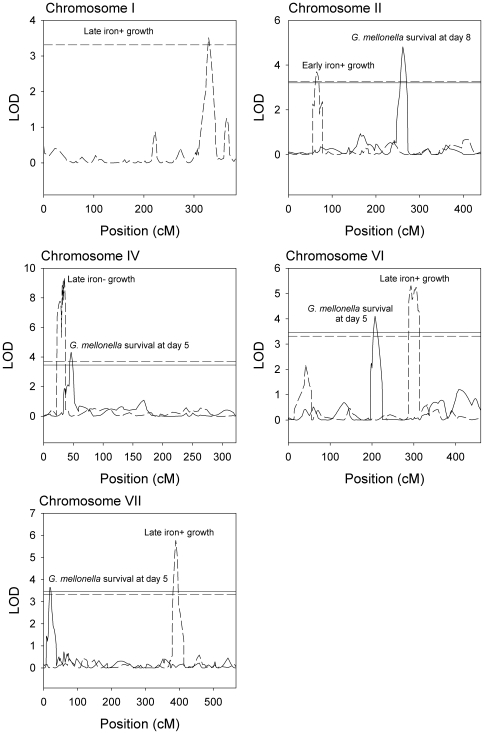
We identified QTL affecting all traits (Table 2). The proximal region of Chromosome IV was associated with variation in all growth traits as well as the number of G. mellonella larvae alive at 5 days post-injection (Fig. 4). However, the growth QTL appeared to be distinct from the virulence QTL; there was no overlap in the 2-LOD support interval (Table 2), which is a conservative estimate of the 95% confidence interval [37]. Markers in this region showed extremely skewed segregation ratios with an excess of A4 alleles (Fig. 1B). At the estimated location of the growth QTL, only 2–4 strains carry the A91 allele, making the support for this QTL somewhat suspect. However, the estimated effects of the QTL are consistent with effects of selection on this locus; the A4 allele increases growth, and so the growth QTL may have caused the skewed marker ratio.
Table 2. Summary of QTL positions and effects.
| Trait | Chromosome | Estimated position (cM) | Estimated effect sizea | LOD score at peak | % variance explained by QTL | 2-LOD support interval (cM) | Proximal marker flanking 2-LOD interval (position in kb) | Distal marker flanking 2-LOD interval (position in kb) |
| G.mellonella survival | ||||||||
| Day 5 | IV | 47 | −0.55 | 4.32 | 19 | 43–51 | cntg-126-39521 (420.7) | cntg-124-54688 (492.1) |
| VI | 207 | 0.40 | 4.10 | 18 | 201–221 | cntg-53-38593 (1416.8) | cntg-52-2303 (1534.7) | |
| VII | 19 | 0.35 | 3.65 | 17 | 14–32 | cntg-167-30076 (253.1) | cntg-165-13625 (433.7) | |
| Day 8 | II | 262 | 0.70 | 4.82 | 20 | 255–271 | cntg-65-149896 (2291.3) | cntg-65-55949 (2385.3) |
| Growth | ||||||||
| Early, iron-supplemented | II | 65 | 0.12 | 3.70 | 12 | 56–71 | cntg-135-275672 (354.4) | cntg-138-30822 (484.5) |
| IV | 35 | 0.45 | 5.45 | 26 | 34–37 | cntg-127-64936 (354.4) | cntg-127-8816 (410.5) | |
| VI | 296 | 0.13 | 4.32 | 14 | 286–301 | cntg-51-684909 (1966.4) | cntg-51-460598 (2190.8) | |
| Late, iron-supplemented | I | 329 | -0.11 | 3.51 | 11 | 319–341 | cntg-112-222346 (3030.3) | cntg-113-57473 (3219.3) |
| IV | 35 | 0.44 | 6.92 | 30 | 34–37 | cntg-127-64936 (354.4) | cntg-127-8816 (410.5) | |
| VI | 293 | 0.14 | 5.31 | 19 | 288–313 | cntg-51-684909 (1966.4) | cntg-51-372274 (2279.1) | |
| VII | 390 | -0.14 | 5.77 | 20 | 383–396 | cntg-36-61980 (3247.2) | cntg-38-171201 (3530.6) | |
| Early, iron-limited | IV | 35 | 0.59 | 13.72 | 44 | 34–37 | cntg-127-64936 (354.4) | cntg-127-8816 (410.5) |
| Late, iron-limited | IV | 35 | 0.46 | 9.39 | 38 | 34–37 | cntg-127-64936 (354.4) | cntg-127-8816 (410.5) |
A positive effect size indicates that the A4 allele increases the value of the trait compared to the A91 allele and vice versa. Effect sizes are in the units of the trait (i.e., number of G. mellonella larvae in the case of virulence and cm in the case of growth).
Figure 4. LOD plots from composite interval mapping of growth and virulence for Chromosomes I, II, IV, VI, and VII.
Horizontal lines show the genome-wide significance thresholds obtained by permutation. For LOD plots and thresholds, dashed lines denote growth traits and solid lines denote virulence traits. For clarity, we have omitted plots for early and late iron-supplemented growth and early iron-limited growth on Chromosome IV and plots for early iron-supplemented growth on Chromosome VI.
Elsewhere in the genome, QTL affecting virulence were distinct from QTL affecting growth. In addition to the Chromosome IV QTL, two other QTL affecting the number of G. mellonella larvae alive at 5 days post-injection were detected on Chromosomes VI and VII, and one QTL affecting the number of larvae alive at 8 days post-injection was detected on Chromosome II.
Virulence QTL regions contain no known candidate genes
The markers flanking the 2-LOD support intervals of the virulence QTL on Chromosomes II, VI, VII span regions of 93.9, 117.9 and 180.6 kb, respectively (Table 2). However, we were able to exclude large parts of these regions where parental strains A4 and A91 share identical sequence since regions without sequence variation cannot be responsible for the effects of QTL; a gene may be important for virulence, but if there is no sequence variation in that gene between the parental strains, it will not contribute to quantitative variation among progeny strains. Excluding genes that are not within 100 bp of a SNP or other sequence variation, or which harbor synonymous SNP only, the QTL on Chromosomes II, VI, VII contain 7, 16 and 24 genes, respectively [38], which are listed in Table 3. Almost half (20/47) of these genes are annotated as “conserved hypothetical protein”, and to our knowledge none have previously been associated with virulence.
Table 3. Genes within virulence QTL regions, excluding genes with no sequence variation or with synonymous SNP only. No sequence variation other than SNP (e.g., indels) was detected in these genes.
| Locus | Gene description | Number of non-synonymous SNP a | Other SNP b |
| Chromosome II | |||
| ANID_03982 | conserved hypothetical protein (calcineurin-like phosphoesterase)c | 2 conservative | 0 |
| ANID_03985 | MFS transporter | 2 radicald | 0 |
| ANID_03988 | conserved hypothetical protein | 2 conservative, 3 radical | 1 intronic |
| ANID_03995 | delta-aminolevulinic acid dehydratase | 0 | 1 intronic |
| ANID_03998 | conserved hypothetical protein | 2 conservative, 2 radical | 1 upstream, 3 downstream |
| ANID_04005 | conserved hypothetical protein | 4 conservative, 2 radical | 2 intronic, 3 downstream |
| ANID_04006 | conserved hypothetical protein (GMC oxidoreductase)c | 6 conservative | 1 upstream, 4 intronic |
| Chromosome VI | |||
| ANID_03176 | ATP-dependent rRNA helicase spb4 | 0 | 1 intronic |
| ANID_03178 | deacetylase complex subunit Sds3 | 1 conservative | 0 |
| ANID_03179 | conserved hypothetical protein | 1 radical | 0 |
| ANID_03184 | aldose 1-epimerase | 0 | 1 downstream |
| ANID_03186 | conserved hypothetical protein (XPG-I and XPG-N terminal domains)c | 1 conservative | 0 |
| ANID_03193 | conserved hypothetical protein | 0 | 1 UTR |
| ANID_03196 | glycosyl hydrolase family 88 protein | 0 | 1 downstream |
| ANID_03200 | glycoside hydrolase family 2 protein | 1 conservative, 1 radical | 1 upstream,1 intronic, 3 UTR, 1 downstream |
| ANID_03201 | beta-galactosidase | 3 conservative, 1 radical | 2 upstream, 2 downstream |
| ANID_03204 | MFS alpha-glucoside transporter | 1 radical | 0 |
| ANID_03205 | aldehyde dehydrogenase | 3 conservative | 1 upstream, 2 intronic,1 downstream |
| ANID_03209 | high affinity copper transporter | 1 conservative | 0 |
| ANID_10380 | dicer-like protein 2 | 1 radical | 0 |
| ANID_10383 | conserved hypothetical protein (glycosyl hydrolase family 2, sugar binding domain)c | 0 | 1 upstream |
| ANID_10384 | C6 transcription factor | 0 | 1 downstream |
| ANID_12377 | conserved hypothetical protein | 1 radical | 0 |
| Chromosome VII | |||
| ANID_08919 | cytochrome P450 monooxygenase | 1 conservative, 1 radical | 1 intronic |
| ANID_08920 | cytochrome b5 | 0 | 1 downstream |
| ANID_08921 | Dehydrogenase | 0 | 1 intronic, 1 downstream |
| ANID_08923 | conserved hypothetical protein (heterokaryon incompatibility protein)c | 0 | 1 intronic |
| ANID_08925 | conserved hypothetical protein | 1 radical | 1 upstream |
| ANID_08926 | conserved hypothetical protein | 0 | 2 intronic |
| ANID_08928 | ABC multidrug transporter | 1 radical | 0 |
| ANID_08931 | conserved hypothetical protein | 0 | 1 upstream |
| ANID_08932 | TIM-barrel enzyme family protein | 0 | 1 upstream, 2 downstream |
| ANID_08933 | conserved hypothetical protein | 0 | 1 intronic |
| ANID_08937 | 3-oxoacyl-(acyl-carrier-protein) reductase 2 | 1 conservative | 0 |
| ANID_08940 | conserved hypothetical protein | 1 radical | 0 |
| ANID_08941 | Na/K ATPase alpha 1 isoform | 0 | 1 intronic |
| ANID_08945 | TAM domain methyltransferase | 0 | 1 intronic |
| ANID_08951 | conserved hypothetical protein | 0 | ?e |
| ANID_08953 | alpha-glucosidase B | 1 conservative | 0 |
| ANID_08957 | multidrug resistance-associated protein | 2 conservative, 1 radical | 0 |
| ANID_08958 | conserved hypothetical protein | 1 radicalf | 0 |
| ANID_08962 | conserved hypothetical protein | ?e | 0 |
| ANID_08968 | isoflavone reductase | 1 conservative | 0 |
| ANID_08970 | conserved hypothetical protein | 1 conservative | 0 |
| ANID_08971 | integral membrane protein | 0 | 2 intronic |
| ANID_11152 | 1,3-beta-glucanosyltransferase | 1 conservative | 0 |
| ANID_12385 | hypothetical protein | 0 | 1 upstream, 2 intronic |
Non-synonymous SNP were classified as conservative if the BLOSUM80 score was 0 or higher for the substitution, or radical if the BLOSUM80 score was negative [74].
Other SNP include SNP in introns, untranslated regions (UTR) and within 100 bp upstream or downstream of the transcript. Synonymous SNP in coding regions are not included.
Protein domains for conserved hypothetical proteins were identified by a BLAST search of the Broad Institute database [38].
There are 3 SNP, but 2 affect the same codon.
No clear polymorphism, but ambiguity due to low sequence coverage for strain A91.
Premature stop codon.
Early and late growth share one QTL but are also affected by distinct QTL
In addition to the Chromosome IV QTL, we observed QTL affecting iron-supplemented growth on Chromosomes I, II, VI with one QTL affecting both early and late growth, one affecting early but not late growth, and two affecting late but not early growth (Table 2). Apart from the Chromosome IV QTL, no QTL affecting iron-limited growth were detected.
Variation in spore colour is associated with the wA gene
The parental strains used in this study differed in spore colour, and among the unique progeny genotypes, 32 had the wild-type green colour of strain A4, and 28 had the beige colour of strain A91 (spore colour was not recorded for one strain), a ratio that was not significantly different from 50∶50 (χ2 = 0.27, P = 0.61). Single marker analysis revealed almost perfect correspondence between the Chromosome II markers cntg-142-37489and cntg-143-2465 and spore colour, with only one unique genotype having the A4 green colour but carrying the A91 allele. At flanking markers cntg-142-11335 and cntg-143-25286, there were two progeny for which spore colour did not match genotype, indicating that the mutation affecting spore colour is located between these two markers that are located 57.8 kb apart. There are 17 genes within this region including the wA gene, in which mutations are known to cause white conidia [39], [40]. The flanking markers cntg-142-11335 and cntg-143-25286 are at 165 cM and 168 cM, respectively, indicating that the colour locus does not overlap with the virulence and growth QTL detected on Chromosome II (Table 2).
Discussion
QTL mapping has potential to identify novel virulence genes in Aspergillus
Our study is the first to map QTL affecting the virulence of a fungal pathogen in an animal host. We identified separate QTL affecting different measures of virulence: the number of G. mellonella larvae alive at 5 days and 8 days post-inoculation, which may reflect virulence factors that act at different stages of infection. Importantly, we identified QTL that affected virulence but not growth, indicating that the underlying genes are true virulence factors as opposed to genes affecting general vigour.
The three virulence QTL regions contain 7–24 genes, many of which are hypothetical genes that were identified using computational methods but have received no study. A. fumigatus orthologues have been identified for many of these genes, suggesting that A. nidulans may be a useful model for identifying A. fumigatus virulence genes.
To our knowledge, none of the genes in our QTL target regions have been previously associated with virulence in Aspergillus. Nevertheless, some are stronger candidates than others. β-glucans of the cell wall are involved in triggering innate immune responses against A. fumigatus [41], [42] and deletion of a β-1,3-glucanosyltransferase, GEL2, reduced virulence of A. fumigatus in a murine model [43]. However, GEL2 is not orthologous to the β-1,3-glucanosyltransferase in the Chromosome VII QTL region. Deletion of CaCWH41 or CaROT2, encoding α-glucosidase I, and α-glucosidase II catalytic subunit, respectively, attenuated the virulence of the pathogenic yeast, Candida albicans in a murine model [44]. However, deletion of α-glucosidase I did not affect the virulence of A. fumigatus [45] and none of these genes are orthologous to the α-glucosidase B within the Chromosome VII QTL region.
Four of the candidate genes in our QTL regions are members of the ATP-binding cassette (ABC) and major facilitator superfamily (MFS) families of transporters. The high representation of these families within our target regions is not surprising given that these genes are very common in fungal genomes; there are 45 ABC transporters and 356 MFS transporters in the A. nidulans genome [46]. ABC and MFS transporters are thought to contribute to virulence by facilitating the export of mycotoxins from fungal cells, and by removing host defence compounds [46]. Although a number of genes from these families have been implicated in plant pathogenesis, only one ABC transporter has been shown to contribute to fungal virulence in a mammalian host [47]. This C. albicans gene, MLT1, is not orthologous to either of the ABC transporters in the Chromosome VII QTL region. The lack of obvious candidates and the large proportion of hypothetical genes within our QTL regions illustrate the power of QTL mapping to identify a short list of unknown genes for further investigation.
Our study demonstrates that the effects of some virulence QTL are sufficiently large, and that quantitative variation in virulence can be measured with sufficient precision, that it is possible to map QTL affecting in vivo virulence in fungal pathogens. Although we phenotyped and genotyped 94 progeny, genotyping revealed only 61 unique genotypes; the presence of clones among progeny has been previously reported in Cryptococcus neoformans [48], [49]. Despite this substantial reduction in sample size, we were still able to map QTL to relatively small regions. A larger sample size would allow still greater resolution, i.e., fewer genes per QTL.
We found QTL affecting virulence even though the difference in this trait between parental strains was very modest. This is not unexpected, since one strain may harbour some alleles that increase virulence, and others that decrease virulence compared with the other strain. We selected these parental strains because they differed in spore colour, which was necessary to identify an outcrossed cleistothecium. Had we used parental strains with a greater difference in virulence, we expect that we would have identified more and/or larger QTL. A. fumigatus is heterothallic and shows quantitative variation in virulence related traits [12], [13], [14], [15], including virulence in G. mellonella [50], and thus it will be possible to cross strains differing in virulence in this species.
This is the first study to use infection of G. mellonella with A. nidulans as a model of Aspergillus virulence. A previous study injected a much lower inoculum of A. nidulans (3000 conidia compared with over 10000 in the present study) into G. mellonella but observed no mortality, whereas A. flavus was found to be virulent [51]. Although there are parallels between insect and mammalian immune responses [24] and correlation between virulence in insect and mammalian models has been observed [25], [26], [27], there are obviously limitations in extrapolating results from our G. mellonella model to human disease due to the adaptive immune response of vertebrates, among other factors. For instance, while some conidial colour mutants of A. fumigatus show reduced virulence in mammalian models, colour mutants were found to have increased virulence in G. mellonella [52]. Ultimately, virulence genes that we identify through subsequent work must be tested in immunocompromised rodents.
We observed significant variation in virulence between the parental strains and among the progeny, but this variation was subtle. Accurate measurement of small differences in virulence required taking many steps to reduce experimental error: suspensions were based on the number of viable conidia and were double checked for accuracy, the weight and age of larvae were kept within a narrow range, and three replicates were performed. Quantitative genetic variation in radial growth and the production of cleistothecia has previously been documented in a cross between two wild-type isolates of A. nidulans [53], but this early work did not examine traits related to virulence.
QTL affecting growth
Several studies have suggested that A. fumigatus is the most common pathogen among Aspergillus species because of its rapid growth rate. However, the identification of distinct QTL affecting radial growth and virulence results suggest that loci contributing to variation in growth do not contribute to variation in virulence. Although different QTL would likely be found in A. fumigatus or different crosses of A. nidulans, our results do not support the hypothesis that growth rate is an important virulence factor.
Distinct QTL affecting early and late growth were detected. Surprisingly, we detected QTL affecting iron-supplemented growth and not iron-limited growth, but not vice-versa. However, given the small number of QTL identified, it is not clear whether this reflects a real difference in genetic architecture between iron-supplemented and iron-limited growth. Although we did not expect early radial growth to be lower on iron-supplemented than iron-limited medium, similar results have been observed in A. fumigatus [54], perhaps due to low-level iron toxicity to germinating conidia [55]. Alternatively, iron-limitation may have led to longer but thinner hyphae, such that radial growth but not biomass was increased.
A. nidulans has low ratio of physical distance to genetic distance, which is correlated with GC content over large scales
Despite the widespread use of A. nidulans as a genetic model, this is the first SNP-based linkage map for this species. We estimated the size of the genome to be 3705 cM. Our estimate of the average ratio of physical distance to genetic distance across the genome is 8.1 kb/cM, similar to previous estimates for A. nidulans [56], and slightly lower than for a number of other fungi [49]. We observed a positive correlation between recombination rate and GC content. While this pattern is widespread in a variety of taxa, we know of no related studies in fungal species other than in Saccharomyces cerevisiae [57]. We observed this relationship at a range of scales from the intervals between markers up to windows of 450 kb. In contrast, in S. cerevisiae, Marsolier-Kergoat and Yeramian found that the strength of this relationship decreased substantially between windows of 5 kb and 100 kb [58].
Many markers showed skewed segregation ratios in our cross, with an excess of A4 alleles in some regions and an excess of A91 alleles in others. A number of other fungal linkage mapping studies have also found large proportions of markers with skewed ratios [48]. We suspect that at least some of the skew was due to selection, whereby one allele conferred more rapid growth and/or germination of ascospores, making progeny carrying this allele more likely to be isolated. In particular, there is a substantial skew towards A4 alleles at the proximal end of Chromosome IV, and in this region there is a QTL of which the A4 allele enhances growth. Although the A91 parental strain showed a higher growth rate than A4, other loci may have compensated for the Chromosome IV locus. Furthermore, the distribution of growth among the progeny (Fig. 3) suggests there was epistasis among loci affecting growth. If all the QTL affecting growth acted in an additive manner, we would expect the distribution of progeny values to be symmetrical around the parents.
Mapping further traits
Our genotyped panel of progeny represents a resource for the entire Aspergillus research community, since it is now possible to map QTL on any trait that varies among the strains, without the need for further genotyping. These progeny will be analogous to “recombinant inbred lines” which in other taxa have been recognized as powerful tools for QTL mapping, particularly for the study of genotype by environment interactions [37], [59].
Materials and Methods
Mapping population
A. nidulans strains A4 and A91 were obtained from the Fungal Genetics Stock Center [60]. A4 is the wild-type strain that has been sequenced [23], while A91 is a spore-colour mutant obtained by ultraviolet irradiation of a different wild-type environmental isolate [61]. It was necessary to use a spore-colour mutant to identify a cleistothecium produced by crossing the two strains; A. nidulans is homothallic, and genetically distinct strains are much more likely to self than to cross fertilize [62], [63]. Crosses were conducted between A4 and A91 on MYPD agar (3 g malt extract, 3 g yeast extract, 5 g peptone, 10 g glucose and 18 g agar in 1L) at 30°C and cleistothecia were screened for out-crossing by plating ascospores from a single cleistothecium on Neiland's agar plates (described below) and looking for colonies with different pigmentation. Over 100 cleistothecia were screened, but only one was found to be out-crossed, which provided the ascospores for the mapping population described below.
Marker discovery and genotyping
Mycelia grown on half-strength liquid MYPD medium for 24 to 30 hours at 37°C were harvested by filtration and DNA was extracted using an Epicentre MasterPure™ Yeast DNA Purification Kit [64]. A91 DNA was sent to the Genome Sciences Centre at the British Columbia Cancer Agency (Vancouver, Canada) for library construction and paired-end tag sequencing on the Illumina Genome Analyzer.
A91 sequences were aligned against the reference A. nidulans genome sequence [23], [65], obtained from the Broad Institute [38] using MAQ v 0.7.1 [66] with default parameters, except for the maximum outer distance for a correct pair (-a, set to 1500) and the maximum number of mismatched that can always be found (-n, set to 3). Pairs with identical outer coordinates were removed using rmdup, following the suggestion in the MAQ manual for accurate SNP calling. The published genome sequence is based on strain A4, and so differences between the A91 and reference sequences allowed us to identify SNP markers for our population. We selected 768 SNPs spread across the genome for which we had at least 20 times coverage for A91, and at least 90% of the A91 sequence reads supported the presence of a SNP (Table S2). DNA samples from 94 progeny strains, as well as A4 and A91 were sent to the Centre for Applied Genomics at The Hospital for Sick Children (Toronto, Canada) for genotyping of SNPs using the Illumina GoldenGate® Assay.
Linkage map construction
The construction of a genetic map, which describes the distance between loci in terms of how frequently recombination occurs, was performed using MapDisto version 1.7.0 [67] considering our population to be doubled haploid. We used the Haldane mapping function to translate recombination frequency into map distance (centimorgans, cM), since there is no evidence of crossover interference in A. nidulans [68]. We initially required LOD (logarithm of odds) scores of 6 or more and a maximum recombination frequency of 0.2 to establish linkage, but subsequently relaxed the criteria to combine linkage groups known to be on the same chromosome (described in Results section). To calculate the total length of each chromosome, we added two times the average intermarker distance for that chromosome to account for chromosome ends [69].
Growth media
Strains were grown on Neiland's agar at 37°C (18 g of agar, 20 g of sucrose, 1 g of K2SO4, 3 g of (NH4)2SO4, 1 g of citric acid, 3 g of K2HPO4, 3 g of K2HPO4, 810 mg of MgSO4·7H2O, 2 mg of thiamine hydro-chloride, 962 µg of MnSO4, 20 µg of CuSO4, 5.5 mg of ZnSO4, per liter of solution with pH adjusted to 6.8–7.0) [70]. For measurement of iron-limited growth, traces of iron were removed from glassware by overnight treatment with 5% HCl and thorough rinsing with deionised water prior to media preparation. For measurement of iron-supplemented growth, 1 mg of FeCl3 per litre (3.7 µM) was added to the medium.
Conidia harvesting and preparation
Conidia from 7-day cultures on iron-limited medium were harvested with 0.05% (v/v) Tween 20 (Sigma Chemica Co., St. Louis, USA) in phosphate buffered saline (pH. 7.4) (PBST), and filtered through Miracloth (Calbiochem) to remove hyphae. Harvested conidia were centrifuged, resuspended in PBST, centrifuged again and resuspended in fresh PBST. The concentration of conidia was determined using a haemocytometer (Hausser Scientific, Horsham, PA).
Radial growth measurements
To obtain isolated colonies, dilute conidial suspensions were inoculated onto either iron-limited or iron-supplemented Neiland's agar. After approximately 24 hours, mats from single germinated conidia were isolated and transferred to the centre of 10 cm Petri dishes, and at least two germinated conidia of each strain were transferred to two different Petri dishes. Plates with isolated colonies were incubated at 37°C and colony diameter was measured every 24 hours from 3 to 6 days after transfer. Three experiments with at least two replicates per experiment were performed for recombinant strains, and conidia were grown and harvested independently for each experiment. Because the parental strains (A4 and A91) were included with each group of strains measured, there are 34 and 21 replicates of each of the parental strains on iron-limited and iron-supplemented growth, respectively. Early growth was defined as colony diameter at day 3 and late growth was defined as the difference in colony diameter between days 3 and 6.
We initially attempted to measure growth in terms of mass in liquid medium, but switched to measuring radial growth because of large variation between replicates. These initial measures of mass showed the same pattern as subsequent measures of radial growth (i.e., A91>A4; data not shown).
Virulence in G. mellonella larvae
G. mellonella larvae were reared on baby mixed cereal (1200 ml) (H.J. Heinz Company, Canada) supplemented with glycerol (119 ml), water (98 ml), sucrose (100 ml) and multi-vitamins (Enfamil, Poly-vi-sol) (1 ml) at 28–30°C with 50–60% relative humidity and a 12L:12D light cycle as described previously [71]. G. mellonella larvae 40 to 50 days of age (in their final instar stage) weighing 0.25–0.30 g were selected for injection.
The goal of this study was to examine variation in virulence among strains, and therefore we needed an inoculum that was not so high that all of the larvae died rapidly, but not so low that none of the larvae died. Preliminary work established that injection of 5 µl of 2080 colony forming units (CFU)/µl yielded intermediate mortality rates, and therefore this was used as the inoculum. To determine the concentration of CFU, conidial suspensions were diluted and inoculated on Neiland's solid agar medium plates (5 plates/strain), which were counted after 36 hours. Suspensions containing 2080 CFU/µl were prepared and used for injection. The concentration of viable conidia was re-confirmed by plate counts of the suspensions used for injection.
Conidial suspensions (5 µl of 2080 CFU/µl) were injected into the hemocoel via the last left proleg using a 25 µl Hamilton syringe (part # 7636 – 01 702RN, Hamilton) with an inner barrel diameter of 0.72 mm and 33 gauge removable needle (part # 7762-06, Hamilton). For each strain and each replicate, one strain was injected into 10 larvae. In addition to all of the strains injected on a given day, the two parental strains, A4 and A91, and two negative controls were included: 10 larvae received 5 µl of PBST, and another 10 larvae received no injection. Between injections, the syringe was washed once with 70% ethanol and twice with PBST to avoid cross contamination. Injected larvae were placed in Petri dishes containing pine wood chips, and dishes were left at 37°C in the dark. Mortality was monitored once a day for 8 days. We performed three replicates for each strain and for each replicate, conidia were grown, harvested, and counted independently, and all replicates were injected on different days. Because the parental strains were always included with the strains injected on a given day, there are 32 replicates of A4 and A91.
Conidia of A. nidulans killed by heating at 100°C for 1 hour were also injected into wax moth larvae to see if non-viable conidia contributed to virulence. Lack of viability of heat killed conidia was confirmed by plating on solid medium. This negative control was repeated three times.
QTL mapping
We mapped QTL by testing the association between phenotype (growth or virulence) and the genotypes of SNPs located throughout the genome. We used composite interval mapping, which scans the genome for QTL while using additional markers as cofactors to account for effects of QTL outside the focal interval, increasing the power to detect QTL and the precision with which positions are estimated [37]. Composite interval mapping was performed using Windows QTL Cartographer Version 2.5 [72], with a walk speed of 1 cM. Genome-wide significance thresholds were determined empirically by permuting the marker data [73], using 1000 permutations. Because the significance thresholds are calculated from an empirical distribution of the test statistic under the null hypothesis that there is no QTL, the analysis does not make any assumptions about the distribution of the phenotypic traits (i.e., traits do not have to be normally distributed).
Supporting Information
Aspergillus nidulans linkage map, including genetic and physical positions and segregation data for each marker.
(XLS)
Sequence flanking SNP markers.
(XLS)
Acknowledgments
We thank the staff of the Michael Smith Genome Sciences Centre, particularly Yongjun Zhao, and the Centre for Applied Genomics, particularly Christian Marshall, for sequencing strain A91 and SNP genotyping, respectively. Sunpreet Bains, Saira Chaudhry, Amar Dhillon, Martha Essak, Balveer Mandar, Joshua Ogden and Anthony Tang assisted with the phenotyping of the progeny strains. Pilar Cepada provided guidance in rearing G. mellonella, and Fabrice Gravelat provided useful advice regarding the use of G. mellonella as a model of aspergillosis.
Footnotes
Competing Interests: The authors have declared that no competing interests exist.
Funding: This study was funded by a BC Transplant Research Foundation Venture Grant to JKC and MMM, Natural Sciences and Engineering Research Council (NSERC) operating grants to JKC, MMM and NC and a Simon Fraser University President's Research Grant to JKC. NC is a Michael Smith Foundation for Health Research (MSFHR) Scholar and a Canadian Institutes of Health Research (CIHR) New Investigator. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Perfect JR, Cox GM, Lee JY, Kauffman CA, de Repentigny L, et al. The impact of culture isolation of Aspergillus species: A hospital-based survey of aspergillosis. Clin Infect Dis. 2001;33:1824–1833. doi: 10.1086/323900. [DOI] [PubMed] [Google Scholar]
- 2.Singh N, Pursell KJ. Combination therapeutic approaches for the management of invasive aspergillosis in organ transplant recipients. Mycoses. 2008;51:99–108. doi: 10.1111/j.1439-0507.2007.01479.x. [DOI] [PubMed] [Google Scholar]
- 3.Munoz P, Singh N, Bouza E. Treatment of solid organ transplant patients with invasive fungal infections: should a combination of antifungal drugs be used? Curr Opin Infect Dis. 2006;19:365–370. doi: 10.1097/01.qco.0000235164.70678.97. [DOI] [PubMed] [Google Scholar]
- 4.Brakhage AA. Systemic fungal infections caused by Aspergillus species: Epidemiology, infection process and virulence determinants. Curr Drug Targets. 2005;6:875–886. doi: 10.2174/138945005774912717. [DOI] [PubMed] [Google Scholar]
- 5.Latge JP. Aspergillus fumigatus and aspergillosis. Clin Microbiol Rev. 1999;12:310–350. doi: 10.1128/cmr.12.2.310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Perfect JR, Casadevall A. Fungal Molecular Pathogenesis: What Can It Do and Why Do We Need It? In: Heitman J, Filler SG, Edwards JE, Mitchell AP, editors. Molecular Principles of Fungal Pathogenesis. Washington, DC: ASM Press; 2006. pp. 3–12. [Google Scholar]
- 7.Hohl TM, Feldmesser M. Aspergillus fumigatus: principles of pathogenesis and host defense. Eukaryot Cell. 2007;6:1953–1963. doi: 10.1128/EC.00274-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Keller NP. Aspergillus nidulans: a Model for Elucidation of Aspergillus fumigatus Secondary Metabolism. In: Heitman J, Filler SG, Edwards JE, Mitchell AP, editors. Molecular Principles of Fungal Pathogenesis. Washington, DC: ASM Press; 2006. pp. 235–244. [Google Scholar]
- 9.Latge JP. The pathobiology of Aspergillus fumigatus. Trends Microbiol. 2001;9:382–389. doi: 10.1016/s0966-842x(01)02104-7. [DOI] [PubMed] [Google Scholar]
- 10.Wasylnka JA, Moore MM. Adhesion of Aspergillus species to extracellular matrix proteins: Evidence for involvement of negatively charged carbohydrates on the conidial surface. Infect Immun. 2000;68:3377–3384. doi: 10.1128/iai.68.6.3377-3384.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wasylnka JA, Simmer MI, Moore MM. Differences in sialic acid density in pathogenic and non-pathogenic Aspergillus species. Microbiology. 2001;147:869–877. doi: 10.1099/00221287-147-4-869. [DOI] [PubMed] [Google Scholar]
- 12.Blanco JL, Hontecillas R, Bouza E, Blanco I, Pelaez T, et al. Correlation between the elastase activity index and invasiveness of clinical isolates of Aspergillus fumigatus. J Clin Microbiol. 2002;40:1811–1813. doi: 10.1128/JCM.40.5.1811-1813.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kothary MH, Chase T, Macmillan JD. Correlation of elastase production by some strains of Aspergillus fumigatus with ability to cause pulmonary invasive aspergillosis in mice. Infect Immun. 1984;43:320–325. doi: 10.1128/iai.43.1.320-325.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pastor A, Gafa V, Boutonnat J, Grillot R, Ambroise-Thomas P, et al. Intracellular oxidative response of human monocytes and granulocytes to different strains of Aspergillus fumigatus. Mycoses. 2006;49:73–79. doi: 10.1111/j.1439-0507.2006.01188.x. [DOI] [PubMed] [Google Scholar]
- 15.Reeves EP, Messina CGM, Doyle S, Kavanagh K. Correlation between gliotoxin production and virulence of Aspergillus fumigatus in Galleria mellonella. Mycopathologia. 2004;158:73–79. doi: 10.1023/b:myco.0000038434.55764.16. [DOI] [PubMed] [Google Scholar]
- 16.Steinmetz LM, Sinha H, Richards DR, Spiegelman JI, Oefner PJ, et al. Dissecting the architecture of a quantitative trait locus in yeast. Nature. 2002;416:326–330. doi: 10.1038/416326a. [DOI] [PubMed] [Google Scholar]
- 17.Lin XR, Huang JC, Mitchell TG, Heitman J. Virulence attributes and hyphal growth of C. neoformans are quantitative traits and the MAT alpha allele enhances filamentation. PLoS Genet. 2006;2:1801–1814. doi: 10.1371/journal.pgen.0020187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.O'Gorman CM, Fuller HT, Dyer PS. Discovery of a sexual cycle in the opportunistic fungal pathogen Aspergillus fumigatus. Nature. 2009;457:471–474. doi: 10.1038/nature07528. [DOI] [PubMed] [Google Scholar]
- 19.Araujo R, Pina-Vaz C, Rodrigues AG. Susceptibility of environmental versus clinical strains of pathogenic Aspergillus. Int J Antimicrob Ag. 2007;29:108–111. doi: 10.1016/j.ijantimicag.2006.09.019. [DOI] [PubMed] [Google Scholar]
- 20.Kontoyiannis DP, Lewis RE, May GS, Osherov N, Rinaldi MG. Aspergillus nidulans is frequently resistant to amphotericin B. Mycoses. 2002;45:406–407. doi: 10.1046/j.1439-0507.2002.00797.x. [DOI] [PubMed] [Google Scholar]
- 21.Akpogheneta O, Gil-Lamaignere C, Maloukou A, Roilides E. Antifungal activity of human polymorphonuclear and mononuclear phagocytes against non-fumigatus Aspergillus species. Mycoses. 2003;46:77–83. doi: 10.1046/j.1439-0507.2003.00852.x. [DOI] [PubMed] [Google Scholar]
- 22.Segal BH, DeCarlo ES, Kwon-Chung KJ, Malech HL, Gallin JI, et al. Aspergillus nidulans infection in chronic granulomatous disease. Medicine. 1998;77:345–354. doi: 10.1097/00005792-199809000-00004. [DOI] [PubMed] [Google Scholar]
- 23.Galagan JE, Calvo SE, Cuomo C, Ma LJ, Wortman JR, et al. Sequencing of Aspergillus nidulans and comparative analysis with A. fumigatus and A. oryzae. Nature. 2005;438:1105–1115. doi: 10.1038/nature04341. [DOI] [PubMed] [Google Scholar]
- 24.Mylonakis E, Casadevall A, Ausubel FM. Exploiting amoeboid and non-vertebrate animal model systems to study the virulence of human pathogenic fungi. PLoS Pathog. 2007;3:859–865. doi: 10.1371/journal.ppat.0030101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chamilos G, Bignell EM, Schrettl M, Lewis RE, Leventakos K, et al. Exploring the concordance of Aspergillus fumigatus pathogenicity in mice and Toll-deficient flies. Med Mycol. 2010;48:506–510. doi: 10.3109/13693780903225813. [DOI] [PubMed] [Google Scholar]
- 26.Gravelat FN, Ejzykowicz DE, Chiang LY, Chabot JC, Urb M, et al. Aspergillus fumigatus MedA governs adherence, host cell interactions and virulence. Cell Microbiol. 2010;12:473–488. doi: 10.1111/j.1462-5822.2009.01408.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Steinbach WJ, Cramer RA, Perfect BZ, Asfaw YG, Sauer TC, et al. Calcineurin controls growth, morphology, and pathogenicity in Aspergillus fumigatus. Eukaryot Cell. 2006;5:1091–1103. doi: 10.1128/EC.00139-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Brennan M, Thomas DY, Whiteway M, Kavanagh K. Correlation between virulence of Candida albicans mutants in mice and Galleria mellonella larvae. FEMS Immunol Med Mic. 2002;34:153–157. doi: 10.1111/j.1574-695X.2002.tb00617.x. [DOI] [PubMed] [Google Scholar]
- 29.Champion OL, Cooper IAM, James SL, Ford D, Karlyshev A, et al. Galleria mellonella as an alternative infection model for Yersinia pseudotuberculosis. Microbiology. 2009;155:1516–1522. doi: 10.1099/mic.0.026823-0. [DOI] [PubMed] [Google Scholar]
- 30.Hissen AHT, Wan ANC, Warwas ML, Pinto LJ, Moore MM. The Aspergillus fumigatus siderophore biosynthetic gene sidA, encoding L-ornithine N-5-oxygenase, is required for virulence. Infect Immun. 2005;73:5493–5503. doi: 10.1128/IAI.73.9.5493-5503.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schrettl M, Beckmann N, Varga J, Heinekamp T, Jacobsen ID, et al. HapX-Mediated Adaption to Iron Starvation Is Crucial for Virulence of Aspergillus fumigatus. PLoS Pathog. 2010;6:e1001124. doi: 10.1371/journal.ppat.1001124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Clutterbuck AJ. The validity of the Aspergillus nidulans linkage map. Fungal Genet Biol. 1997;21:267–277. doi: 10.1006/fgbi.1997.0984. [DOI] [PubMed] [Google Scholar]
- 33.Clutterbuck AJ. The Aspergillus nidulans Linkage Map. Available: http://www.gla.ac.uk/ibls/molgen/aspergillus/index.html. Accessed 2010 June. [DOI] [PubMed]
- 34.CADRE. The Central Aspergillus Data REpository, Aspergillus nidulans database. Available: www.cadre-genomes.org.uk/Aspergillus_nidulans/. Accessed J2011 January.
- 35.Clutterbuck AJ, Farman M. Aspergillus nidulans linkage map and genome sequence: closing gaps and adding telomeres. In: Goldman GH, Osmani SA, editors. The Aspergilli: genomics, medical aspects, biotechnology, and research methods. Boca Raton: CRC Press; 2008. pp. 57–73. [Google Scholar]
- 36. Ensembl Genomes. Available: ftp://ftp.ensemblgenomes.org/pub/fungi/release-5/fasta/aspergillus_nidulans/dna/. Accessed 2009 October.
- 37.Lynch M, Walsh B. Sunderland: Sinauer Associates, Inc; 1998. Genetics and Analysis of Quantitative Traits. [Google Scholar]
- 38. Broad Institute of Harvard and MIT Aspergillus Comparative Sequencing Project. Available: www.broadinstitute.org/annotation/genome/aspergillus_group/MultiDownloads.html. Accessed 2009 October.
- 39.Mayorga ME, Timberlake WE. Isolation and molecular characterization of the Aspergillus nidulans wA gene. Genetics. 1990;126:73–79. doi: 10.1093/genetics/126.1.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tilburn J, Roussel F, Scazzocchio C. Insertional inactivation and cloning of the wA gene of Aspergillus nidulans. Genetics. 1990;126:81–90. doi: 10.1093/genetics/126.1.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hohl TM, Van Epps HL, Rivera A, Morgan LA, Chen PL, et al. Aspergillus fumigatus triggers inflammatory responses by stage-specific beta-glucan display. PLoS Pathog. 2005;1:232–240. doi: 10.1371/journal.ppat.0010030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Steele C, Rapaka RR, Metz A, Pop SM, Williams DL, et al. The beta-glucan receptor dectin-1 recognizes specific morphologies of Aspergillus fumigatus. PLoS Pathog. 2005;1:323–334. doi: 10.1371/journal.ppat.0010042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mouyna I, Morelle W, Vai M, Monod M, Lechenne B, et al. Deletion of GEL2 encoding for a beta(1-3)glucanosyltransferase affects morphogenesis and virulence in Aspergillus fumigatus. Mol Microbiol. 2005;56:1675–1688. doi: 10.1111/j.1365-2958.2005.04654.x. [DOI] [PubMed] [Google Scholar]
- 44.Mora-Montes HM, Bates S, Netea MG, Diaz-Jimenez DF, Lopez-Romero E, et al. Endoplasmic reticulum alpha-glycosidases of Candida albicans are required for N glycosylation, cell wall integrity, and normal host-fungus interaction. Eukaryot Cell. 2007;6:2184–2193. doi: 10.1128/EC.00350-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhang L, Zhou H, Ouyang HM, Li YJ, Jin C. Afcwh41 is required for cell wall synthesis, conidiation, and polarity in Aspergillus fumigatus. FEMS Microbiol Lett. 2008;289:155–165. doi: 10.1111/j.1574-6968.2008.01376.x. [DOI] [PubMed] [Google Scholar]
- 46.Coleman JJ, Mylonakis E. Efflux in Fungi: La Piece de Resistance. PLoS Pathog. 2009;5:e1000486. doi: 10.1371/journal.ppat.1000486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Theiss S, Kretschmar M, Nichterlein T, Hof H, Agabian N, et al. Functional analysis of a vacuolar ABC transporter in wild-type Candida albicans reveals its involvement in virulence. Mol Microbiol. 2002;43:571–584. doi: 10.1046/j.1365-2958.2002.02769.x. [DOI] [PubMed] [Google Scholar]
- 48.Sun S, Xu JP. Genetic analyses of a hybrid cross between serotypes A and D strains of the human pathogenic fungus Cryptococcus neoformans. Genetics. 2007;177:1475–1486. doi: 10.1534/genetics.107.078923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Marra RE, Huang JC, Fung E, Nielsen K, Heitman J, et al. A genetic linkage map of Cryptococcus neoformans variety neoformans serotype D (Filobasidiella neoformans). Genetics. 2004;167:619–631. doi: 10.1534/genetics.103.023408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Cheema MS, Christians JK. Virulence in an insect model differs between mating types in Aspergillus fumigatus. Med Mycol Sep. 6, Epub ahead of print. 2010 doi: 10.3109/13693786.2010.512301. [DOI] [PubMed] [Google Scholar]
- 51.Leger RJS, Screen SE, Shams-Pirzadeh B. Lack of host specialization in Aspergillus flavus. Appl Environ Microb. 2000;66:320–324. doi: 10.1128/aem.66.1.320-324.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Jackson JC, Higgins LA, Lin XR. Conidiation color mutants of Aspergillus fumigatus are highly pathogenic to the heterologous insect host Galleria mellonella. PLoS One. 2009;4:e4224. doi: 10.1371/journal.pone.0004224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Croft JH, Jinks JL. Aspects of the Population Genetics of Aspergillus nidulans. In: Smith JE, Pateman JA, editors. Genetics and Physiology of Aspergillus. London: Academic Press; 1977. pp. 339–360. [Google Scholar]
- 54.Schrettl M, Bignell E, Kragl C, Sabiha Y, Loss O, et al. Distinct roles for intra- and extracellular siderophores during Aspergillus fumigatus infection. PLoS Pathog. 2007;3:1195–1207. doi: 10.1371/journal.ppat.0030128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Haas H, Eisendle M, Turgeon BG. Siderophores in fungal physiology and virulence. Annu Rev Phytopathol. 2008;46:149–187. doi: 10.1146/annurev.phyto.45.062806.094338. [DOI] [PubMed] [Google Scholar]
- 56.Aleksenko A, Nielsen ML, Clutterbuck AJ. Genetic and physical mapping of two centromere-proximal regions of chromosome IV in Aspergillus nidulans. Fungal Genet Biol. 2001;32:45–54. doi: 10.1006/fgbi.2001.1251. [DOI] [PubMed] [Google Scholar]
- 57.Birdsell JA. Integrating genomics, bioinformatics, and classical genetics to study the effects of recombination on genome evolution. Mol Biol Evol. 2002;19:1181–1197. doi: 10.1093/oxfordjournals.molbev.a004176. [DOI] [PubMed] [Google Scholar]
- 58.Marsolier-Kergoat MC, Yeramian E. GC content and recombination: Reassessing the causal effects for the Saccharomyces cerevisiae genome. Genetics. 2009;183:31–38. doi: 10.1534/genetics.109.105049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Keurentjes JJB, Bentsink L, Alonso-Blanco C, Hanhart CJ, Vries HBD, et al. Development of a near-isogenic line population of Arabidopsis thaliana and comparison of mapping power with a recombinant inbred line population. Genetics. 2007;175:891–905. doi: 10.1534/genetics.106.066423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.McCluskey K, Wiest A, Plamann M. The Fungal Genetics Stock Center: a repository for 50 years of fungal genetics research. J Biosciences. 2010;35:119–126. doi: 10.1007/s12038-010-0014-6. [DOI] [PubMed] [Google Scholar]
- 61.Grindle M. Heterokaryon compatibility of unrelated strains in the Aspergillus nidulans group. Heredity. 1963;18:191–204. doi: 10.1038/hdy.1963.21. [DOI] [PubMed] [Google Scholar]
- 62.Bruggeman J, Debets AJM, Swart K, Hoekstra RF. Male and female roles in crosses of Aspergillus nidulans as revealed by vegetatively incompatible parents. Fungal Genet Biol. 2003;39:136–141. doi: 10.1016/s1087-1845(03)00016-1. [DOI] [PubMed] [Google Scholar]
- 63.Butcher AC. The relationship between sexual outcrossing and heterokaryon incompatibility in Aspergillus nidulans. Heredity. 1968;23:443–452. doi: 10.1038/hdy.1968.55. [DOI] [PubMed] [Google Scholar]
- 64.Jin J, Lee YK, Wickes BL. Simple chemical extraction method for DNA isolation from Aspergillus fumigatus and other Aspergillus species. J Clin Microbiol. 2004;42:4293–4296. doi: 10.1128/JCM.42.9.4293-4296.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wortman JR, Gilsenan JM, Joardar V, Deegan J, Clutterbuck J, et al. The 2008 update of the Aspergillus nidulans genome annotation: A community effort. Fungal Genet Biol. 2009;46:S2–S13. doi: 10.1016/j.fgb.2008.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Li H, Ruan J, Durbin R. Mapping short DNA sequencing reads and calling variants using mapping quality scores. Genome Res. 2008;18:1851–1858. doi: 10.1101/gr.078212.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Lorieux M. 2006. MapDisto, a tool for easy mapping of genetic markers, Poster P886. Plant and Animal Genome XIV conference, San Diego, CA. Available: http://mapdisto.free.fr/. Accessed 2010 July.
- 68.Egel-Mitani M, Olson LW, Egel R. Meiosis in Aspergillus nidulans: Another example for lacking synaptonemal complexes in the absence of crossover interference. Hereditas. 1982;97:179–187. doi: 10.1111/j.1601-5223.1982.tb00761.x. [DOI] [PubMed] [Google Scholar]
- 69.Tripathi N, Hoffmann M, Willing EM, Lanz C, Weigel D, et al. Genetic linkage map of the guppy, Poecilia reticulata, and quantitative trait loci analysis of male size and colour variation. P Roy Soc B-Biol Sci. 2009;276:2195–2208. doi: 10.1098/rspb.2008.1930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Neilands JB. Microbial iron transport compounds (siderophores) as chelating agents. In: Martell AE, Anderson WF, Badman DG, editors. Development of Iron Chelators for Clinical Use. New York: Elsevier/North Holland; 1981. pp. 13–31. [Google Scholar]
- 71.Dutsky SR, Thompson JV, Cantwell GE. A technique for mass rearing the greater wax moth (Lepidoptera, Galleridae). P Entomol Soc Wash. 1962;64:56–58. [Google Scholar]
- 72.Wang S, Basten CJ, Zeng ZB. 2010. Windows QTL Cartographer 2.5. Department of Statistics, North Carolina State University, Raleigh, NC. Available: http://statgen.ncsu.edu/qtlcart/WQTLCart.htm. Accessed: 2010 August.
- 73.Churchill GA, Doerge RW. Empirical threshold values for quantitative trait mapping. Genetics. 1994;138:963–971. doi: 10.1093/genetics/138.3.963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Henikoff S, Henikoff JG. Amino acid substitution matrices from protein blocks. P Natl Acad Sci USA. 1992;89:10915–10919. doi: 10.1073/pnas.89.22.10915. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Aspergillus nidulans linkage map, including genetic and physical positions and segregation data for each marker.
(XLS)
Sequence flanking SNP markers.
(XLS)