Abstract
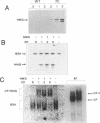
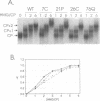
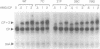
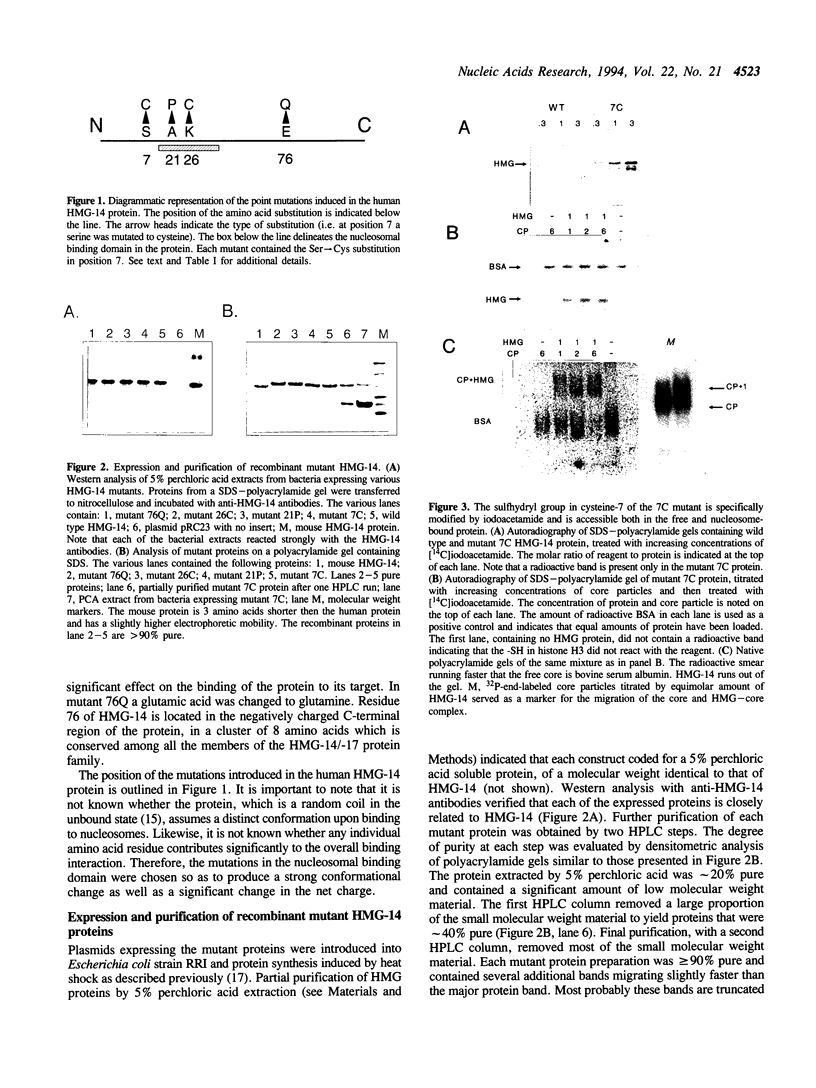
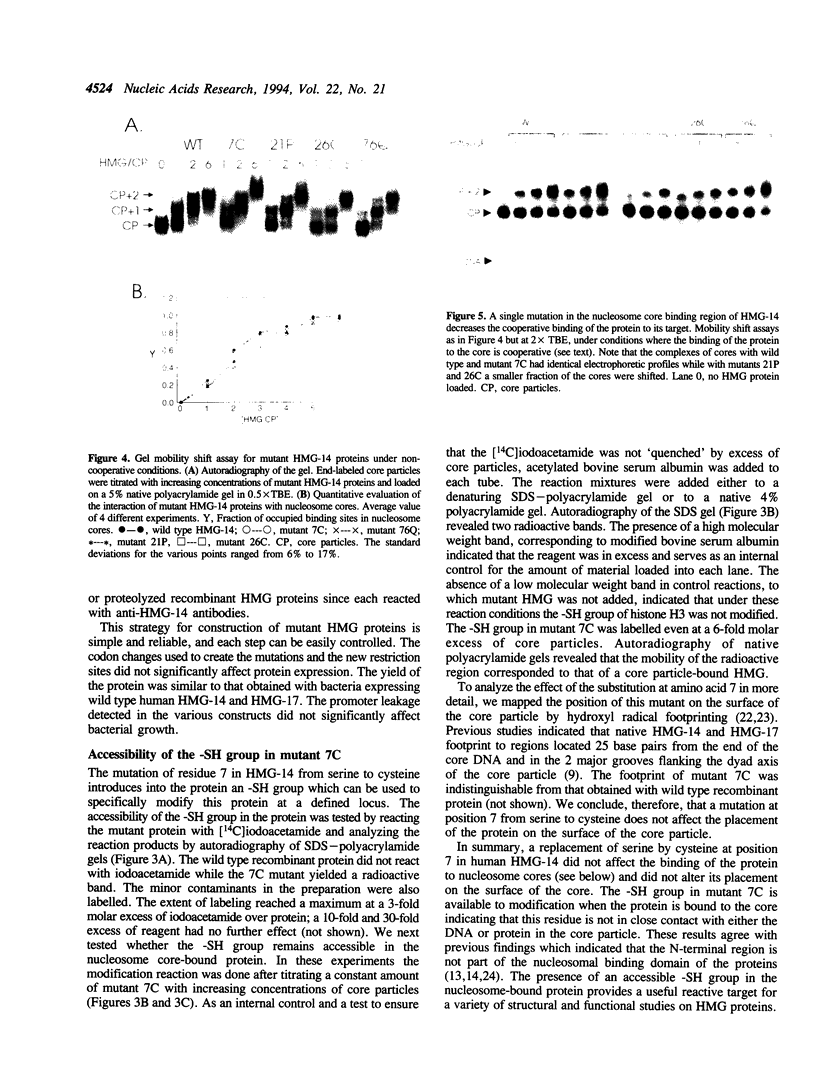
Mutants of human chromosomal protein HMG-14 were generated by site directed mutagenesis and used to study functional domains in this protein. A replacement of serine by cysteine at position 7 did not affect the binding of the protein to nucleosome cores. The sulfhydryl group in the nucleosome-bound protein is accessible to modifying agents suggesting that position 7 in the protein is not in close contact with either the DNA or the histones in the core particles. Under cooperative binding conditions, replacements of alanine by proline at position 21, or of lysine by cysteine at position 26, decreased the affinity of the protein for nucleosome cores 6.7- and 3-fold respectively. In contrast, the non-cooperative mode of binding was only minimally affected. A replacement of glutamic acid by glutamine at position 76 caused only minor changes in the binding of the protein to the cores. The results indicate that single point mutations, which change either the conformation or change in the nucleosomal binding domain of the protein, significantly reduce the ability of the HMG-14 protein to bind to nucleosome cores. We suggest that in chromatin the protein binds to nucleosomes in a cooperative manner and that upon binding to nucleosomes the protein acquires a distinct conformation.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Albright S. C., Wiseman J. M., Lange R. A., Garrard W. T. Subunit structures of different electrophoretic forms of nucleosomes. J Biol Chem. 1980 Apr 25;255(8):3673–3684. [PubMed] [Google Scholar]
- Alfonso P. J., Crippa M. P., Hayes J. J., Bustin M. The footprint of chromosomal proteins HMG-14 and HMG-17 on chromatin subunits. J Mol Biol. 1994 Feb 11;236(1):189–198. doi: 10.1006/jmbi.1994.1128. [DOI] [PubMed] [Google Scholar]
- Ausio J., Dong F., van Holde K. E. Use of selectively trypsinized nucleosome core particles to analyze the role of the histone "tails" in the stabilization of the nucleosome. J Mol Biol. 1989 Apr 5;206(3):451–463. doi: 10.1016/0022-2836(89)90493-2. [DOI] [PubMed] [Google Scholar]
- Bustin M., Becerra P. S., Crippa M. P., Lehn D. A., Pash J. M., Shiloach J. Recombinant human chromosomal proteins HMG-14 and HMG-17. Nucleic Acids Res. 1991 Jun 11;19(11):3115–3121. doi: 10.1093/nar/19.11.3115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bustin M., Crippa M. P., Pash J. M. Immunochemical analysis of the exposure of high mobility group protein 14 and 17 surfaces in chromatin. J Biol Chem. 1990 Nov 25;265(33):20077–20080. [PubMed] [Google Scholar]
- Bustin M., Lehn D. A., Landsman D. Structural features of the HMG chromosomal proteins and their genes. Biochim Biophys Acta. 1990 Jul 30;1049(3):231–243. doi: 10.1016/0167-4781(90)90092-g. [DOI] [PubMed] [Google Scholar]
- Cook G. R., Minch M., Schroth G. P., Bradbury E. M. Analysis of the binding of high mobility group protein 17 to the nucleosome core particle by 1H NMR spectroscopy. J Biol Chem. 1989 Jan 25;264(3):1799–1803. [PubMed] [Google Scholar]
- Crippa M. P., Alfonso P. J., Bustin M. Nucleosome core binding region of chromosomal protein HMG-17 acts as an independent functional domain. J Mol Biol. 1992 Nov 20;228(2):442–449. doi: 10.1016/0022-2836(92)90833-6. [DOI] [PubMed] [Google Scholar]
- Crippa M. P., Trieschmann L., Alfonso P. J., Wolffe A. P., Bustin M. Deposition of chromosomal protein HMG-17 during replication affects the nucleosomal ladder and transcriptional potential of nascent chromatin. EMBO J. 1993 Oct;12(10):3855–3864. doi: 10.1002/j.1460-2075.1993.tb06064.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du W., Thanos D., Maniatis T. Mechanisms of transcriptional synergism between distinct virus-inducible enhancer elements. Cell. 1993 Sep 10;74(5):887–898. doi: 10.1016/0092-8674(93)90468-6. [DOI] [PubMed] [Google Scholar]
- Ferrari S., Harley V. R., Pontiggia A., Goodfellow P. N., Lovell-Badge R., Bianchi M. E. SRY, like HMG1, recognizes sharp angles in DNA. EMBO J. 1992 Dec;11(12):4497–4506. doi: 10.1002/j.1460-2075.1992.tb05551.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayes J. J., Tullius T. D., Wolffe A. P. The structure of DNA in a nucleosome. Proc Natl Acad Sci U S A. 1990 Oct;87(19):7405–7409. doi: 10.1073/pnas.87.19.7405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landsman D., McBride O. W., Soares N., Crippa M. P., Srikantha T., Bustin M. Chromosomal protein HMG-14. Identification, characterization, and chromosome localization of a functional gene from the large human multigene family. J Biol Chem. 1989 Feb 25;264(6):3421–3427. [PubMed] [Google Scholar]
- Landsman D., Mendelson E., Druckmann S., Bustin M. Exchange of proteins during immunofractionation of chromatin. Exp Cell Res. 1986 Mar;163(1):95–102. doi: 10.1016/0014-4827(86)90561-6. [DOI] [PubMed] [Google Scholar]
- Mardian J. K., Paton A. E., Bunick G. J., Olins D. E. Nucleosome cores have two specific binding sites for nonhistone chromosomal proteins HMG 14 and HMG 17. Science. 1980 Sep 26;209(4464):1534–1536. doi: 10.1126/science.7433974. [DOI] [PubMed] [Google Scholar]
- Paton A. E., Wilkinson-Singley E., Olins D. E. Nonhistone nuclear high mobility group proteins 14 and 17 stabilize nucleosome core particles. J Biol Chem. 1983 Nov 10;258(21):13221–13229. [PubMed] [Google Scholar]
- Paull T. T., Haykinson M. J., Johnson R. C. The nonspecific DNA-binding and -bending proteins HMG1 and HMG2 promote the assembly of complex nucleoprotein structures. Genes Dev. 1993 Aug;7(8):1521–1534. doi: 10.1101/gad.7.8.1521. [DOI] [PubMed] [Google Scholar]
- Sandeen G., Wood W. I., Felsenfeld G. The interaction of high mobility proteins HMG14 and 17 with nucleosomes. Nucleic Acids Res. 1980 Sep 11;8(17):3757–3778. doi: 10.1093/nar/8.17.3757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schröter H., Bode J. The binding sites for large and small high-mobility-group (HMG) proteins. Studies on HMG-nucleosome interactions in vitro. Eur J Biochem. 1982 Oct;127(2):429–436. doi: 10.1111/j.1432-1033.1982.tb06890.x. [DOI] [PubMed] [Google Scholar]
- Thanos D., Maniatis T. The high mobility group protein HMG I(Y) is required for NF-kappa B-dependent virus induction of the human IFN-beta gene. Cell. 1992 Nov 27;71(5):777–789. doi: 10.1016/0092-8674(92)90554-p. [DOI] [PubMed] [Google Scholar]
- Tullius T. D., Dombroski B. A., Churchill M. E., Kam L. Hydroxyl radical footprinting: a high-resolution method for mapping protein-DNA contacts. Methods Enzymol. 1987;155:537–558. doi: 10.1016/0076-6879(87)55035-2. [DOI] [PubMed] [Google Scholar]
- Walker J., Chen T. A., Sterner R., Berger M., Winston F., Allfrey V. G. Affinity chromatography of mammalian and yeast nucleosomes. Two modes of binding of transcriptionally active mammalian nucleosomes to organomercurial-agarose columns, and contrasting behavior of the active nucleosomes of yeast. J Biol Chem. 1990 Apr 5;265(10):5736–5746. [PubMed] [Google Scholar]
- Westermann R., Grossbach U. Localization of nuclear proteins related to high mobility group protein 14 (HMG 14) in polytene chromosomes. Chromosoma. 1984;90(5):355–365. doi: 10.1007/BF00294162. [DOI] [PubMed] [Google Scholar]