Abstract
A parallel microfluidic cytometer (PMC) uses a high-speed scanning photomultiplier-based detector to combine low-pixel-count 1-D imaging with flow cytometry. The 384 parallel flow channels of the PMC decouple count rate from signal-to-noise ratio. Using 6-pixel 1-D images, we investigated protein-localization in a yeast model for a human protein-misfolding disease and demonstrated the feasibility of a nuclear-translocation assay in Chinese-hamster-ovary (CHO) cells expressing a NFκB-GFP reporter.
Applications of high-content screening (HCS) 1–7 are circumscribed by several practical aspects, including low sample throughput and absence of sorting capability. Moreover high-resolution 2-D images consume limited detector bandwidth, introduce a data acquisition delay that is a barrier for real-time decisions needed for sorting, and introduce noise via inaccuracies in image segmentation. Imaging flow cytometers based on wide-field CCD imagers 8 have similar throughput limitations to microscopes. To address these restrictions we have developed a multi-channel parallel microfluidic cytometer (PMC) that is based on analog detection combined with parallel microfluidics. By taking approximately 1/1000 the amount of data per cell compared to a CCD image, we are able to drastically reduce the data-buffering and storage requirements and can simplify the classification algorithm to a fraction of a microsecond. Furthermore, parallel microfluidics overcome the sample-changeover bottleneck of a single-channel flow cytometer (FCM). Therefore the new architecture circumvents many of the throughput limitations of both HCS and FCM, but combines many of the best features of each technology.
While multi-color 1-D imaging reduces the data load of HCS, the corollary disadvantage is a sparser image and a greater number of potentially ambiguous images. The question is how well can a sparse 1-D image work in high-content screening?
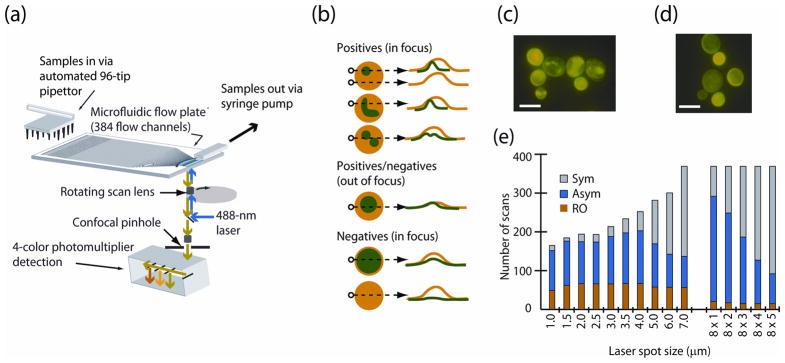
The concept of the PMC is shown in Figure 1. As the cells cross the detection window a confocal laser scanner records fluorescence values on photomultiplier detectors (PMTs) every 1 μm across the 100-μm-wide flow channels (Fig. 1a). A multicolor 1-D image representing the cell (Fig. 1b) is sent to a classification algorithm, which measures cell features and classifies each 1-D image. Since the statistical distribution of classified images varies as the population of cells changes, differences in this distribution can be used to create a high-content assay. Image classification is complicated by intrinsically non-discriminatory images and similar problems have been addressed in 2-D HCS using data filters and thresholds 1. Several thousand events per second can be processed on the PMC, but for a simple binary assay the number of discriminating objects required is often as few as 100 objects.
Figure 1.
High-content screening on a PMC: (a) schematic of the device (b) simulated 2-D microscope images. The dashed arrow shows the location of the single 1-D scan with a reporter in green and a whole cell marker in orange. The two-color 1D images are converted to multicolor 1D images (shown to the right of each image). Simulation of phenotyping with 1-D images of (c) the positive S. cerevesiae phenotype exhibiting induced α-Syn-GFP focal aggregates and (d) the negative phenotype with diffuse and membrane localized α-Syn-GFP (5-μm scale bars). (e) Simulation model counts of 1-D image classes (Sym, Asym, and RO) from a total of 400 scans, when a cell is scanned with the indicated dimensions of the laser spot (Supplementary Fig. 10).
We decided to test the principle of 1-D flow imaging with a HCS assay for amyloid aggregation in S. cerevisiae. Re-arrangements of the amyloid proteins in this organism are marked by distinct condensate patterns and comprise models for protein misfolding diseases 10, 11 (Fig. 1c,d). We first simulated our method using four high-content features, (1) green symmetrical with red (Sym), (2) green asymmetrical with red (Asym), (3) red only (RO) and (4) red overlapping green (R=G) (Fig. 1e). Even for relatively small (5.5-μm-diameter) S. cerevisiae cells, it was possible to efficiently distinguish the idealized phenotypes using a laser focal diameter of 4–5μm. Somewhat surprisingly, the efficiency of the PMC detector actually increased as the diameter of a laser spot was increased from 1μm to 4μm (Fig. 1e, Online Methods).
Based on these simulations an optical scanner was developed and set to an effective optical resolution of 4.3μm. We cultured strains of S. cerevisiae expressing an α–Syn-GFP fusion protein, , and mCherry as a whole cell marker, as described previously, 10, 11 fixed and analyzed the cells in the PMC.
In the first step of data reduction, cell events were extracted, converted into (i) raw, (ii) three-point-smoothed, normalized, and (iii) Gaussian-fit 1-D images, before being analyzed as a two-color 1-D images. A total of 82 feature calculations (such as max signal strength, max signal position, and total signal area and perimeter) were made on each cell event using the raw, smoothed, Gaussian-fitted two-color signals (Online Methods and Supplementary Table 1).
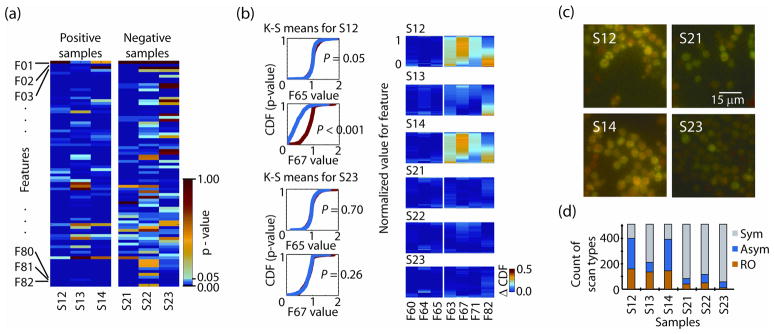
For group statistical analysis, we first thresholded the sample to a minimum green signal. We evaluated the 82 feature values in negative-to-negative and negative-to-positive samples using the Kolmogorov-Smirnov test (K-S test) 6, 7. From 82 features (Fig. 2a) we identified those that separated control and positives with p-values < 0.05, and did not separate control and negative samples (p-values > 0.05). We found the greatest separation for features that were based on GFP signal symmetry 5.
Figure 2.
Phenotyping α-Syn-GFP aggregation by PMC imaging. (a) K-S test of 82 features for three positive (S12–S14) and three negative (S21–S23) samples displayed as a p-value heat map with increasing probability from blue to red. (b) Plots of cumulative distribution functions for the K-S test, shown for 2 features for a positive and negative sample (left). K-S heat map signatures (right) show the difference in the CDF plots generated for 6 yeast samples across 7 features. Control red features; F60 - total intensity ratio around signal peak, F64 - intensity perimeter around signal peak, F65 - red perimeter ratio around calculated object center. Discriminating features; F63 – green area around red peak, F67 - green perimeter ratio around object center, F71 – ratio F67:F65, F82 – green P2A/red P2A (P2A=perimeter2/2π*area). (c) aggregated (positive) [correct?] S. cerevisiaeα-Syn-GFP samples (S12, S14) and non-aggregated (negative) samples (S209, S22). (d) Single-cell event distribution across three 1-D image classes; Sym, Asym, RO for positive and negative samples.
We subsequently selected 4 features based on GFP signal symmetry around the center of the cell and on three red-channel control features. For each test sample and selected feature we created a difference map by subtracting the two cumulative distribution functions (CDF’s) for the K-S test calculated for each sample and feature (Fig 2b, left column). In this heat map (Fig. 2b, right column) separation from zero (blue) indicates lack of homogeneity and failure of the K-S hypothesis that the samples are the same. We see separation in the positive sample features based on green symmetry while the remainder appears homogeneous. Using a 95% probability threshold for homogeneity we can reject all of the positive samples using all four green symmetry features, but only feature F67 (green perimeter ratio around the calculated object center) accepts homogeneity for the negative samples. For our three control features most samples show homogeneity (p-value < 0.001), but only feature F65 (red perimeter ratio around the calculated object center) shows all samples to be from the same group (p-value = 0.05). Therefore, we could reduce the assay to only features F67 (indicating positive) and F65 (control). Positive and negative samples identified using this analysis were confirmed using fluorescence microscopy (Fig 2c). Image classification distributions across three 1-D image classes (Sym, Asym, RO) for three positive (Fig 2d, left) and three negative samples (Fig 2d, right), revealed that positive samples show higher percentages of RO and Asym 1-D images (500 cells per sample, n=6). While reproducibility was verified separately (Online Methods), we conclude that several hundred 1-D cell events per well are sufficient for the amyloid aggregation assay, which is comparable to the number of events required for 2-D HCS using a microscope 1.
For a second assay we chose the NFκB nuclear-translocation assay using a mammalian CHOhIR cell line stably expressing human NFκB-GFP and tumor necrosis factor alpha receptor 1 (TNFαR1). Translocation of NFκB-GFP from the cytoplasm into the nucleus can be induced by interleukin-1β (IL-1β) 12, 13. We modified the standard protocol to create suspension samples (Supplementary Fig. 1–2). Cells after various levels of IL-1β induction were detached, fixed, counter stained (CyTRAK Orange), then scanned with the same settings used for the S. cerevisiae assay. After K-S analysis, we found four features to separate induced (NFκB translocated) and non-induced (NFκB non-translocated) samples. We show the minimum sample size for stable sample separation to be 100 events and demonstrated a rudimentary IC-50 dose response (Supplementary Figs. 1–5). Other experiments with fixed murine leukocytes (Supplementary Figs. 6–8) and live human osteocytes 9 have demonstrated the ability of the PMC to detect rare cells approaching 0.01% of the population.
Because S. cerevisiae is small (3–6μm diameter), it is challenging as a model system for proof of low-pixel-count imaging. However, we found 6-pixel 1-D images to be sufficient to resolve prion aggregate localization. Moreover, the number of data dimensions for 1D imaging increases according to f = nFi, where Fi is the number of independent image features used on n color channels. For example, with just four uncorrelated image features, we can add 16 potential data dimensions to four-color flow cytometry. This additional spatially-encoded information is achieved with highly efficient use of the scarce resources of the data acquisition system, namely digitization rate and buffering capacity.
While much can be borrowed from the methods of 2-D HCS 2–4, 7, the 1-D algorithms are fundamentally different. Most importantly, microscope algorithms usually start by drawing boundaries around “primary and secondary objects” (also called segmentation 1). The user-defined aspect of segmentation is a source of assay variability and is often considered the most challenging and time-consuming step 1, 2. In 1-D imaging on the PMC, we set the resolution in hardware and thus eliminate potential segmentation issues by accepting any resolution element as an “object”. With this approach, we are able to develop efficient HCS algorithms using relatively low resolution 1-D images.
The PMC has fundamentally different architecture from either a single-channel cytometer (FCM) or a microscope-based HCS system. The multiple microfluidic flow channels not only enable miniaturization but also allow the independent optimization of cell count rate, samples per minute, and signal-to-noise ratio. In our slow-flow regime, a PMC is often able to quantify fluorescence more carefully than a conventional FCM system (Ref. 9 and Supplementary Fig. 8) yet is still able to collect 3000 four-color cell profiles distributed across 16, 32, or 384 simultaneous flow channels (Supplementary Fig. 9). Furthermore, by flowing the samples into the focal volume, the PMC eliminates the focusing and stage motions that limit HCS on a microscope to about 2–6 wells per minute. Therefore where a 1-D image can suffice, the PMC throughput is limited primarily by its digitization rate and should exceed microscope-based HCS by several orders of magnitude. Fraction sorting is generally not possible on microscope-based HCS instruments, but could be achieved on the PMC via integration of a competent flow switch into the microfluidics 12. The current (2-msec) Matlab algorithms should optimally be implemented in electronics firmware to support an image-based sub-μs switch.
Online Methods
Data Acquisition on the PMC
A parallel microfluidic network sweeps cells past a confocal laser-induced-fluorescence detector that scans transversely at a velocity much faster than the flow (Fig. 1a). The laser focus is adjusted to be circular or oblong with an X dimension small enough to resolve intracellular structure. A 1-D image is generated for each active PMT channel and in our yeast aggregate assay yields two: one for the reporter protein (Fig. 1b, green) aggregates and the other for the whole cell label (Fig. 1b, orange). The aggregated phenotypes are designated “positives”. The fluorescence values recorded by the separate photomultipliers (PMTs) are converted to 16-bit digital values and then assembled in computer memory as multicolor 1-D images.
Data Analysis
Raw data was saved in binary files (one for each PMT) in an “x by n” format where 320 bytes per “x” scan and “n” total number of scans (n usually set at 10,000). The instrument saves the data in a “raw” binary file containing values for all PMT’s annotated by scan position and by designated scan number. Once the data acquisition is complete, this data file is archived off the instrument. To process results, an initial data reduction program reads through the raw file and identifies lane positions and noise levels, then creates smaller lane- (sample-) specific binary files with multiple “empty” scans eliminated. These files are then are then read by our assay-specific algorithm file, which identifies objects using one “dominant” PMT channel, rejects objects due to some simple criteria (e.g., cell at edge of scan window, values off scale, object FWHM width too small or large too be a single cell) and sends the remaining identified objects to the assay specific algorithm, which will evaluate “feature” values for each object. The number of total events rejected by all of the threshold filters is less than 20%. Feature values are saved in a CSV file for further ad-hoc evaluation by Matlab, R statistical software, Excel, or converted to FCS 3.0 File format for import to cytometry software such as FlowJo, using available CSV to FCS file converters such as TextToFCS. For specific uses we have also created a direct FCS file conversion program and an optional image generator that converts the raw scan image data (in PMT x lane width x number of scans format) into multiple sets of 4 (PMT specific) 1024 × 1024 TIFF images for evaluation by microscope image analysis software such as Cell Profiler (Broad Institute) or MetaMorph (Molecular Devices).
The first step of post processing is initial data reduction. This step identifies scans with sufficient red PMT signal to possibly contain a good scan (signal above average noise). This scan, as well as a blank scan before it (to separate single-scanned objects from multi-scanned objects), was moved to a reduced, but still “raw” data set. If a sample run had been split among multiple raw data files, they were reduced to one sample file.
In the next step this reduced file is analyzed for identified good scan objects by evaluating the red PMT channel for objects that met minimal criteria: maximum signal strength above a set fixed threshold, below the A/D maximum value, and with a calculated raw FWHM measured between 2 – 12 microns. The scans before and after identified objects were evaluated to ensure that the object was scanned only once. From each sample we collected the first 500 scans that met these basic criteria.
Next, the selected scan objects and the 50 microns around the max signal points were cut from the 320-micron scan, then “digitally zoomed” in width by 10x with a straight line interpolation process used to draw the line between the original points and the zoomed points. This step was done primarily to allow for better visual interpretation of the data and use of simpler integer math in calculations. From this zoomed scan, three more representations of this scan were created – three-point smoothing was performed on the raw data, the smoothed data was normalized and, a Gaussian version of the scan was created from the normalized data. This was completed for both the red and green channels.
For each of the resulting eight 1-D images some basic calculations were made, including max signal strength, max signal position, FWHM, center, total signal area and perimeter of signal. Next, channel vs. channel calculations were made using those basic calculations including green signal area/red signal area, green FWHM/red FWHM, etc. For the yeast assay we then created a few other calculations that measured the distribution of green signal and perimeter on each side of the cell “center”, which we designated several ways using red channel values. All of these values, called scan features, were collected for each cell and were used to 1) reject 1-D images as not being useful in data interpretation and 2) show that this group was either the same or different. For some measurements we would expect each sample to be similar (i.e. if both samples use the same whole cell red stain they should have comparable red channel measurements). For other features the groups should be different (i.e. if the green GFP marker is aggregated or not, we should see variance in the different GFP-related features for different samples). In all, we calculated ~82 features for each 1-D image (Supplementary Table 1). As there is little time penalty in computing each additional feature, most of this list will have measurements that will not be used for the given assay, but included for future algorithm development.
Using this feature data, two approaches to evaluate the samples were taken. The first approach, used for our simulated data, was to classify each 1-D image and then evaluate the sample based on the proportions of each class observed in the sample. This method typically requires a well-characterized standard. The second method was to use group statistics to evaluate the features of one sample vs. another sample to determine the likelihood that they were both “identical” samples.
For 1-D image classification, the same algorithm used for the model data was applied to the PMC data. Changes were made to the threshold values used to define “red only” -as the absolute signal strengths were determined differently, and the overlap threshold used to define “asymmetry” was increased for the PMC data (compared to the simulation) to account for the higher noise level in the data.
Using group statistics we first identified and removed data objects with insufficient green signal (i.e. red-only 1-D images). This reduced the data set from the original 500 objects. We then equalized the data sets to the number of objects in control or sample data set, which ever was smaller. The number of objects used for Kolmogorov-Smirnov (K-S) tests were between 380 and 420. We then evaluated the various features values in one negative sample versus the other negative and positive samples using the K-S test to determine the probability that the samples were different. Using Matlab and the K-S function ktest2, we created a program that looked at one test sample and one control sample and calculated the K-S value and p-value for each of the 82 features. This was done for three positive and three negative samples. The results were combined in an excel spreadsheet and an array of score values (feature vs. sample) were evaluated to identify a) features remained constant for all samples and b) features which separated between positive and negative samples. These results are presented as Fig. 2d in the paper.
For the example in the paper, we chose three features that should remain constant for all samples and four that should vary between positive and negative samples. We concentrated on features that were based on symmetry calculations and were independent of signal intensity. The constant check features we selected were: F60 - red total intensity ratio around the maximum signal point, F64 - red intensity perimeter around the maximum signal point and F65 - red intensity perimeter around the calculated object center.
The differing features we selected were: F63 - green intensity area around calculated red object center, F67 - ratio of green perimeter before and after the calculated red object center, F71 - ratio of F67/F65 and F81 - green P2A/red P2A (P2A = perimeter2/2π*area).
To visually identify sample separation, we created a K-S heat map of the selected features for each sample. We generated the cumulative distribution function used by the K-S test for the reference and test sample and then calculated the difference over F(x) by subtracting the two CDF’s. We were not concerned with direction so we used absolute values for easier visualization. In this particular example, the most robust measure of sample homogeneity was F65 and the most robust differentiating feature was F67. A full list of the 82 features used is presented in Supplementary Table 1.
α-Syn-GFP Aggregation and NFκB Nuclear Translocation Assays
We cultured strains of S. cerevisiae requiring adenine-supplemented growth medium (ADE-) or not (ADE+), each expressing an α–Syn-GFP fusion protein, as previously described. 10, 11 The samples were stained with mCherry as a whole cell marker, fixed and then analyzed in the PMC. Cells injected into the PMC were drawn at a rate of 20μL/hr/lane past the detector at a digitizing interval of 1 μm per data point, 4 scans/s.
The NFκB p65 redistribution assay was performed using a CHOhIR cell line (#092-01, Dharmacon, Inc., Chicago, IL). Cells were grown on fibronectin (10 μg/ml) coated flasks overnight at 37°C, 5% CO2. To induce NFκB translocation into the nucleus, an assay buffer containing IL-1β (ranging from 0 to 20 ng/ml) was added to the flask. Cells were incubated at 37°C, 5% CO2 for 40 minutes, detached with a 4 minute treatment with Accutase (Invitrogen, Carlsbad, CA) and then fixed with 4% formaldehyde (15 minutes). Cells were transferred to centrifuge tubes, spun down, and stained with CyTRAK Orange (eBioscience, Inc., San Diego, CA) at room temperature for 45 minutes. Cho cells were drawn into the PMC at the same rate as for S. cerevisiae cells.
Verification of Reproducibility for α-Syn-GFP Assay
To verify reproducibility of the α-Syn-GFP assay (Fig. 2) we prepared two controls; a positive (aggregated phenotype) and a pure negative phenotype in which the α-Syn-GFP protein was localized to the cell membrane. Twelve positive and twelve negative samples were run over 10 days. One feature, F82, a measure of the green signal roundness stood out. The negative control mean value was 1.17 with a standard deviation of 0.2234, indicating that 99.7% of samples should be below 1.84. The positive mean value was 2.61 with a standard deviation of 0.1776 indicating that 99.7% of samples would have a mean above 2.07. The lack of overlap (or “presence of a window”) indicates that this feature alone can reliably determine whether an unknown sample is negative or positive.
Simulation Models
Optical settings and 1-D image classification of cell phenotypes were simulated using a geometrical model (Supplementary Fig. 10). The simulated laser scan with a shape of adjustable dimensions, was stepped vertically at 1 μm increments and the simulation of the cell was rotated through 360° to generate data that represented random two-color scans on the PMT detectors. The simulation then counted the number of 1-D images collected, with various laser dimensions, which fell into four high-content feature-based classes; green symmetrical with red (Sym), green asymmetrical with red (Asym), red only (RO) and red overlapping green (R=G).
PMC System Overview
The PMC used in the study is shown in Supplementary Figs. 11–12. The microfluidic flow devices are loaded from 384-well microtiter plates and maintained with a gantry robot. The sample deck includes positions for nutrient trays that can also be accessed by the pipettor. As a result, the system has some provision to maintain live-cell cultures. All 384 channels can be loaded from a microtiter plate in <30 seconds. Flow is actuated by suction using syringe pumps. The optical detector is a photomultiplier-based rotary scanner located under the microfluidics (Supplementary Figs. 11,12). The system is operated from a graphical user interface that displays data in real time.
Microfluidics
Microdevices were fabricated in aluminasilicate glass (Corning, EagleTM, 0.7-mm-thick plates of 25×50 and 25×25 cm size). Fabrication is by unaligned contact lithography, laser drilling, and high-temperature fusion bonding. The microchannels had a hemispherical cross section with a radius of 60 μm and converged to a density of 5-channels/mm in the scan zone. Flow devices of 32-channel and 384-channel complexity were fabricated for the study (Supplementary Fig. 11a).
Automation
The 96-tip robot head accesses water and buffer reservoirs, an ultra sound washing station, a microtiter plate elevator with up to 32 sample plates for continuous operations (Packard Instrument Co, Meriden, CT). The pipettor head has programmable suction and injection capability for volumes between 2–20 μl, and is driven by a DC brushless servo amplifier (Model 503, Copley Controls Corp, Westwood, MA).
Detector
A 100-mW multi-line argon-ion laser beam (Melles Griot #532-MA-A04, Melles Griot, Carlsbad, CA) is passed through a rotating head that rotates a 0.5-numerical-aperture (NA) aspheric lens (# 350240, Thor Labs, Corp., Newton, NJ) in an arc. The laser focus is adjusted between NA 0.01 and NA 0.50. The fluorescence is collected at NA 0.5 through the rotating head (Supplementary Fig. 11c), is re-imaged through a pinhole, then is separated into four wavelength bands using dichroics and bandpass filters and distributed onto four PMTs (H957-8 Hamamatsu, Bridgewater, NJ) (Supplementary Fig. 13).
The constancy and reproducibility of the speed profile have been measured and show a standard deviation of less than 1% from the target velocity (10,000 scans, all flow channels). The sensitivity of the system was evaluated with fluorescence standards and shown to have a 10 pM (fluorescein) detection limit in a 60-μm-deep channel, which is near the state of the art for on-column laser-induced fluorescence (LIF) detectors.
Determining the effective optical resolution of the scanner
Using the various scanning speed profiles we collected data for 2.5-μm-dia. beads and calculated the average observed bead FWHM. The values (average FWHM) increase with faster scan programs indicating that there is an electronic component to our observed object size. We calculated the effective X-dimension of the optical pixel for the 4 Hz program to be 4.30 microns, as determined by collecting an average observed FWHM of 2.5 micron beads then extracting the effective broadening factor due to the optics and electronics. Experimental data is shown in Supplementary Fig. 14.
Supplementary Material
Acknowledgments
We thank Brooke Bevis and Susan Lindquist of the Whitehead Institute for helpful conversations and for supplying the S. cerevisiae samples. This work was supported by the National Institutes of Health under grant R01 HG-001389.
References
- 1.Taylor DL, Haskins JR, Giuliano KA. High Content Screening. Humana Press; Totowa, NJ: 2007. [Google Scholar]
- 2.De Vos WH, Van Neste L, Dieriks B, Joss GH, Van Oostveldt P. Cytometry Part A. 2010;77A:64–75. doi: 10.1002/cyto.a.20807. [DOI] [PubMed] [Google Scholar]
- 3.Perlman ZE, et al. Science. 2004;306:1194–1198. doi: 10.1126/science.1100709. [DOI] [PubMed] [Google Scholar]
- 4.Ng AYJ, et al. J Biomol Screen. 2010;15:858–868. doi: 10.1177/1087057110372256. [DOI] [PubMed] [Google Scholar]
- 5.Comeau JWD, Costantino S, Wiseman PW. Biophysical Journal. 2006;91:4611–4622. doi: 10.1529/biophysj.106.089441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lapan P, et al. PLoS. 2009;4:e6822. doi: 10.1371/journal.pone.0006822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Feng Y, Bender TJ, Young DW, Tallarico JA. Nat Rev Drug Disc. 2009;8:567–578. doi: 10.1038/nrd2876. [DOI] [PubMed] [Google Scholar]
- 8.George TC, Fanning SL, Fitzgerald-Bocarsly P, Medeiros RB. J Immun Meth. 2006;311:117–129. doi: 10.1016/j.jim.2006.01.018. [DOI] [PubMed] [Google Scholar]
- 9.McKenna BK, Salim H, Bringhurst FR, Ehrlich DJ. Lab on a Chip. 2009;9:305–310. doi: 10.1039/b811889b. [DOI] [PubMed] [Google Scholar]
- 10.Krishnan R, Lindquist SL. Nature. 2005;435:765–72. doi: 10.1038/nature03679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shorter J, Lindquist SL. Nat Rev Genet. 2005;6:435–450. doi: 10.1038/nrg1616. [DOI] [PubMed] [Google Scholar]
- 12.Ding GJ, et al. J Biol Chem. 1998;273:28897–905. doi: 10.1074/jbc.273.44.28897. [DOI] [PubMed] [Google Scholar]
- 13.Schmidt JA, Birbach A, Hofer-Warbinek R. J Biol Chem. 2000;275:17035–17042. doi: 10.1074/jbc.M000291200. [DOI] [PubMed] [Google Scholar]
- 14.Bohm S, Gilbert J, Deshpande M. 7, 157, 274. US Patent. 2007
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.