Abstract
We reexamine the individual components for human ES and iPS cell culture, and formulate a cell culture system in which all protein reagents for liquid media, attachment surfaces, and splitting are chemically defined. A major improvement is the lack of a serum albumin component, as variations in either animal or human sourced albumin batches have previously plagued human ES and iPS cell culture with inconsistencies. Using this new medium (E8) and vitronectin-coated surfaces, we demonstrate improved derivation efficiencies of vector-free human iPS cells with an episomal approach. This simplified E8 medium should facilitate both the research use and clinical applications of human ES and iPS cells and their derivatives, and should be applicable to other reprogramming methods.
Human ES cells and human induced pluripotent stem (iPS) cells can proliferate without limit and yet maintain the potential to generate derivatives of all three germ layers. These properties make them useful for understanding the basic biology of the human body, for drug discovery and testing, and for transplantation therapies1–6.
The culture conditions used to support the derivation and expansion of human iPS cells have been based on conditions developed for human embryonic stem cells over the last decade, which have also been extensively compared and summarized by The International Stem Cell Initiative Consortium7. Our lab previously described the development of a medium (TeSR) for human ES cell culture, which has more recently been used for the derivation and culture of human iPS cells8. However, although we demonstrate that TeSR could be used to derive human ES cells in the complete absence of animal proteins, the inclusion of human serum albumin and human sourced matrix proteins makes those conditions prohibitively expensive, impractical for routine use, and not truly completely defined. Although cloned human serum albumin is available, and defined surfaces have now been described, because of the relative costs involved, many laboratories, including our own, continue to culture human embryonic stem and iPS cells routinely in media that includes bovine serum albumin (BSA) on Matrigel, a complex mixture of matrix proteins derived from Engelbreth-Holm-Swarm mouse tumors. However, the variation in sources of these media components is substantial, making extensive quality control necessary for all new batches. Because of the batch variation in media components, different labs making the same medium often report substantially different results9,10. The batch variability of albumin is particularly problematic, both because of the unusually high concentrations used in the culture media compared to other proteins and because of its ability to bind lipids and other impurities11.
Media optimization is a daunting challenge in combinatorics. TeSR medium has 18 components added to a DMEM/F12 base medium that itself has 52 components. In the initial development of TeSR, we demonstrated that subtracting albumin or any of the growth factors from the medium led to a statistically significant decline in human embryonic stem cell culture performance. However, because of the combinatoric complexity involved, we did not originally examine pair wise interactions between each factor. Here we showed that removal of albumin (Bovine Serum Albumin, BSA, in this study) from the medium leads to toxicity by a second component, β-mercaptoethanol (BME), and we demonstrated that in the absence of BME, BSA is no longer necessary for human ES or iPS cell culture. We then re-optimized the basic components of human ES and iPS cell culture in the absence of BSA and BME, and developed practical, completely defined E8 (eight components, including the DMEM/F12) medium and surfaces that support established human embryonic stem and iPS cells, and which greatly improve the efficiency of human iPS cell derivation from dermal biopsy samples. Using the E8-based medium, defined conditions can be used for all stages of iPS cell derivation and culture.
RESULTS
Albumin-free E8 medium for human ES and iPS cell culture
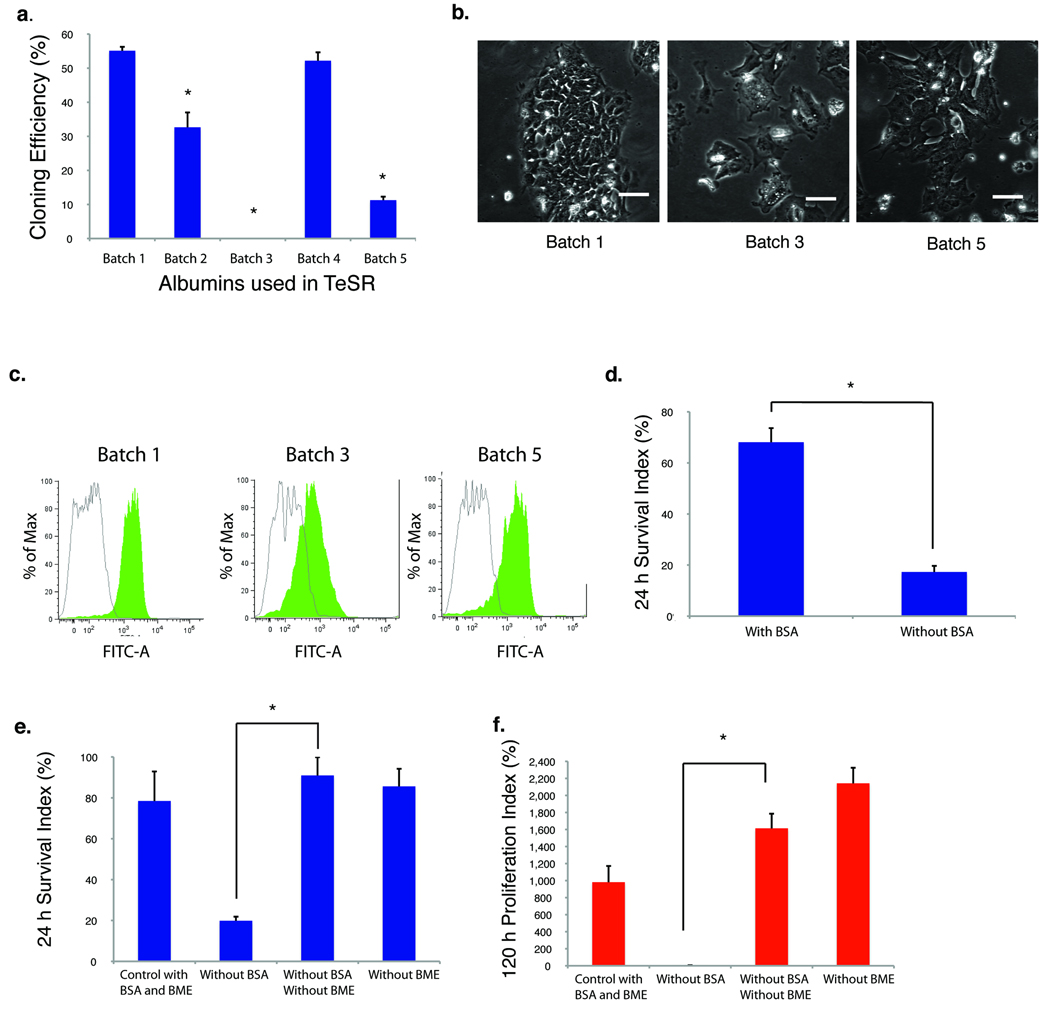
In addition to the components of DMEM/F12 (Supplementary Table 1), TeSR has 18 components, the major protein component being BSA (~1% in weight). Tremendous variability exists in the ability of different batches of BSA to support the undifferentiated proliferation of human ES cells (Fig. 1a, b, c). The absence of several growth factors (TGFβ, LiCl, GABA and Pipecolic acid; see TeSR core in Supplementary Table 1) did not affect short-term cell survival and proliferation (Supplementary Fig. 1a). However, the removal of BSA led to cell death of dissociated human ES cells (Fig. 1d). This suggests that either BSA contributes directly to ES cell survival, or that BSA counteracts the toxic effects of other medium components. Pair wise dropout experiments revealed that human ES cells survived without BSA when β-mercaptoethanol (BME) was also removed from the medium (Fig. 1e), and that cells subsequently proliferated well in such conditions (Fig. 1f).
Fig. 1. Albumin is not required for human ES cell culture.
(a) The plots show cloning efficiency (day 5) of H1 ES cells plated at clonal density (500 cells/well) with ROCK inhibitor on a Matrigel-coated surface in TeSR media made with different commercial batches of BSA. (*p < 0.05, n = 3, relative to BSA batch 1, the only batch supporting long-term self-renewal). (b) Micrographs show colonies after 72 hours of incubation in TeSR with different batches of BSA. Scale bar = 70 µm. (c) FACS analysis of OCT4 expression of cells depicted in (b). Green peak: OCT4 staining; unshaded peak: mouse IgG control. (d) The plots show survival of dissociated H1 ES cells 24 hours after plating into simplified medium (TeSR core) with or without BSA (Supplementary Table 1, Supplementary Fig. 1A). The “Survival Index” represents the number of surviving cells divided by the number of input cells (*p < 0.05, n = 3). (e) Plots show cell survival of dissociated cells 24 hours after plating into TeSR-based medium with the indicated combinations of BSA and β-mecaptoethanol (BME). (The control medium is TeSR core, Supplementary Table 1). (*p < 0.05, n = 3). (f) In the cell culture described in (e), cell proliferation was measured 120 hours after plating (*p < 0.05, n = 3). The “Proliferation Index” represents the cell number at a specific time point divided by the number of input cells at time 0.
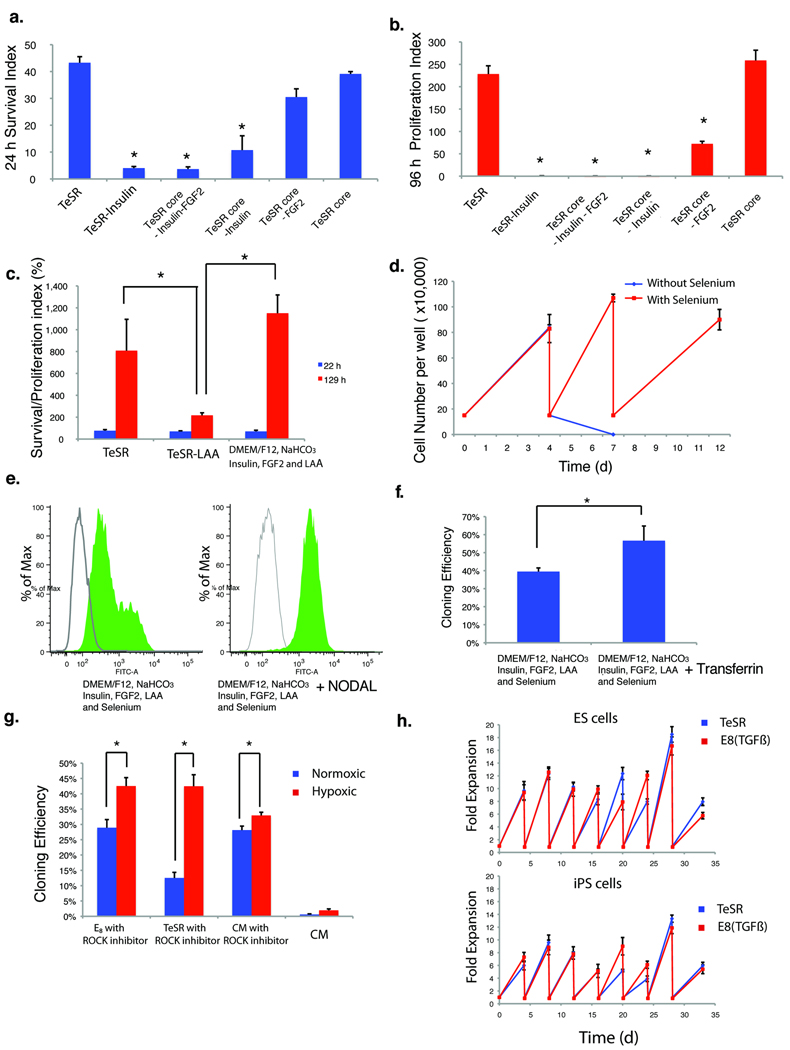
We then reexamined the other medium components of TeSR in the absence of BSA and BME. Insulin and FGF2 remained important for cell survival and proliferation (Fig. 2a and 2b). In these conditions, we found that L-ascorbic acid (Vitamin C, LAA) promoted ES cell proliferation (Fig. 2c), and that selenium was essential for sustained culture expansion (Fig. 2d). A comparative analysis of 12 different base media failed to identify a base medium that performed better than DMEM/F12 (Supplementary Fig. 1c). We found that human ES and iPS cells could be expanded in a simple medium consisting only of insulin, FGF2, L-ascorbic acid, and selenium in DMEM/F12 with pH adjusted with NaHCO3, but that cultures were often prone to sporadic differentiation after long-term passage (Fig. 1e). The addition of NODAL (100 ng ml−1) or TGFβ (2 ng ml−1) increased NANOG expression levels (Supplementary Fig. 1b) and led to consistent long-term culture stability of both human ES and iPS cells (Fig. 2e and Supplementary Figs. 1d, e). The inclusion of either a ROCK inhibitor (HA100 or Y27632)12 or blebbistatin13 improved initial survival and supported a high cloning efficiency (Supplementary Fig. 1f, g), which was further increased by the addition of transferrin (Fig. 2f) and by culture in hypoxic conditions (Fig. 2g).
Fig. 2. Essential media components for human ES cell survival and proliferation.
(a) The plots show survival of dissociated H1 ES cells 24 hours after plating into the indicated media (Supplementary Table 1) on Matrigel-coated plates. (*p < 0.05, n = 3, relative to survival in TeSR) (b) The plots show proliferation of cells from (a) 96 hours after plating and culture in the same media with daily media change.(*p<0.05, n=3, relative to proliferation in TeSR) (c) The plots show survival (blue) and proliferation (red) of human ES cells dissociated and plated in the indicated media, 22 h and 129 h after plating, respectively. (*p < 0.05, n = 3). (d) The plots show cell numbers over time of H1 cells maintained in defined media (DMEM/F12, NaHCO3, Insulin, FGF2 and LAA) for multiple passages with or without selenium. 150,000 starting cells were seeded in each passage on day 0, 4 and 7. (e) FACS analysis of OCT4 expression in H1 cells grown in the indicated media for 4 passages. Green peak, OCT4 staining; unshaded peak: mouse IgG control. (f) The plots show cloning efficiency of human foreskin iPS cells29 grown in the indicated defined media, with ROCK inhibitor HA100, in hypoxic conditions (*p < 0.05, n = 3). Similar results were obtained for human ES and other iPS cell lines. (g) The plots show cloning efficiency of ES cells in the indicated media under hypoxic (red) and normoxic (blue) conditions. (*p < 0.05, n = 3). Similar results were obtained from iPS cell lines. (h) The plots show fold expansion of ES (H1) and iPS cells15 cultured under hypoxic conditions in the indicated media. 200,000 cells were plated at each passage.
After this optimization, the final E8 medium consisted just of insulin, selenium, transferrin, L-ascorbic acid, FGF2, and TGFβ (or NODAL) in DMEM/F12 with pH adjusted with NaHCO3. This simplified medium supported undifferentiated proliferation of both human ES and iPS cells comparably to TeSR (Fig. 2h), and maintained pluripotency markers and normal karyotypes for over 25 passages and for more than three months in two ES and five iPS cell lines (Supplementary Fig. 1h, i). Both ES and all five iPS cell lines tested also formed teratomas in immunocompromised mice. Global gene expression also demonstrated that cells maintained in E8 media have an expression pattern similar to cells maintained in TeSR (Supplementary Fig 3a). Even without albumin, E8 medium is suitable for most common cell culture practices, and has a two week shelf life in regular storage (Supplementary Fig. 4).
Vitronectin-coated surfaces support growth in E8 media
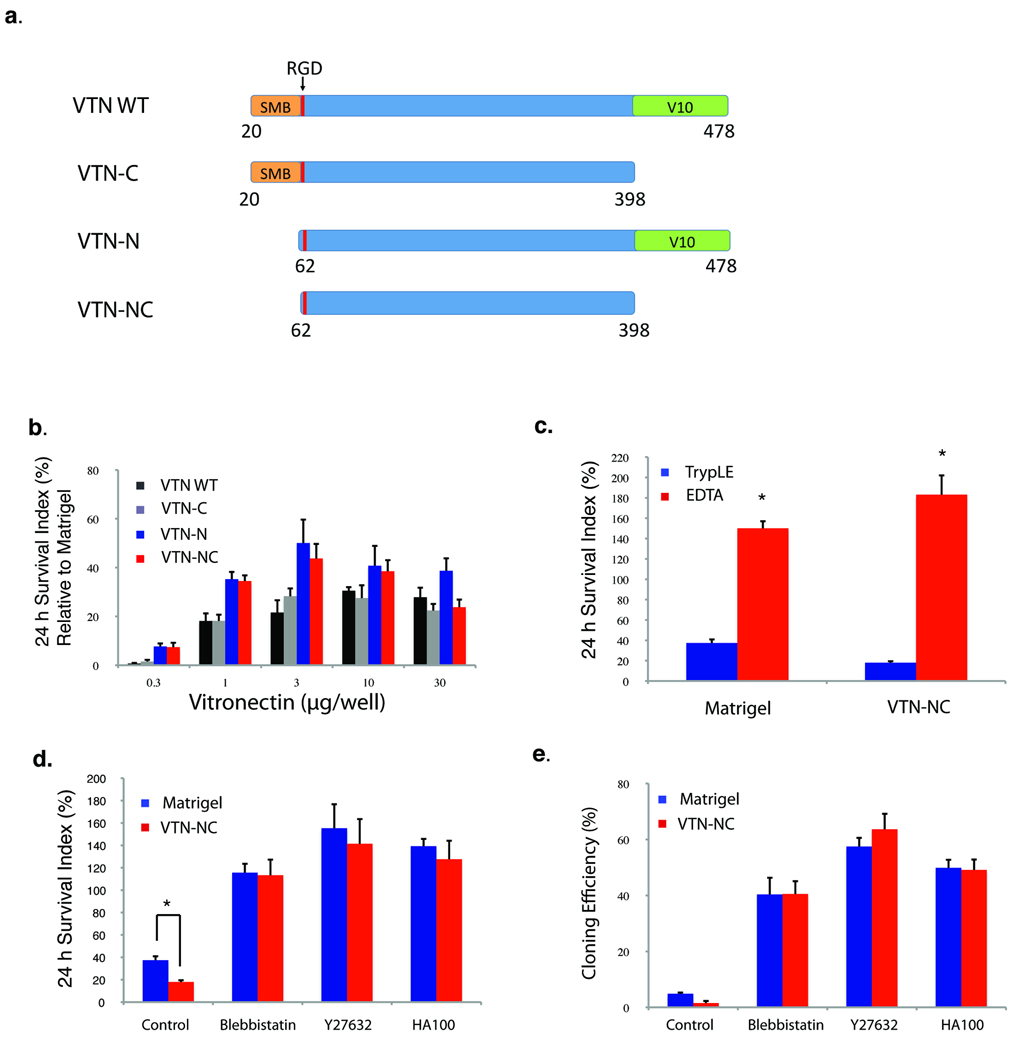
Multiple matrix proteins, such as laminin, vitronectin and fibronectin, support human ES cell growth. Synthetic surfaces have also been developed for human ES cells. Most of these materials are too expensive for large-scale usage. Because vitronectin is relatively easy to over-express and purify14, we tested several vitronectin variants and identified two (VTN-NC and VTN-N) that supported human ES cell attachment and survival better than wild type in E8 medium (Fig. 3a, b). We used VTN-NC for the rest of this report, demonstrating that VTN-NC supports initial attachment and survival of human ES cells well in E8 medium when cells were passaged in small clumps using EDTA (see Methods), but less efficiently than Matrigel when cells were passaged as single cells (Figure 3c). However, when a ROCK inhibitor or blebbistatin was added, VTN-NC supported both initial human ES cell survival and cloning efficiency as effectively as Matrigel (Fig. 3d, e).
Fig. 3. Vitronectin coated surfaces support human ES and iPS cells cultured in E8 medium.
(a) The schematics show the four vitronectin variants (VTN-WT, VTN-C, VTN-N, VTC-NC) that were expressed and purified as coating materials for ES cells. SMB (Somatomedin B domain30) and V10 are functional domains31 of wild type vitronectin; RGD, integrin-binding site. (b) The plots show cell survival on the four vitronectin variants in E8 media after 24 hours. The survival index was normalized to cell survival on Matrigel (*p < 0.05, n = 3). (c) The plots show cell survival after 24 hours on VTN-NC (right) and matrigel (left) in E8(TGFβ) media after passaging with EDTA (red) and TrypLE (blue) (*p < 0.05, n = 3). (d, e) The plots show cell survival after 24 hours (d) or cloning efficiency (e) on VTN-NC (red) and matrigel (blue) surfaces in the presence of the indicated small chemical inhibitors (*p < 0.05, n = 3).
E8 medium improves the efficiency of iPS cell derivation
Serum-containing media are often used during the derivation of dermal fibroblasts from patient biopsy samples, so we next examined whether E8 medium and vitronectin coated surfaces could be used both for this purpose and for subsequent iPS cell derivation. FGF2 alone, which is already present in the E8 medium, supported the proliferation of dermal fibroblasts as effectively as any other FGF we tested, but not as well as fetal bovine serum (Supplementary Fig. 2a). The addition of hydrocortisone and TGFβ further improved fibroblast growth (Supplementary Fig. 2b).
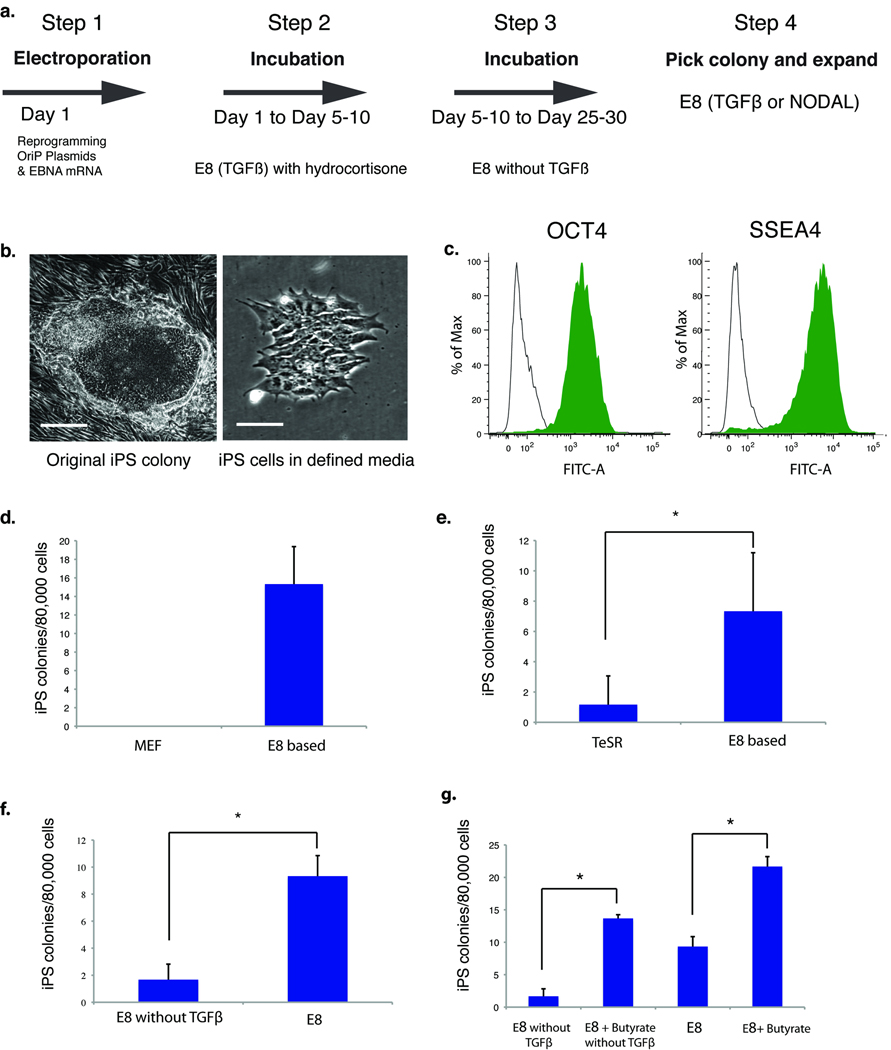
We then tested whether these defined fibroblast culture conditions would support subsequent iPS cell derivation by an episomal approach15 from established fibroblast cell lines. We found that co-electroporation of EBNA mRNA with oriP plasmids greatly improved subsequent expression by the oriP plasmids (Supplementary Fig. 2c)16, and thus EBNA mRNA was also subsequently used during reprogramming with previously described oriP vectors15. Hydrocortisone was added to E8 initially to facilitate robust fibroblast proliferation, and was later (5–10 days) removed, along with TGFβ, to inhibit fibroblast overgrowth of the iPS cells (Fig. 4a). iPS cell colonies (Fig. 4b) intermingled with partially reprogrammed colonies (Supplementary Fig. 2d), appeared at 20–30 days after transfection. Individual iPS cell colonies were selected and subsequently passaged and maintained in complete E8 medium for long-term expansion. Because fibroblast overgrowth was reduced by hydrocortisone and TGFβ withdrawal, we were able to obtain iPS cell colonies without secondary passage (Supplementary Table 2). Individual iPS cell clones were confirmed to be integration-free by PCR and, after about twenty passages, cells had normal karyotypes, expressed pluripotency markers (Fig. 4c and Supplementary Fig. 2e), and formed teratomas. E8-based medium supported improved reprogramming efficiencies compared to co-culture with mouse fibroblast feeder cells, FBS-containing medium, or TeSR medium (Figs. 4d, e, and Supplementary Fig. 2f).
Fig. 4. Reprogramming fibroblast cells in fully defined condition.
(a) Schematic of procedure to derive integration-free iPS cells from fibroblast cells in fully defined conditions. (b) The micrographs show a typical iPS colony 25 days after reprogramming (left, scale bar = 200 µm.) and prior to picking, and after first passage (right, scale bar = 25 µm.). This particular iPS clone was maintained in E8 (NODAL) on Matrigel. (c) FACS analysis of OCT4 and SSEA4 of a typical iPS cell line derived from foreskin fibroblasts and maintained in E8 for 20 passages. Green peak: OCT4 staining; unshaded peak: mouse IgG control. (d – g) Human foreskin fibroblasts were reprogrammed in the indicated media (see Methods for details). The plots show reprogramming efficiency scored after 30 days. In (d), sodium butyrate (100 µM) was added to both conditions to improve efficiency. (In (d, f, g), *p < 0.05, n = 3, experiments were repeated twice (d, f) and five times (g), with similar results. In (e), due to the inconsistency of reprogramming efficiency in TeSR, four independent experiments were each repeated three times, *p < 0.05, n = 12).
TGFβ is a common growth factor in defined fibroblast growth media17,18, and its presence in the first 6–8 days increased the number of iPS colonies compared with reprogramming in the absence of TGFβ (Fig. 4f). It was previously reported that TGFβ has an inhibitory effect when present at later stages of reprogramming19, consistent with our observation that fibroblasts tended to overgrow and inhibit or obscure the emergence of iPS colonies when TGFβ was present throughout the reprogramming procedure. To slow down fibroblast proliferation and to allow time for the emergence of iPS cells without fibroblast overgrowth, TGFβ and hydrocortisone were both removed at 5–10 days until iPS cell colonies were selected and passaged into complete E8 with TGFβ or NODAL (Fig. 4a). Similar to the findings of others20–22, we found that butyrate further improved the efficiency of iPS cell colony formation, which had an additive effect with TGFβ (Fig. 4g).
We have used E8 medium to derive iPS cell lines with both episomal and lentiviral methods from five banked fibroblast cell lines from commercial sources, obtained from individuals ranging in age from neonatal to 38 years of age (Supplementary Table 2). Using an episomal approach, all these fibroblast cell lines tested were successfully reprogrammed at efficiencies ranging from four to 200 iPS cell colonies per 106 fibroblasts initially transfected. iPS cell colonies were counted on the initial transfected fibroblast plates without additional passage, so the numbers represent independent clones. Global gene expression also demonstrated that cells derived in E8 media have an expression pattern similar to ES cells and iPS cell derived on feeder cells (Supplementary Fig 3b–d).
Derivation of human iPS cells in defined conditions from biopsy
We next developed fully defined conditions for the processing of patient dermal biopsies. We found that vitronectin (VTN-NC) has a stimulatory effect on the growth of adult fibroblast cells (Fig. 5a). We also formulated a defined fibroblast medium based on the E8 medium (Fig. 5b) to further promote fibroblast proliferation from fresh biopsy samples. Combining all of these conditions, we derived integration-free iPS cells in totally defined conditions from the initial patient biopsy.
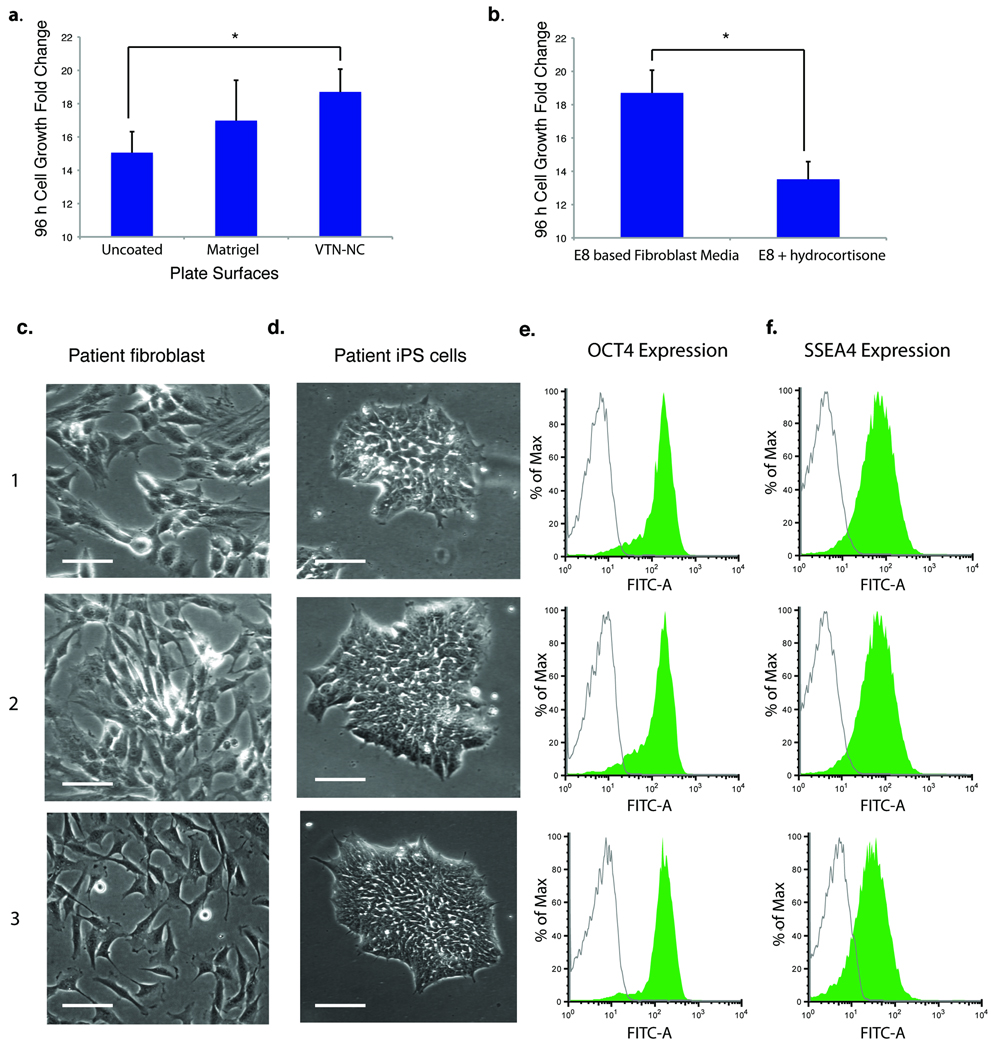
Fig. 5. Derivation of human iPS cells directly from biopsy samples in chemically defined conditions.
(a, b) The plots show growth (fold change in cell number) of adult fibroblast cells (a) plated onto the indicated plate coating materials in E8-based fibroblast media (see Methods) and counted after 4 days (*p < 0.05, n = 3), or (b) cultured in the indicated media on vitronectin (see Methods) and counted 96 hours after plating (*p < 0.05, n = 3). (c) The micrographs show three fibroblast cell lines derived from skin biopsies in defined fibroblast medium on vitronectin-coated plates. Scale bar = 100 µm. (d) The micrographs show representative iPS colonies obtained by reprogramming fibroblasts from (c) according to the procedure in Figure 4a, in the presence of butyrate. Colonies are shown after multiple passages in E8 (TGFβ). Scale bar = 100 µm. (e and f) FACS analysis of pluripotency markers OCT4 and SSEA4 in iPS cells after 10 passages.
The initial skin biopsy was treated with recombinant enzymes, and the dermis portion was dissociated to derive skin fibroblast in E8-based fibroblast medium (see Methods). E8-based medium on vitronectin effectively supported initial fibroblast derivation from biopsy samples with robust proliferation (Fig. 5c). After expansion, the fibroblasts were subjected to the episomal reprogramming procedure described above (Fig. 4a). iPS colonies appeared after around 25 days, and were passaged onto vitronectin-coated surfaces in E8 (Fig. 5d). Individual iPS cells were clonally expanded under hypoxic condition on vitronectin-coated plates with a ROCK inhibitor at splitting, and were subsequently screened for lack of vector integration. Pluripotency markers OCT4 and SSEA4 were maintained at high levels in iPS cells derived in these defined conditions (Fig. 5e, f).
Fibroblast cells freshly isolated from biopsy samples in E8-based fibroblast medium on vitronectin were consistently reprogrammed at a higher efficiency (60 to ~1,000 iPS cell colonies per 106 transfected fibroblasts) than the established cell lines we had previously obtained from commercial sources (four to 30 iPS cell colonies per 106 transfected fibroblasts, excluding the neonatal foreskin fibroblasts; see Supplementary Table 2). In the experiments performed in Figure 5, individual clones (10–24 clones for each patient sample) were expanded and analyzed for genomic integration after iPS cell derivation. Two integration-free clones of each patient sample were confirmed with normal karyotypes and then injected into immune-deficient mice. Teratomas were formed in all tested cell lines.
DISCUSSION
During the course of these studies, we were most surprised by what human ES and iPS cells do not need, and by how challenging it is to remove something from a medium once its inclusion becomes routine and accepted. Albumin, for example, has had many cell culture roles attributed to it, from lipid carrier and blocking reagent to physical protection against sheer force11,23. However, our results clearly demonstrated that the dominant role of BSA in TeSR is to prevent a toxic effect of β-mercaptoethanol and that, in the absence of β-mercaptoethanol, BSA is not required (Fig. 1e, f). β-mercaptoethanol’s inclusion in our previous TeSR medium for human ES cells traces its origin back to a positive effect on cloning efficiency of mouse EC cells described in 197824. Because of those results, β-mercaptoethanol was included in mouse ES cell media25 and subsequently in early human ES cell media1.
The finding of an interaction between BSA and β-mercaptoethanol underscores the challenge of optimizing media, as it is not sufficient to just examine the effects of the addition or subtraction of individual components. We thus reexamined all other components of TeSR medium made without BSA or β-mercaptoethanol and found that several (i.e., pipocolic acid, GABA, LiCl, chemically defined lipids, trace elements B, trace elements C, glutathione, and additional Thiamine and L-Glutamine) no longer had a positive effect. In spite of the much greater simplicity of E8 medium, it supports the long-term undifferentiated proliferation of both human ES and iPS cells comparably to TeSR (Fig. 2h).
E8 medium supports higher reprogramming efficiencies for both viral and episomal approaches (Fig. 4d. e, and Supplementary Fig 2f). In every attempt, from both established, banked fibroblasts (5) and from independent biopsy samples (4), integration free iPS cell lines were successfully derived in E8-based medium on vitronectin (Supplementary Table 2). With the exception of the banked neonatal fibroblasts that reprogrammed at a high efficiency, the efficiencies obtained directly from biopsy samples grown in E8-based medium were consistently higher than those of established cell lines obtained from commercial sources, suggesting that fibroblast passage history is important. Although these efficiencies could be further improved, the number of clones obtained per biopsy sample in these defined conditions using an episomal approach (60 - ~1,000 iPS cell colonies / 106 fibroblasts) already greatly exceeds what is typically used for subsequent characterization.
It is important to distinguish mechanistically between culture components that improve reprogramming itself, and those that merely improve the survival and proliferation of the resulting iPS cell clones19,26. Because reprogramming occurs over time, it is often difficult to cleanly distinguish between these two effects, and indeed, they do not need to be mutually exclusive. We found, for example, that L-ascorbic acid promotes ES cell proliferation and expansion (Fig. 2c, d). L-ascorbic acid has previously been reported to promote reprogramming27, but our results are consistent with L-ascorbic acid only promoting iPS cell growth and survival, with reprogramming itself being driven by other factors. It is also possible that ascorbic acid leads to epigenetic modifications essential to both reprogramming and cell survival in serum-free culture conditions28. Similarly, most derivation procedures have a splitting step before iPS colonies are visible, and reprogramming efficiency is often calculated based on the final iPS colony number. When splitting is required for the emergence of iPS cells in some situations (e.g. fibroblast overgrowth) it can lead to confusion between factors that improve reprogramming itself, and those that improve survival after splitting. Importantly, if the iPS cell colony number is only assessed after splitting, an independent clonal origin of distinct iPS cell colonies cannot be confirmed, making calculations of true reprogramming efficiency problematic. Because our method removes TGFβ and hydrocortisone from the defined medium after 5–10 days, fibroblast over-growth is inhibited and truly independent iPS cell clones can be isolated and counted without splitting.
Because the E8 medium reduces medium cost and simplifies quality control, it is now used for all routine culture of both human ES and iPS cells in this laboratory. This simplified, defined medium also provides a much cleaner background for examining specific pathways in self-renewal, cell death, and differentiation13 and it supports substantially improved reprogramming efficiencies. Although we have only demonstrated improved efficiencies for viral and episomal reprogramming approaches, these conditions should be equally useful for other non-integrative reprogramming approaches. Finally, since E8 medium is highly defined, it should also help facilitate the transfer of basic research on human pluripotent stem cells to the clinic.
METHODS
Human ES Cell Culture
Human ES cells (H1 and H9) were usually maintained in specific media on Matrigel-coated tissue culture plates32. Cells were passaged routinely with EDTA as described previously13. Briefly, cells were washed twice with PBS/EDTA medium (0.5 mM EDTA in PBS, osmolarity 340 mOsm), then incubated with PBS/EDTA for 5 minutes at 37°C. PBS/EDTA was removed, and cells were washed off swiftly with a small volume of corresponding media.
E8 media composition: Media contained DMEM/F12, L-ascorbic acid-2-phosphate magnesium (64 mg/l), sodium selenium (14 µg/l), FGF2 (100 µg/l), insulin (19.4 mg/l), NaHCO3 (543 mg/l) and transferrin (10.7 mg/l), TGFβ1(2 µg/l) or NODAL (100 µg/l). Osmolarity of all media was adjusted to 340 mOsm at pH7.4. All the media were stored at 4°C, and were used within 2 weeks of production. L-ascorbic acid-2-phosphate magnesium is the stable form of L-ascorbic acid in cell culture.
Reagents
Reagents: HA100 (Sigma), Blebbistatin (Sigma), Y27632 (Tocris), Sodium Butyrate (Sigma), Hydrocortisone (Sigma) Sodium Bicarbonate (Sigma), L-Ascorbic acid 2 phosphate magnesium salt (Sigma), Sodium Selenite (Sigma), Holo-transferrin (Sigma), DMEM/F12 (Invitrogen), Insulin (Sigma), TGFβ1 (R&D), NODAL (R&D), and FGF232.
BSA: Batch 1- Sigma A2153-066K0738, Batch 2-Sigma A2153 049K1585, Batch 3 – Sigma A7906-069K1653, Batch 4-Sigma A2153-018K0665, Batch 5-Hyclone SH30574-090205068A.
Antibodies: anti-OCT4 (Santa Cruz), anti-SSEA4 (Millipore) and Alexa-488 anti-mouse IgG (Invitrogen).
Survival Assay
Survival assays followed the procedure previously described unless specified13. All experiments were done on 12-well plates, usually in triplicate for each treatment. Prior to the addition of cells, 500 µl of medium was loaded into each well. Cells were dissociated with TrypLE (Invitrogen) for 5 minutes or until fully detached from the plate, neutralized with equal volumes of media, counted, washed, and diluted to 300,000 to 1,000,000 cells/ml−1, and 100 µl of cells were added into each well. Plates were normally placed into 5% O2 and 10% CO2 (hypoxic condition), 37°C incubator, unless specified. Small chemical compounds or proteins were added or washed away according to the specified procedure. At each time point, cells were again dissociated with 0.4 ml TrypLE, neutralized with equal volumes of 10% FBS in DMEM, harvested with pipettes, and counted by flow cytometry. As an internal control, 5,000 Count-bright beads (Invitrogen) were added to each sample, and usually ~200 beads were counted for each sample. For proliferation experiments, media were changed daily till the day of analysis, and cells were counted as described above.
Cloning Assay
The cloning assay was described previously13. Briefly, triplicates were prepared in a 12-well plate format for each treatment. Prior to the addition of cells, 500 µl media were loaded in each well. Cells were dissociated with TrypLE for 5 minutes or until fully detached from the plate, neutralized with equal volumes of basic media, counted, washed and then diluted to 5,000 cells/ml−1. Finally, a 100µl suspension (500 cells) was added into each well. Plates were then placed into 5% O2 and 10% CO2 (hypoxic condition), 37°C incubator unless otherwise specified. Small chemical compounds or proteins were added or washed away according to the specified procedure. Media were changed every 1–2 day(s) if not specified. After 5–6 days, colonies were stained with an APS kit using a standard procedure (Vector Lab) and counted. Chemical concentrations for cloning assays in this report: 10 µM Blebbistatin, 10 µM Y27632, or 10 µM HA100.
Oxygen and Carbon Dioxide Control
Cells were maintained in a water-jacketed CO2 incubator (Forma Series II), where O2 and CO2 were controlled through the injection of nitrogen and carbon dioxide. There are two conditions discussed in this paper: hypoxic condition (5% O2, 10% CO2, red column) and normoxic condition (5% CO2). The hypoxic condition was used for most experiments in this report unless otherwise specified.
Statistics
In cloning or survival assays, triplicate data points were obtained for each condition. A t-test was performed to calculate p-values for the difference between the means of the experimental conditions and control. Error bars in each figure represents the standard error of three individual experiments. Each finding was confirmed by independent biological replicates, unless specified.
iPS Cell Derivation in Defined Condition
Episomal plasmids and methods were described previously15. Plasmid combination #19 (pEP4-E-O2S-E-T2K, pEP4-E-O2S-E-N2K and pCEP4M2L) was used for most reprogramming unless mentioned otherwise. Plasmids and EBNA mRNA were electroporated into fibroblast cells on Amaxa apparatus according to company instructions. One million cells were used in each electroporation, which were then plated into two 6-well plates. E8 + hydrocortisone media were used for the first 5–10 days, according to cell survival and proliferation after electroporation. When confluency was reached ~20%, hydrocortisone was removed. ES-like iPS cell colonies usually appear after ~25 days. Cells were then picked into individual wells with E8 (TGFβ or NODAL). Cells were passaged for ~ 15 passages before subcloning with Y27632 on Matrigel or vitronectin.
Operation for Skin Biopsy
The area of biopsy on the left arm was prepped in a sterile fashion using 70% ethanol, 1% lidocaine with epinephrine (1:100,000 dilution) was used for local anesthesia, and the skin biopsy was obtained with a 4 mm punch. All specimens were obtained following standard IRB protocol approved by University of Wisconsin-Madison. For underage patients, informed written consent was obtained from the participant's guardian.
Fibroblast Derivation in Defined Conditions
After a skin punch biopsy was obtained from the patient, it was maintained in defined fibroblast media (DMEM/F12 (Invitrogen), 64 mg/l L-Ascorbic acid 2 phosphate magnesium salt (Sigma), 20 mg/l insulin, 14 µg/l sodium selenite, 10 mg/l transferrin (Sigma), 1 unit ml−1 thrombin (Sigma), 100 nM hydrocortisone (Sigma), 100 µg/l EGF, 100 µg/l hFGF2 and 2 µg/l TGFβ), and later immersed in TrypLE Select (Invitrogen) enzyme solution at 4°C overnight. The epidermis was then peeled off the dermis, and the dermis was cut into small pieces before being incubated in enzyme mix (HEPES containing RPMI with 1 mM pyruvate, 10 mg ml−1 Collagenase (Sigma), 0.5 AMP ml−1 Hyaluronidase (Sigma) and 140 units ml−1 DNase I (Roche)) for 30 minutes at room temperature. Cells were spun by centrifugation and washed twice with fibroblast media, and then were plated onto vitronectin-coated tissue culture plates with fibroblast media. After 3–7 days, fibroblast cells emerged and proliferated. TypLE was used for regular splitting to expand the cells prior to reprogramming.
Derivation of iPS Cells from Fibroblast Cells in Defined Conditions
A schematic diagram of the procedure is described in Figure 4. Episomal plasmids and methods were described previously15. Plasmid combination #19 (pEP4-E-O2S-E-T2K, pEP4-E-O2S-E-N2K and pCEP4M2L) was used for most reprogramming unless mentioned otherwise. Plasmids and EBNA mRNA were electroporated into fibroblast cells on Amaxa apparatus according to company instructions. One million cells were used in each electroporation, which were then plated into two 6-well plates. E8 + hydrocortisone media were used for the first 5–10 days, according to cell survival and proliferation after electroporation. When confluency was reached ~20%, hydrocortisone was removed. ES-like iPS cell colonies usually appear after ~25 days. Cells were then picked into individual wells with E8 (TGFβ or NODAL). Cells were passaged for ~ 15 passages before subcloning with Y27632 on Matrigel or vitronectin.
EBNA mRNA for Electroporation
EBNA mRNA was synthesized and purified as previously reported16. A plasmid containing oriP sequence and GFP gene was used to evaluate the improvement made by EBNA mRNA during transfection. The electroporation was performed with Bio-Rad Gene Pulser II (Supplementary Fig. 2c). Unless specifically mentioned, all electroporation experiments mentioned in this paper used Amaxa Nucleofector with Human Dermal Fibroblast Nucleofector Solution (Lonza). Program U-020 was used for foreskin fibroblast, and program U-023 was used for adult or patient samples.
Vitronectin Expression and Purification
Coding sequences of human vitronectin and various truncation mutants (Fig. 3a) were amplified from a cDNA clone purchased from Origene and cloned into NdeI and BamHI sites of pET3c (Novagen). All constructs were verified by sequencing. Protein expression was done in Rosetta2 (DE3) pLysS cells (Novagen) using Magic Media (Invitrogen) at 37°C for 24 hours. Purification of vitronectin and its variants was done using a protocol modified from a previous report33. Briefly, the E coli pellet was resuspended in PBS and lysed with FastBreak cell lysis reagent (Promega). Insoluble material was pelleted by centrifugation at 10,000 g. The pellet was washed once with PBS + 0.5 M NaCl and then solubilized in urea buffer (8 M urea, 20 mM Tris pH 7.6, 150 mM NaCl and 3 mM DTT). Urea solubilized vitronectin was loaded onto a Heparin sepharose column and the column was then washed extensively with urea buffer. Protein was eluted with urea buffer + 500 mM NaCl and then dialyzed into PBS overnight.
Global Gene Expression Analysis
The expression levels from ES, iPS and foreskin samples were measured by RNA SEQ via Illumina’s Genome Analyzer GAIIx. The RNA SEQ libraries were built following Sengupta et al34. The sequencing results were then normalized via the RSEM package by Bo et al35. Transcripts Per Million (TPM) values were obtained to measure the RNA expression. Spearman Correlation coefficient (R values) was calculated in order to get a general overview of the expression profiles from each sample.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by The Charlotte Geyer Foundation, the Morgridge Institute for Research, NIH grant UO1ES017166 (to J.A.T.) and NIH contract RR-05-19 (to J.A.T.). We thank Krista Eastman for editorial assistance. We also thank Colin Dewey, Ron Stewart, and Angela Elwell for their assistance with gene expression analysis.
Footnotes
AUTHOR’S CONTRIBUTIONS
G.C. and J.A.T. conceived the experiment; G.C., D.R.G., J.M.B., K.S.O, and S.E.H. performed the reprogramming; Z.H., G.C., and N.E.P. produced vitronectin; G.C., D.R.G., J.M.B., N.R.D., G.O.L., and J.A.B. performed the cell culture test; G.C., M.D.P., and R.W. derived fibroblasts; J.M.C.T. obtained the skin biopsy; V.R. and G.C. analyzed global expression. G.C. and J.A.T. wrote the paper.
COMPETING INTERESTS STATEMENT
The authors declare competing financial interests: J.A.T. is a founder, stockowner, consultant and board member of Cellular Dynamics International (CDI). He also serves as scientific advisor to and has financial interests in Tactics II Stem Cell Ventures.
REFERENCES
- 1.Thomson JA, et al. Embryonic stem cell lines derived from human blastocysts. Science. 1998;282:1145–1147. doi: 10.1126/science.282.5391.1145. [DOI] [PubMed] [Google Scholar]
- 2.Reubinoff BE, Pera MF, Fong CY, Trounson A, Bongso A. Embryonic stem cell lines from human blastocysts: somatic differentiation in vitro. Nat Biotechnol. 2000;18:399–404. doi: 10.1038/74447. [DOI] [PubMed] [Google Scholar]
- 3.Yu J, et al. Induced pluripotent stem cell lines derived from human somatic cells. Science. 2007;318:1917–1920. doi: 10.1126/science.1151526. [DOI] [PubMed] [Google Scholar]
- 4.Takahashi K, et al. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007;131:861–872. doi: 10.1016/j.cell.2007.11.019. [DOI] [PubMed] [Google Scholar]
- 5.Park IH, et al. Reprogramming of human somatic cells to pluripotency with defined factors. Nature. 2008;451 doi: 10.1038/nature06534. 141-U141. [DOI] [PubMed] [Google Scholar]
- 6.Lowry WE, et al. Generation of human induced pluripotent stem cells from dermal fibroblasts. P Natl Acad Sci USA. 2008;105:2883–2888. doi: 10.1073/pnas.0711983105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Akopian V, et al. Comparison of defined culture systems for feeder cell free propagation of human embryonic stem cells. In Vitro Cell Dev Biol Anim. 2010;46:247–258. doi: 10.1007/s11626-010-9297-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sun N, et al. Feeder-free derivation of induced pluripotent stem cells from adult human adipose stem cells. Proc Natl Acad Sci U S A. 2009;106:15720–15725. doi: 10.1073/pnas.0908450106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Skottman H, Hovatta O. Culture conditions for human embryonic stem cells. Reproduction. 2006;132:691–698. doi: 10.1530/rep.1.01079. [DOI] [PubMed] [Google Scholar]
- 10.Akopian V, et al. Comparison of defined culture systems for feeder cell free propagation of human embryonic stem cells. In Vitro Cell Dev-An. 2010;46:247–258. doi: 10.1007/s11626-010-9297-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Garcia-Gonzalo FR, Izpisua Belmonte JC. Albumin-associated lipids regulate human embryonic stem cell self-renewal. PLoS One. 2008;3:e1384. doi: 10.1371/journal.pone.0001384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Watanabe K, et al. A ROCK inhibitor permits survival of dissociated human embryonic stem cells. Nature Biotechnology. 2007;25:681–686. doi: 10.1038/nbt1310. [DOI] [PubMed] [Google Scholar]
- 13.Chen G, Hou Z, Gulbranson DR, Thomson JA. Actin-myosin contractility is responsible for the reduced viability of dissociated human embryonic stem cells. Cell Stem Cell. 2010;7:240–248. doi: 10.1016/j.stem.2010.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Braam SR, et al. Recombinant vitronectin is a functionally defined substrate that supports human embryonic stem cell self-renewal via alphavbeta5 integrin. Stem Cells. 2008;26:2257–2265. doi: 10.1634/stemcells.2008-0291. [DOI] [PubMed] [Google Scholar]
- 15.Yu JY, et al. Human Induced Pluripotent Stem Cells Free of Vector and Transgene Sequences. Science. 2009;324:797–801. doi: 10.1126/science.1172482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Howden SE, et al. Chromatin-binding regions of EBNA1 protein facilitate the enhanced transfection of Epstein-Barr virus-based vectors. Hum Gene Ther. 2006;17:833–844. doi: 10.1089/hum.2006.17.833. [DOI] [PubMed] [Google Scholar]
- 17.Assoian RK, Frolik CA, Roberts AB, Miller DM, Sporn MB. Transforming growth factor-beta controls receptor levels for epidermal growth factor in NRK fibroblasts. Cell. 1984;36:35–41. doi: 10.1016/0092-8674(84)90071-0. [DOI] [PubMed] [Google Scholar]
- 18.Roberts AB, et al. Type beta transforming growth factor: a bifunctional regulator of cellular growth. Proc Natl Acad Sci U S A. 1985;82:119–123. doi: 10.1073/pnas.82.1.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lin T, et al. A chemical platform for improved induction of human iPSCs. Nat Methods. 2009;6:805–808. doi: 10.1038/nmeth.1393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liang G, Taranova O, Xia K, Zhang Y. Butyrate promotes induced pluripotent stem cell generation. J Biol Chem. 2010;285 doi: 10.1074/jbc.M110.142059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mali P, et al. Butyrate greatly enhances derivation of human induced pluripotent stem cells by promoting epigenetic remodeling and the expression of pluripotency-associated genes. Stem Cells. 2010;28:713–720. doi: 10.1002/stem.402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ware CB, et al. Histone deacetylase inhibition elicits an evolutionarily conserved self-renewal program in embryonic stem cells. Cell Stem Cell. 2009;4:359–369. doi: 10.1016/j.stem.2009.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rothschild MA, Oratz M, Schreiber SS. Regulation of albumin metabolism. Annu Rev Med. 1975;26:91–104. doi: 10.1146/annurev.me.26.020175.000515. [DOI] [PubMed] [Google Scholar]
- 24.Oshima R. Stimulation of the clonal growth and differentiation of feeder layer dependent mouse embryonal carcinoma cells by beta-mercaptoethanol. Differentiation. 1978;11:149–155. doi: 10.1111/j.1432-0436.1978.tb00978.x. [DOI] [PubMed] [Google Scholar]
- 25.Martin GR. Isolation of a pluripotent cell line from early mouse embryos cultured in medium conditioned by teratocarcinoma stem cells. Proc Natl Acad Sci U S A. 1981;78:7634–7638. doi: 10.1073/pnas.78.12.7634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Xu Y, et al. Revealing a core signaling regulatory mechanism for pluripotent stem cell survival and self-renewal by small molecules. Proc Natl Acad Sci U S A. 2010;107:8129–8134. doi: 10.1073/pnas.1002024107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Esteban MA, et al. Vitamin C enhances the generation of mouse and human induced pluripotent stem cells. Cell Stem Cell. 2010;6:71–79. doi: 10.1016/j.stem.2009.12.001. [DOI] [PubMed] [Google Scholar]
- 28.Chung TL, et al. Vitamin C Promotes Widespread Yet Specific DNA Demethylation of the Epigenome in Human Embryonic Stem Cells. Stem Cells. 2010;28:1848–1855. doi: 10.1002/stem.493. [DOI] [PubMed] [Google Scholar]
- 29.Yu JY, et al. Induced pluripotent stem cell lines derived from human somatic cells. Science. 2007;318:1917–1920. doi: 10.1126/science.1151526. [DOI] [PubMed] [Google Scholar]
- 30.Seiffert D, Loskutoff DJ. Evidence that type 1 plasminogen activator inhibitor binds to the somatomedin B domain of vitronectin. The Journal of Biological Chemistry. 1991;266:2824–2830. [PubMed] [Google Scholar]
- 31.Hayman EG, Pierschbacher MD, Ohgren Y, Ruoslahti E. Serum spreading factor (vitronectin) is present at the cell surface and in tissues. Proc Natl Acad Sci U S A. 1983;80:4003–4007. doi: 10.1073/pnas.80.13.4003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ludwig TE, et al. Feeder-independent culture of human embryonic stem cells. Nature Methods. 2006;3:637–646. doi: 10.1038/nmeth902. [DOI] [PubMed] [Google Scholar]
- 33.Wojciechowski K, Chang CH, Hocking DC. Expression, production, and characterization of full-length vitronectin in Escherichia coli. Protein Expr Purif. 2004;36:131–138. doi: 10.1016/j.pep.2004.04.004. [DOI] [PubMed] [Google Scholar]
- 34.Sengupta S, et al. Highly consistent, fully representative mRNA-Seq libraries from ten nanograms of total RNA. Biotechniques. 2010;49:898–904. doi: 10.2144/000113556. [DOI] [PubMed] [Google Scholar]
- 35.Li B, Ruotti V, Stewart RM, Thomson JA, Dewey CN. RNA-Seq gene expression estimation with read mapping uncertainty. Bioinformatics. 2010;26:493–500. doi: 10.1093/bioinformatics/btp692. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.