Abstract
Caenorhabditis elegans AFF-1 and EFF-1 (CeFFs) proteins are essential for developmental cell-to-cell fusion and can merge insect cells. To study the structure and function of AFF-1, we constructed Vesicular Stomatitis Virus (VSV) displaying AFF-1 on the viral envelope, substituting the native fusogen VSVG. Electron microscopy and tomography revealed that AFF-1 formed distinct supercomplexes resembling pentameric and hexameric flowers on pseudoviruses. Viruses carrying AFF-1 infected mammalian cells only when CeFFs were on the target cell surface. Furthermore, we identified Fusion Family proteins (FFs) within and beyond nematodes and divergent members from the human parasitic nematode Trichinella spiralis and the chordate Branchiostoma floridae could also fuse mammalian cells. Thus FFs comprise an ancient family of cellular fusogens that can promote fusion when expressed on a viral particle.
Membrane fusion is critical for many biological processes such as fertilization, development, intracellular trafficking and viral infection (1–6). Current models of the molecular mechanisms of membrane fusion rely upon experimental and biophysical analyses performed on viral and intracellular, minimal, fusion-mediating machineries. However, how well these models correspond to the mechanisms of cell-cell fusion is unknown (4, 5). CeFFs were identified as C. elegans fusogens that are expressed at the time and place of cell fusion in vivo (7, 8). Expression of CeFFs is essential for developmental cell fusion via hemifusion and sufficient to fuse cells in vivo and in insect cell cultures (8–10).
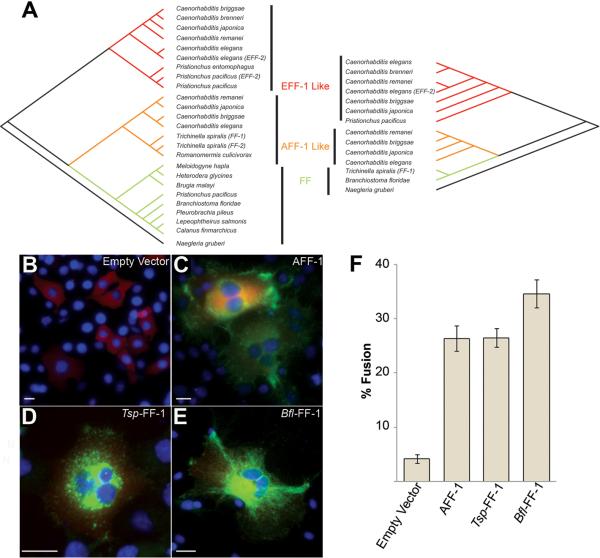
To identify putative FF members in other species we conducted sequence comparisons (4, 11). These comparisons yielded putative members in thirty-five nematodes, two arthropods (Calanus finmarchicus and Lepeophtheirus salmonis), a ctenophore (Pleurobrachia pileus), a chordate (Branchiostoma floridae) and a protist (Naegleria gruberi) (Fig. 1A). FF proteins are putative members of the `mostly beta sheet super family' and share a pattern of cysteines implying that they are conserved at the level of structure (Fig. S1).
Fig. 1.
A family of eukaryotic cell-cell fusogens: FF orthologs from two phyla fuse mammalian BHK cells
(A) Two trees produced using maximum parsimony analysis show phylogenic relationships of 25 taxa (left; based on the TGFβ-RI like domain, Fig. S1B) and 14 taxa (right; based on the full length extracellular domain), FF proteins are classified into three subgroups EFF-1-like (red), AFF-1-like (orange) and FF (green) (Table S1). Consistency, retention and composite indexes are in (11).
(B%#x2013;D) Immunofluorescence with anti-Flag antibodies (green), and nuclei DAPI staining (blue) on BHK cells transfected with (B) empty vector, (C) aff-1, (D) Tsp-ff-1 and (E) Bfl-ff-1. Cotransfection marker (red). Scale bars are 20 μm.
(F) Fusion index for BHKs expressing FF proteins and negative control (empty vector).
Data are means ± SE. Empty vector, n=14, aff-1, n=14, Tsp-ff-1, n=8, Bfl-ff-1, n=9; n represents number of experiments.
To determine whether divergent FFs maintained their function as fusogens through evolution we expressed FFs from the human parasitic nematode Trichinella spiralis (Tsp-ff-1) and the chordate B. floridae (Bfl-ff-1) in Baby Hamster Kidney (BHK) cells and compared their fusogenic activity to AFF-1 (Fig. 1B to F). These orthologs share 26% and 22% sequence identity with AFF-1, respectively. We observed by immunofluorescence 28±4% and 37±7% multinucleation in cells transfected with Tsp-ff-1 and Bfl-ff-1, compared to 26±2% and 4±3% multinucleation in controls transfected with aff-1 and empty vector, respectively (Fig. 1F; (11)). In addition, when we expressed the EFF-1 paralog from the nematode Pristionchus pacificus in C. elegans embryos we detected ectopic fusion of cells that normally do not fuse (Fig. S2). Thus FFs represent a conserved family of cellular fusogens.
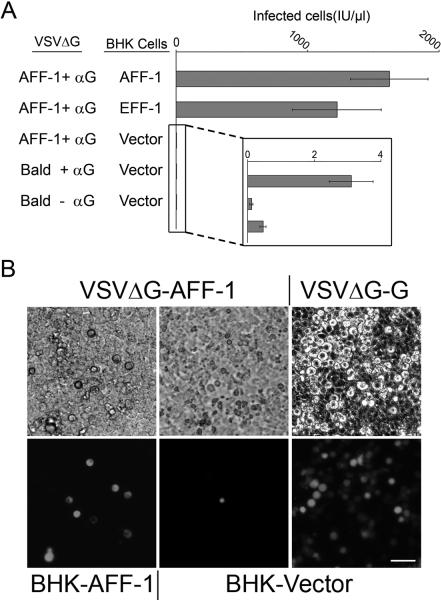
To explore whether FFs can functionally substitute for viral fusogens, we complemented VSVΔG pseudoviruses with AFF-1 (Fig. 2 and S3). Recombinant VSV named VSVΔG in which the glycoprotein G (VSVG) gene was replaced by a Green Fluorescent Protein (GFP) reporter was used initially to infect BHK cells over-expressing VSVG (11–16). The resulting VSVΔG-G viruses were capable of only a single round of infection, manifested by production of GFP. We achieved complementation with AFF-1 by VSVΔG-G infection of BHK cells expressing AFF-1 (BHK-AFF-1), which generated pseudotyped particles carrying the nematode fusogen (VSVΔG-AFF-1). We biochemically validated incorporation of AFF-1 into VSVΔG pseudotypes by SDS-PAGE, Coomassie, silver staining, immunoblotting and mass spectrometry (11). We found that the major proteins on VSVΔG-AFF-1 were the viral proteins N, P, L, M and AFF-1. For comparison we also analyzed VSVΔG-G and VSVΔG (Fig. S4 and Table S5). Infection of BHK-AFF-1 cells with VSVΔG-AFF-1 showed a 600-fold increase compared to infection of BHK control cells not expressing AFF-1 (Fig. 2A). Although infection due to residual VSVG complemented VSVΔG (VSVΔG-G) was negligible (Fig. 2), we performed inoculations in the presence of neutralizing anti-G antibody mAb I1 (17) to assure that we only measured AFF-1-mediated infection (Fig. S5). Thus, AFF-1 can replace the viral fusogen VSVG and can mediate virus-cell binding and fusion.
Fig. 2.
AFF-1 can complement the infection of a fusion deficient VSVΔG
(A) Titers of VSVΔG pseudoviruses. The type of protein on the viral membrane (Bald or AFF-1) and on the BHK cell membrane (Vector, AFF-1 or EFF-1) is indicated (Fig. S3). Anti-VSVG antibody (αG) was used to neutralize any residual VSVΔG-G virus (11) (Fig. S5). Titers are in infectious units (IU) representing the number of cells expressing GFP per microliter 24 hours after virus inoculation. Data are mean ± SE (n=3 experiments). The inset shows background infection. We found no significant difference between infection of BHKAFF-1 and BHK-EFF-1 (Two-tailed paired t test, P=0.5841).
(B) Infection of BHKs monitored as GFP expression; phase contrast (top panels), fluorescence (bottom panels). VSVΔG-G served as positive control (Fig. S5). Scale bar is 50 μm.
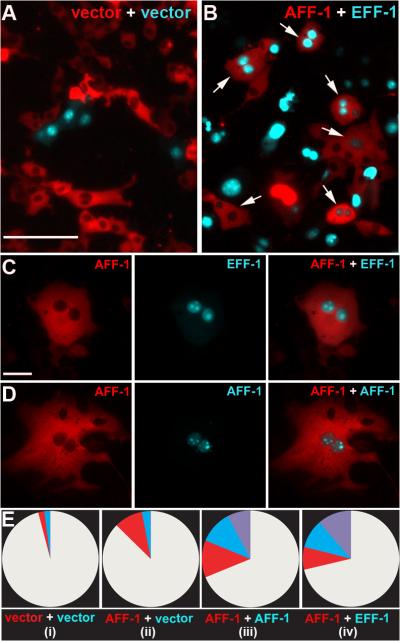
VSVΔG-AFF-1 could also infect cells expressing EFF-1 (BHK-EFF-1, Fig. 2A) with comparable efficiency, suggesting that different CeFFs can functionally interact to mediate membrane fusion. To test this hypothesis we evaluated cytoplasmic mixing between cells. We co-expressed aff-1 with a red fluorescent protein containing a nuclear export signal (RFPnes; Fig. 3) and mixed them with cells co-expressing eff-1 and a cyan fluorescent protein containing a nuclear localization signal (CFPnls) (18). We co-cultured the two cell populations and observed multinucleated cells expressing both markers (Fig. 3). In contrast, we did not observe cells expressing both markers in co-cultured cells co-transfected with empty vector (Fig. 3A). Thus AFF-1 and EFF-1 can promote heterotypic membrane fusion. To show independently that these results were a consequence of fusion, we recorded time-lapse images of BHK-AFF-1 cells (movies S1 and S2, Fig. S6), supporting the conclusion that AFF-1 expression was enough to fuse cells. Thus AFF-1 and EFF-1 can mediate cell-cell fusion as well as viral-cell fusion by a CeFF-mediated mechanism. However the VSVΔGAFF-1 infection mechanism is fundamentally different from that of native VSV. While the infection of VSV is mediated by VSVG only on the viral membrane, infection mediated by VSVΔG-AFF-1 requires an FF protein on both the viral and the cell membrane.
Fig. 3.
BHK-AFF-1 can fuse with BHK-EFF-1 cells
(A) Negative control. Mixed cells co-transfected with empty vector and RFPnes or CFPnls. Scale bar is 100 μm.
(B) Red, BHK-AFF-1 and Cyan, BHK-EFF-1 cells were mixed. Hybrids express cyan nuclei and red cytoplasm (Arrows).
(C) BHK cell with red cytoplasm surrounding two cyan nuclei appeared following AFF-1-EFF-1 expression and mixing of the cells. Scale bar is 10 μm.
(D) Hybrid binucleate cell appeared following AFF-1 expression and mixing of the cells.
(E) Quantification of content mixing experiments. Red, cyan and purple-colored pies represent the fraction of multinucleated cells (2 nuclei or higher). Results are mean of four independent experiments (n ≥ 1000 total cells). As indicated:
(i) Empty vector transfected cells only. All multinucleated cells were bi-nucleated (red or blue, not purple); total binucleate cells = 4%, probably dividing cells.
(ii) AFF-1 expressing cells (red) mixed with empty vector transfected cells (cyan). Elevation in multinucleation was only observed for AFF-1 expressing cells (red, 11%; cyan, 3%). One cell with a single nucleus expressing both markers (red and cyan) was observed.
(iii) AFF-1 expressing cells (red) mixed with AFF-1 expressing cells (cyan) resulting in four cell populations – mononucleated white (64%), multinucleated red (13%), cyan (12%) and mixed (purple, 11%).
(iv) AFF-1 expressing cells (red, 9%) mixed with EFF-1 expressing cells (cyan, 11%). AFF-1 and EFF-1 expressing cells fuse (purple, 18%).
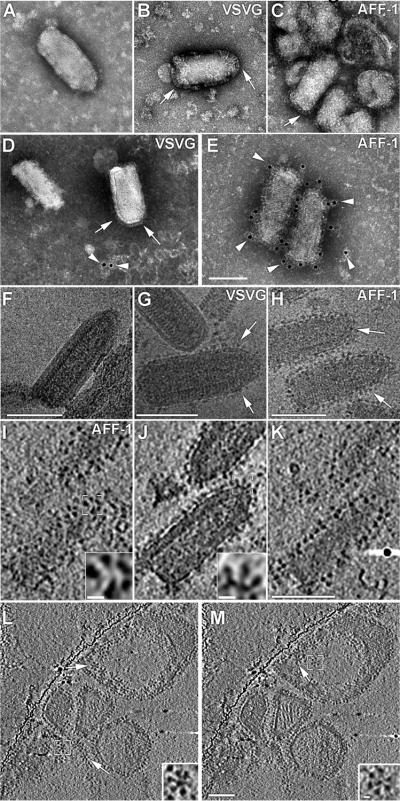
To study the relationship between structure and function of AFF-1 we used transmission electron microscopy (TEM). We compared negatively stained samples of VSVΔG, to VSVG and AFF-1 complemented VSVΔG preparations (11). VSVΔG virions have the typical VSV shape with a smooth membrane, hence termed bald, while both VSVΔG-G (19) and VSVΔG-AFF-1 virions displayed distinct spikes on their envelopes (Fig. 4A to C). In negative stain (pH 5), VSVG forms elongated spikes on VSVΔG-G (19) (Fig. 4B), while VSVΔG-AFF-1 showed shorter spikes (Fig. 4C). The estimated average spike lengths of VSVG and AFF-1 were 145 Å and 109 Å, respectively (Table S2). To confirm that the observed spikes were indeed AFF-1, we performed immunogold labeling using anti-AFF-1 polyclonal antibodies. We observed specific immunoreactivity on the surface of VSVΔGAFF-1 (Figs. 4D and E, S7 and S8). To further characterize the pseudoviruses at higher resolution and in a more native state they were imaged embedded in vitreous ice by cryoTEM (Fig. 4F to H) and cryo electron tomography (cryoET, Fig. 4I to K and movie S3). CryoTEM projection images showed that AFF-1 proteins coat the pseudoviruses. Side views of individual spikes could be observed at central sections of the tomograms (Fig. 4J). Higher order assemblies of AFF-1 in the form of penta- or hexa- meric “flower” shaped supercomplexes could be observed in slices through the tomogram oriented peripheral to the pseudotyped virus particles (Fig. 4I). These assemblies were more visible in slices through the tomograms of co-purified vesicles (Figs. 4L and M, S9 and movies S4 and S5). The order of these arrays may have a critical function in bending and deforming plasma membranes to mediate fusion.
Fig. 4.
Electron microscopy of VSVΔG-AFF-1 reveals specific bulky surface spikes (A–C) Negative stained particles of (A) VSVΔG; (B) VSVΔG-G; and (C) VSVΔG-AFF-1. (D–E) Anti-AFF-1 polyclonal antibodies followed by immunogold labeling and negative stain of (D) VSVΔG-G and (E) VSVΔG-AFF-1 (Fig. S7 and S8).
(F–H) CryoEM of (F) VSVΔG; (G) VSVΔG-G; and (H) VSVΔG-AFF-1.
(I–K) Top, center and bottom slice from VSVΔG-AFF-1 tomogram (movie S3).
(L–M) Slices from cryoET of vesicles co-purified with VSVΔG-AFF-1 preparations displaying penta- or hexa- meric “flower” shaped assemblies (movie S5).
Scale bars are 100 nm and 10 nm for the insets; Arrows: surface spike assemblies; Arrowheads: gold particles; White Square: indicating area shown magnified in inset.
Here we have presented evidence that FF proteins are functionally conserved in evolution and can restore the infectivity of VSVΔG through interactions with FF proteins on the target cell. Thus, FF, viral and intracellular fusogens converge functionally as minimal fusion machines that function on their own to promote fusion.
Supplementary Material
Acknowledgments
We thank I. Nagano, M. Whitt, A. Fire, C. Giraudo and J. Rothman for reagents, F. Glaser, M. Glickman, A. Harel, O. Kleifeld, T. Schwartz, I. Sharon, R. Sommer and members of the White and Podbilewicz labs for discussions; the Smoler Proteomics Center at the Technion and E. Spooner for mass spectrometry, T. Ziv for proteomics analysis, the Caenorhabditis Genetics Center for nematode strains and I. Yanai, and A. de Silva for critically reading the manuscript. Supported by grants from: The FIRST Program of the ISRAEL SCIENCE FOUNDATION (ISF 1542/07 to BP), the HFSP (RG0079/2009-C to KG); a Wellcome Trust Senior Research Fellowship to KG and the NIH (AI22470 to JMW).
Footnotes
One Sentence Summary: A Caenorhabditis elegans cell surface fusion protein can promote viral fusion with mammalian cells.
Supporting Online Material www.sciencemag.org
References and Notes
- 1.Wickner W, Schekman R. Nat Struct Mol Biol. 2008;15:658. doi: 10.1038/nsmb.1451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Martens S, McMahon HT. Nat Rev Mol Cell Biol. 2008;9:543. doi: 10.1038/nrm2417. [DOI] [PubMed] [Google Scholar]
- 3.White JM, Delos SE, Brecher M, Schornberg K. Crit Rev Biochem Mol Biol. 2008;43:189. doi: 10.1080/10409230802058320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sapir A, Avinoam O, Podbilewicz B, Chernomordik LV. Dev Cell. 2008;14:11. doi: 10.1016/j.devcel.2007.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Oren-Suissa M, Podbilewicz B. Trends Cell Biol. 2007;17:537. doi: 10.1016/j.tcb.2007.09.004. [DOI] [PubMed] [Google Scholar]
- 6.Chen EH, Grote E, Mohler W, Vignery A. FEBS Lett. 2007 doi: 10.1016/j.febslet.2007.03.033. [DOI] [PubMed] [Google Scholar]
- 7.Mohler WA, et al. Dev. Cell. 2002;2:355. doi: 10.1016/s1534-5807(02)00129-6. [DOI] [PubMed] [Google Scholar]
- 8.Sapir A, et al. Dev Cell. 2007;12:683. doi: 10.1016/j.devcel.2007.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Podbilewicz B, et al. Dev Cell. 2006;11:471. doi: 10.1016/j.devcel.2006.09.004. [DOI] [PubMed] [Google Scholar]
- 10.Shemer G, et al. Curr Biol. 2004;14:1587. doi: 10.1016/j.cub.2004.07.059. [DOI] [PubMed] [Google Scholar]
- 11.Materials and methods are available as supporting material on Science Online.
- 12.Zavada J. J Gen Virol. 1972;15:183. doi: 10.1099/0022-1317-15-3-183. [DOI] [PubMed] [Google Scholar]
- 13.Schnell MJ, Buonocore L, Kretzschmar E, Johnson E, Rose JK. Proc Natl Acad Sci U S A. 1996;93:11359. doi: 10.1073/pnas.93.21.11359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Takada A, et al. Proc Natl Acad Sci U S A. 1997;94:14764. doi: 10.1073/pnas.94.26.14764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Matsuura Y, et al. Virology. 2001;286:263. doi: 10.1006/viro.2001.0971. [DOI] [PubMed] [Google Scholar]
- 16.Fukushi S, et al. J Gen Virol. 2005;86:2269. doi: 10.1099/vir.0.80955-0. [DOI] [PubMed] [Google Scholar]
- 17.Lefrancois L, Lyles DS. Virology. 1982;121:157. [PubMed] [Google Scholar]
- 18.Hu C, et al. Science. 2003;300:1745. doi: 10.1126/science.1084909. [DOI] [PubMed] [Google Scholar]
- 19.Libersou S, et al. J Cell Biol. 2010;191:199. doi: 10.1083/jcb.201006116. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.