Erythroid Krüppel-Like Factor (KLF1; previously known as EKLF) is an essential erythroid-specific transcription factor that was first identified by Miller and Bieker in 1993.1 It binds the CACCC motif, an important DNA binding site in the regulatory elements of many erythroid genes including the HBB (β-globin) gene. Mutations in the β-globin CACC box which prevent KLF1 binding are a cause of β-thalassemia.2 KLF1 has three zinc finger domains, which mediate sequence-specific binding to DNA and are, therefore, essential for activation of KLF1 target genes (Figure 1).
Figure 1.
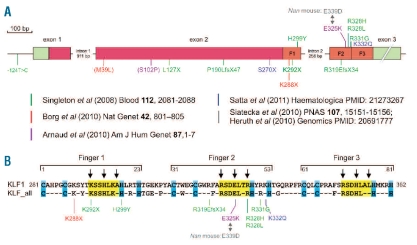
Mutations in human KLF1. All currently reported mutations are shown; the color code refers to the original publications as indicated. (A) Schematic drawing of the KLF1 gene. Exons: green = non-coding regions; red = coding regions, orange = zinc fingers (F1, F2, F3). Mutations between brackets are believed to be neutral substitutions. (B) Amino acid sequence of the zinc fingers of KLF1 (top line). Bottom line: amino acids invariable between the zinc finger domains of all 17 human KLF transcription factors (KLF_all). Blue boxes highlight amino acids involved in coordination of the Zn atom; yellow boxes highlight amino acids directly involved in DNA binding. The arrows indicate amino acids that make base-specific contacts with the DNA double helix.
Functions of KLF1: studies in mice
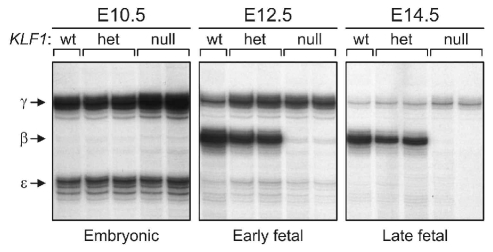
Mouse KLF1 null mutants displayed grossly normal erythropoiesis at the embryonic stage when hematopoiesis takes place in the yolk sac, but they rapidly developed a very severe form of anemia at the fetal stage, when the site of hematopoiesis has shifted to the fetal liver. KLF1 null mutants failed to activate expression of β-globin, which is a fetal/adult globin in the mouse. Thus, inactivation of KLF1 causes lethal β-thalassemia. Remarkably, the expression of embryonic β-like globin genes, ɛy and βh1, and the α-like globin genes, embryonic ζ and α1/α2, appeared to be normal.3–4 This suggested that KLF1 has a role in fetal-to-adult globin gene switching as it occurs in humans. To test this idea, transgenic mice carrying a complete human β-globin locus were used.5–6 Such mice express human γ-globin at the early fetal stages while the switch to β-globin is completed at the late fetal stages. An austere reduction in β-globin expression was observed in KLF1 null fetuses, while expression of γ-globin was not dependent on KLF1 and even extended in its absence5–6 (Figure 2). These data supported a role for KLF1 in globin switching. However, the anemia of KLF1 null mutants was not rescued by expression of γ-globin.7 Genome-wide gene expression profiling studies revealed a global role for KLF1 in the activation of erythroid-specific genes8–10 including globins, membrane- and structural proteins, heme synthesis enzymes and many other proteins involved in red cell metabolism. This explained the particularly severe anemia of KLF1 null mutants (Figure 3).
Figure 2.
Activation of a human β-globin locus transgene in KLF1 null mouse mutants. RNA expression levels of the human β-like globins [embryonic (ɛ), fetal (γ), adult (β)] were assessed by quantitative S1 nuclease protection assays.6 Days of mouse embryonic development are indicated (E10.5, E12.5, E14.5). The status of the mouse KLF1 alleles is shown: wt = wild-type; het = heterozygous for KLF1 null allele, null = homozygous for KLF1 null allele. Data courtesy of Drs. Beatriz Nuez and Frank Grosveld.
Figure 3.
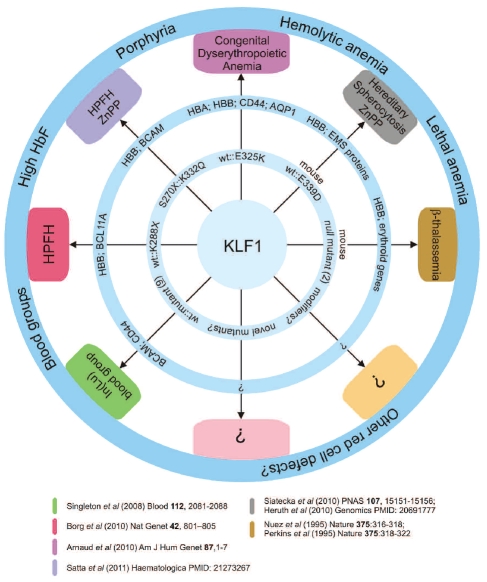
Phenotypes caused by KLF1 mutations The inner ring displays KLF1 mutations and potential modifiers. The number of different mutants reported is shown. The middle ring displays critical KLF1 target genes/loci whose expression is affected by the KLF1 mutation(s). The outer ring displays phenotypes. Clinical conditions are in the boxes; the colors refer to the publications shown below. HBA = α-globin locus; HBB = β-globin locus; EMS = erythrocyte membrane skeleton, ZnPP = zinc protoporphyrin.
KLF1 mutations in humans – inhibitor of Lutheran antigen expression [In(Lu)]
Mutations in human KLF1 were first reported in 2008 by Singleton and colleagues.11 They described 9 different loss-of-function KLF1 mutations which were causative to the rare In(Lu) blood group (Figures 1 and 3). Gene expression profiling revealed a list of more than 650 putative KLF1 target genes which considerably overlapped with those reported in KLF1 null mouse studies.8–11 These target genes included BCAM that carries the Lutheran blood group antigens and CD44 that carries the Indian blood group antigens (Figure 3). The expression of these blood group antigens is suppressed in the In(Lu) individuals.11 No other clinical features were reported.
KLF1 mutations in humans – hereditary persistence of fetal hemoglobin (HPFH)
The direct association of mutations in human KLF1 with hemoglobin regulation came from the study of a large family from Malta.12 Ten out of 27 family members exhibited HPFH due to a single point mutation in KLF1 (p.K288X; Figures 1 and 3). This mutation completely abrogates the DNA binding domain and therefore results in haploinsufficiency for KLF1. The KLF1 p.K288X carriers displayed high HbF levels, although with considerable variation (mean 8.4%; range 3.3–19.5%). Part of this variability could be explained by SNP haplotypes at the BCL11A locus, which encodes a repressor of γ-globin expression.13 Importantly, BCL11A expression was reduced in the KLF1 p.K288X carriers and KLF1 was shown to be a direct activator of BCL11A expression12,14 (Figure 3). Collectively, these data suggested that attenuation of KLF1 activity could be a fruitful approach to raise HbF levels in patients with β-type hemoglobinopathies.
KLF1 mutations in humans – congenital dyserythropoietic anemia
A further very interesting twist came from the study of 2 unrelated patients with congenital dyserythropoietic anemia (CDA15). On one KLF1 allele, these patients carried a missense mutation in the second zinc finger (Figures 1 and 3). The mutation, p.E325K, had a dominant effect and resulted in severe hemolytic anemia. The patients displayed 31.6% and 44% HbF, and also expressed embryonic ζ- and ɛ-globin.15 The erythroid cells were deficient for CD44 and the water channel AQP1, while BCAM expression was reduced in one case and normal in the other15 (Figure 3). Remarkably, an ethylnitrosourea-induced mutation in the homologous position in mouse KLF1 causes the dominant Nan (neonatal anemia) phenotype.16–17 Heterozygous Nan/+ mice displayed hereditary spherocytosis and severe hemolytic anemia. Expression of erythrocyte membrane skeleton proteins and β-globin was reduced, but embryonic globins were present at aberrantly high levels (Figures 1 and 3). The mutation, p.E339D, reduced binding to a subset of KLF1 binding sites, and this selectively affected activation of KLF1 target genes.17 The variant residues have opposite charges: positive in the CAD patients (p.E325K, lysine) and negative in the Nan mouse (p.E339D, aspartic acid). These mutants have different DNA binding properties that determine the impact on the expression of KLF1 target genes.15, 17 Thus, different KLF1 missense mutations that affect DNA binding properties could well lead to distinct red cell disorders15 (Figure 3).
KLF1 mutations in humans – zinc protoporphyrin
Significantly raised HbF levels are not always observed in association with KLF1 mutations. The study of the Maltese family already noted variability in HbF levels amongst individuals carrying the p.K288X mutation. SNP haplotyping of the BCL111A locus accounted for only a small part of the variability, strongly suggesting that as yet unknown modifiers are involved (Figure 3). This idea is further supported by data on a family from Sardinia reported in this issue of Haematologica.18 Three heterozygous carriers of KLF1 mutations (p.S270X or p.K332Q; Figure 1) had HbF levels of 0.9–1.2%, which is at the upper end of the normal range. Interestingly, the two compound heterozygotes (p.S270X::p.K332Q) displayed very high HbF levels (22.1% and 30.9%). In addition, these individuals had high levels of zinc protoporphyrin in their circulation; this was also observed in the Nan mouse16–17 (Figure 3).
KLF1 mutations in humans – known unknowns and unknown unknowns
The recent reports on KLF1 mutations have opened up a novel research area of human erythropoiesis, with a range of questions that urgently need answers. The feasibility of attenuating KLF1 activity to raise HbF levels in patients with β-type hemoglobinopathies remains to be determined. Related to this, we have only begun to understand the phenotypes associated with the different KLF1 mutations, and more in-depth analyses might very well reveal that there is significantly more overlap between these phenotypes than currently appreciated (Figure 3). The impact of missense mutations on the phenotypic outcome also warrants further investigation. For instance, missense mutations of critical residues in the zinc finger domains of KLF1 affect its DNA binding properties and may have a dominant phenotype, as exemplified by the p.E325K mutation in the CDA patients15 and the p.E339D mutation in the Nan mouse.16–17 In contrast, the p.K332Q mutation, which also affects the DNA binding properties of KLF1, does not have a dominant effect.18 Genome-wide assessment of the in vivo binding sites of these mutant KLF1 proteins may help to understand the phenotypic variability. In addition, it appears that some target genes, such as BCL11A, EPB4.9 and CD44, are very sensitive to perturbations in KLF1 activity, while the effects on others, such as BCAM and γ-globin, are much more variable. Studies in the Nan mouse indicate that this might depend on the class of KLF1 binding site responsible for activation of KLF1 target genes.17 In addition, modifier genes may affect the expression of the “variable response” genes, as exemplified by the intricate interplay of KLF1 and BCL11A on γ-globin expression.12,14 Identifying these modifier genes and characterizing the molecular properties of KLF1 mutants are important challenges for further research. In addition, we expect that many novel cases of individuals with KLF1 mutations will be discovered in the near future. Novel mutations may provide insight into the functional domains of KLF1. Most of the reported mutations affect the DNA binding domain (Figures 1 and 3). KLF1 is known to interact with co-factors such as the chromatin remodeling SWI/SNF complex, and it undergoes post-translational modifications such as phosphorylation, acetylation and SUMOylation (M Siatecka and JJ Bieker, submitted manuscript, 2011). Mutations affecting these biochemical properties may provide important clues to the functional roles of protein-protein interactions and post-translational modifications. Furthermore, given the broad impact of KLF1 on erythroid-specific gene expression in the mouse,8–10 the array of human erythroid phenotypes associated with KLF1 mutations will likely be expanded as more cases are described in detail. We note that mutations without an obvious phenotype are equally important to report, but difficult to publish in the peer-reviewed literature. To keep track of all KLF1 mutations and associated phenotypes, we have implemented the microattribution process19 and initiated a collaborative effort for functional analysis of KLF1 mutants (http://www.ithanet.eu/eklf-klf1). We strongly call upon the hematology community to participate in these initiatives, to ensure that valuable information on KLF1 mutations is systematically collected and accessible to both clinical and research scientists.
Acknowledgments
This work has been supported by institutional funding of the University of Malta, and the Malta Department of Health (AEF and JB), a fellowship of the Malta Government Scholarship Scheme (JB), European Commission grants (GEN2PHEN; FP7-200754 and ITHANET; FP6-026539) to GPP, and the Netherlands Genomics Initiative (NGI), Erasmus MC (MRace; 296088), the Landsteiner Foundation for Blood Transfusion Research (LSBR; 1040), and the Dutch organization for scientific research (NWO; DN 82-301 and 40-00812-98-08032) to SP. We apologize to our colleagues whose work could not be cited due to space constraints.
Footnotes
Joseph Borg is an academic staff member of the University of Malta, Faculty of Health Sciences, Department of Applied Biomedical Science, and a researcher on the Thalassaemia Project in the Laboratory of Molecular Genetics. George P. Patrinos is Assistant Professor of Pharmacogenomics at the University of Patras, Department of Pharmacy. He has been involved in globin gene regulation research for over 15 years. His research interests include pharmacogenomics of fetal hemoglobin augmenting agents. Alex. E. Felice is the founder of the Thalassaemia Project at the University of Malta where he is Professor, and the Malta Department of Health, Mater Dei Hospital where he is Visiting Consultant. His interests are in hemoglobin epidemiology and globin gene control. Sjaak Philipsen is Professor of Genomics of Cell Differentiation at the Department of Cell Biology in Erasmus University Medical Center in Rotterdam. His main research interest is how transcription factor networks control erythropoiesis.
Financial and other disclosures provided by the author using the ICMJE (www.icmje.org) Uniform Format for Disclosure of Competing Interests are available with the full text of this paper at www.haematologica.org.
References
- 1.Miller IJ, Bieker JJ. A novel, erythroid cell-specific murine transcription factor that binds to the CACCC element and is related to the Kruppel family of nuclear proteins. Mol Cell Biol. 1993;13(5):2776–86. doi: 10.1128/mcb.13.5.2776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Feng WC, Southwood CM, Bieker JJ. Analyses of beta-thalassemia mutant DNA interactions with erythroid Kruppel-like factor (EKLF), an erythroid cell-specific transcription factor. J Biol Chem. 1994;269(2):1493–500. [PubMed] [Google Scholar]
- 3.Nuez B, Michalovich D, Bygrave A, Ploemacher R, Grosveld F. Defective haematopoiesis in fetal liver resulting from inactivation of the EKLF gene. Nature. 1995;375(6529):316–8. doi: 10.1038/375316a0. [DOI] [PubMed] [Google Scholar]
- 4.Perkins AC, Sharpe AH, Orkin SH. Lethal beta-thalassaemia in mice lacking the erythroid CACCCtranscription factor EKLF. Nature. 1995;375(6529):318–22. doi: 10.1038/375318a0. [DOI] [PubMed] [Google Scholar]
- 5.Perkins AC, Gaensler KM, Orkin SH. Silencing of human fetal globin expression is impaired in the absence of the adult beta-globin gene activator protein EKLF. Proc Natl Acad Sci USA. 1996;93(22):12267–71. doi: 10.1073/pnas.93.22.12267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wijgerde M, Gribnau J, Trimborn T, Nuez B, Philipsen S, Grosveld F, et al. The role of EKLF in human beta-globin gene competition. Genes Dev. 1996;10(22):2894–902. doi: 10.1101/gad.10.22.2894. [DOI] [PubMed] [Google Scholar]
- 7.Perkins AC, Peterson KR, Stamatoyannopoulos G, Witkowska HE, Orkin SH. Fetal expression of a human Agamma globin transgene rescues globin chain imbalance but not hemolysis in EKLF null mouse embryos. Blood. 2000;95(5):1827–33. [PubMed] [Google Scholar]
- 8.Drissen R, von Lindern M, Kolbus A, Driegen S, Steinlein P, Beug H, et al. The erythroid phenotype of EKLF-null mice: defects in hemoglobin metabolism and membrane stability. Mol Cell Biol. 2005;25(12):5205–14. doi: 10.1128/MCB.25.12.5205-5214.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hodge D, Coghill E, Keys J, Maguire T, Hartmann B, McDowall A, et al. A global role for EKLF in definitive and primitive erythropoiesis. Blood. 2006;107(8):3359–70. doi: 10.1182/blood-2005-07-2888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pilon AM, Arcasoy MO, Dressman HK, Vayda SE, Maksimova YD, Sangerman JI, et al. Failure of terminal erythroid differentiation in EKLF-deficient mice is associated with cell cycle perturbation and reduced expression of E2F2. Mol Cell Biol. 2008;28(24):7394–401. doi: 10.1128/MCB.01087-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Singleton BK, Burton NM, Green C, Brady RL, Anstee DJ. Mutations in EKLF/KLF1 form the molecular basis of the rare blood group In(Lu) phenotype. Blood. 2008;112(5):2081–8. doi: 10.1182/blood-2008-03-145672. [DOI] [PubMed] [Google Scholar]
- 12.Borg J, Papadopoulos P, Georgitsi M, Gutierrez L, Grech G, Fanis P, et al. Haploinsufficiency for the erythroid transcription factor KLF1 causes hereditary persistence of fetal hemoglobin. Nat Genet. 2010;42(9):801–5. doi: 10.1038/ng.630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sankaran VG, Menne TF, Xu J, Akie TE, Lettre G, Van Handel B, et al. Human fetal hemoglobin expression is regulated by the developmental stage-specific repressor BCL11A. Science. 2008;322(5909):1839–42. doi: 10.1126/science.1165409. [DOI] [PubMed] [Google Scholar]
- 14.Zhou D, Liu K, Sun CW, Pawlik KM, Townes TM. KLF1 regulates BCL11A expression and gamma- to beta-globin gene switching. Nat Genet. 2010;42(9):742–4. doi: 10.1038/ng.637. [DOI] [PubMed] [Google Scholar]
- 15.Arnaud L, Saison C, Helias V, Lucien N, Steschenko D, Giarratana MC, et al. A dominant mutation in the gene encoding the erythroid transcription factor KLF1 causes a congenital dyserythropoietic anemia. Am J Hum Genet. 2010;87(5):721–7. doi: 10.1016/j.ajhg.2010.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Heruth DP, Hawkins T, Logsdon DP, Gibson MI, Sokolovsky IV, Nsumu NN, et al. Mutation in erythroid specific transcription factor KLF1 causes Hereditary Spherocytosis in the Nan hemolytic anemia mouse model. Genomics. 2010;96(5):303–7. doi: 10.1016/j.ygeno.2010.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Siatecka M, Sahr KE, Andersen SG, Mezei M, Bieker JJ, Peters LL. Severe anemia in the Nan mutant mouse caused by sequence-selective disruption of erythroid Kruppel-like factor. Proc Natl Acad Sci USA. 2010;107(34):15151–6. doi: 10.1073/pnas.1004996107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Satta S, Perseu L, Moi P, Asunis I, Cabriolu A, Maccioni L, et al. Compound heterozygosity for KLF1 mutations associated with remarkable increase of fetal hemoglobin and red cell protoporphyrin. Haematologica. 2011 Jan 27; doi: 10.3324/haematol.2010.037333. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Giardine B, Borg J, Higgs DR, Peterson KR, Philipsen S, Maglott D, et al. Systematic documentation and analysis of human genetic variation in hemoglobinopathies using the microattribution approach. Nat Genet. 2011;43(4):295–301. doi: 10.1038/ng.785. [DOI] [PMC free article] [PubMed] [Google Scholar]