The deletion of 5q (5q-) is a frequent clonal chromosomal abnormality detected in patients with myelodysplastic syndromes (MDS). MDS with 5q- as a sole chromosome alteration is characterized by a more favorable prognosis when compared to other forms of MDS.1 By contrast, when detected in association with other chromosome abnormalities in the context of a complex karyotype, the 5q- aberration is associated in both MDS and acute myeloid leukemia (AML) with adverse prognostic outcome.2,3
The nucleophosmin (NPM1) gene located on chromosome 5q35 encodes a multifunctional nucleolar protein that plays multiple roles in cell growth and proliferation. Grisendi et al. reported that NPM1 is essential to maintain genomic stability in the mouse model. In fact, these authors showed that NPM1 is haploinsufficient for regulating centrosome duplication as NPM1 heterozygous (NPM1 +/−) cells show aberrant centrosome numbers, genomic instability and aneuploidy.4 The 5q35 region where NPM1 maps is deleted in a number of MDS and loss of chromosome 5 is a frequent finding in MDS.5–7 To investigate haploinsufficiency of NPM1 in human myeloid malignancies, we analyzed NPM1 status in terms of deletion, methylation and mutations in 53 patients with MDS or secondary AML (sAML) carrying the 5q- abnormality as a sole chromosomal alteration or associated with additional chromosome defects.
Cytogenetic methanol/acetic acid fixed cell suspensions and DNA samples were available from 53 unselected patients with a diagnosis of MDS (48 cases) or MDS-sAML (5 cases) with 5q abnormalities. Patients were diagnosed at the Genetic Department of the University of Navarra, Spain and at the Department of Biopathology of the University Tor Vergata of Rome, Italy. MDS were classified according to the French-American-British (FAB) system which was in use at the time of the diagnosis. Bone marrow samples were collected for the study in all cases at the time of presentation.
Cytogenetic and FISH studies were performed according to standard methods. NPM1 FISH analysis was carried out with bacterial artificial chromosome (BAC) (RP11-117L6 chr: 170,746,923 – 170,922,033), kindly provided by Prof M. Rocchi (Department of Genetics and Microbiology, University of Bari, Italy; http://www.biologia.uniba.it/rmc/2-YAC-BAC/BAC-Chromosome/BAC-05.html), and directly labeled with spectrum red dUTPs with a Nick Translation Kit (Vysis inc, IL, USA). To investigate whether the NPM1 gene was deleted in cases with karyotypically present 5q-, we also analyzed both interphase nuclei and metaphases with the LSI 5ptel48 spectrum green probe (Cytocell, Cambridge, UK). Detection of NPM1 mutations was carried out by RT-PCR followed by capillary gel electrophoresis as previously described.8 The methylation status of the NPM1 gene promoter was investigated by methylation-specific PCR.9 Bone marrow DNA from healthy donors was used as a negative control and human male genomic DNA universally methylated for all genes was used as a positive control for methylation-specific assays.
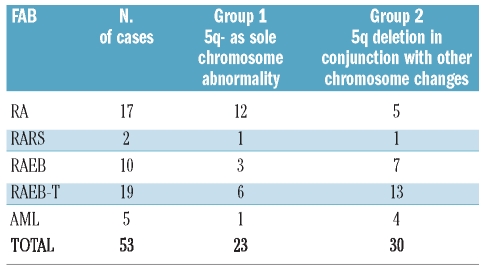
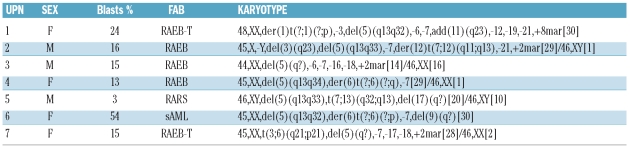
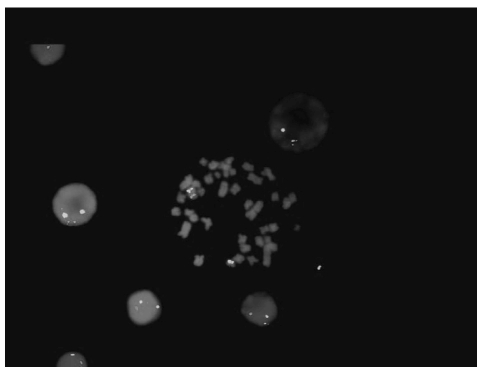
Twenty-three patients presented the 5q- as a sole chromosome abnormality (group 1) while 30 patients presented the 5q- in conjunction with other chromosome changes (group 2). Detailed FAB classification and karyotypic groups are reported in Table 1. Interestingly, in group 1 only 43% (10 of 23) of patients had more than 5% blast infiltration whereas in group 2, the blast count was greater than 5% in 80% (24 of 30) of cases (P=0.009). Methylation analysis of the NPM1 gene promoter and mutational studies of NPM1 exon 12 disclosed unmethylated CpG islands and wild-type NPM1 exon 12 in all patients. FISH analysis of the NPM1 locus, revealed deletion of one copy of the gene in 7 of 30 cases with complex karyotype and in none of 23 of those with isolated 5q- (P=0.01). Six out of these 7 cases with deleted NPM1, 5q- and complex karyotype showed more than 10% marrow blast infiltration at diagnosis (Table 2). A representative case with FISH analysis showing 5q- and NPM1 gene deletion is shown in Figure 1. Neither NPM1 hypermethylation, nor mutations were found in patients with 5q-MDS. After the submission of this study, La Starza et al. reported the occurrence of NPM1 deletion in 40% of patients with complex karyotype MDS/AML with 5q-.10 In our series, none of the 7 cases with NPM1 deletions showed at karyotypic examination a deletion that included the NPM1 gene locus at 5q35. However, it should be noted that the inaccuracy of the deleted region assignment by chromosome banding has been reported for 5q deletions in both MDS and AML.11
Table 1.
FAB classification and partial karyotypic details of 53 patients with 5q- abnormality.
Table 2.
Karyotype analysis of 7 patients with NPM1 deletion.
Figure 1.
Methaphase and interphase FISH analysis of a representative MDS case with 5q- and deletion of NPM1 gene. Spectrum red: NPM1 specific BAC (RP11-117L6). Spectrum green: 5ptel48 probe.
Our findings suggest that NPM1 haploinsufficiency may have a role in myeloid malignancies associated with large 5q- deletions contributing to MDS development likely through genetic instability. This would be in line with the finding that NPM1 is haploinsufficient in the control of centrosome duplication in the mouse model, as well as with evidence that NPM1 hypomorphic mouse embryonic fibroblasts reveal high levels of tetraploidy and aneuploidy.4 Alternatively, NPM1 deletions may represent secondary events linked to the progression of the disease in MDS.
Given the low number of cases and heterogeneity of treatments, we were unable to determine here the clinical significance of NPM1 deletion in MDS with 5q-. Hence a larger series of cases including homogeneously treated patients is needed to investigate the prognostic impact of these abnormalities.
Acknowledgments
The skilled FISH technical assistance of Aroa Irigoyen and Estela Fernández from the Department of Genetics of the University of Navarra, Pamplona, Spain is gratefully acknowledged. We are also grateful to Paola Curzi and Valentina Summa for their generous help in FISH experiments and to Nélida I. Noguera for critical reading of the manuscript.
References
- 1.Nimer SD, Golde DW. The 5q- abnormality. Blood. 1987;70(6):1705–12. [PubMed] [Google Scholar]
- 2.Van den Berghe H, Cassiman JJ, David G, Fryns JP, Michaux JL, Sokal G. Distinct haematological disorder with deletion of long arm of no. 5 chromosome. Nature. 1974;251(5474):437–8. doi: 10.1038/251437a0. [DOI] [PubMed] [Google Scholar]
- 3.Malcovati L, Porta MG, Pascutto C, Invernizzi R, Boni M, Travaglino E, et al. Prognostic factors and life expectancy in myelodysplastic syndromes classified according to WHO criteria: a basis for clinical decision making. J Clin Oncol. 2005;23(30):7594–603. doi: 10.1200/JCO.2005.01.7038. [DOI] [PubMed] [Google Scholar]
- 4.Grisendi S, Bernardi R, Rossi M, Cheng K, Khandker L, Manova K, et al. Role of nucleophosmin in embryonic development and tumorigenesis. Nature. 2005;437(7055):147–53. doi: 10.1038/nature03915. [DOI] [PubMed] [Google Scholar]
- 5.Van den Berghe H, Michaux L. 5q-, twenty-five years later: a synopsis. Cancer Genet Cytogenet. 1997;94(1):1–7. doi: 10.1016/s0165-4608(96)00350-0. [DOI] [PubMed] [Google Scholar]
- 6.Westbrook CA, Hsu WT, Chyna B, Litvak D, Raza A, Horrigan SK. Cytogenetic and molecular diagnosis of chromosome 5 deletions in myelodysplasia. Br J Haematol. 2000;110(4):847–55. doi: 10.1046/j.1365-2141.2000.02285.x. [DOI] [PubMed] [Google Scholar]
- 7.Giagounidis AA, Germing U, Haase S, Hildebrandt B, Schlegelberger B, Schoch C. Clinical, morphological, cytogenetic, and prognostic features of patients with myelodysplatic syndromes and del(5q) including band q31. Leukemia. 2004;18(1):113–9. doi: 10.1038/sj.leu.2403189. [DOI] [PubMed] [Google Scholar]
- 8.Noguera NI, Ammatuna E, Zangrilli D, Lavorgna S, Divona M, Buccisano F, et al. Simultaneous detection of NPM1 and FLT3-ITD mutations by capillary electrophoresis in acute myeloid leukemia. Leukemia. 2005;19(8):1479–82. doi: 10.1038/sj.leu.2403846. [DOI] [PubMed] [Google Scholar]
- 9.Yasuhiro Oki Y, Jelinek J, Beran M, Verstovsek S, Kantarjian HM, Issa JPJ. Mutations and promoter methylation status of NPM1 in myeloproliferative disorders. Haematologica. 2006;91(8):1147–8. [PubMed] [Google Scholar]
- 10.La Starza R, Matteucci C, Gorello P, Brandimarte L, Pierini V, Crescenzi B, et al. NPM1 deletion is associated with gross chromosomal rearrangements in leukemia. PLoS One. 2010;5:e12855. doi: 10.1371/journal.pone.0012855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pedersen B. Anatomy of the 5q-deletion: different sex ratios and deleted 5q bands in MDS and AML. Leukemia. 1996;10(12):1883–90. [PubMed] [Google Scholar]