Abstract
The c-Myc protein, encoded by c-myc gene, in its wild-type form can induce tumors with a high frequency and can induce massive programmed cell death (PCD) in most transgenic mouse models, with greater efficiency than other oncogenes. Evidence also indicates that c-Myc can cause proliferative inhibition, i.e., mitoinhibition. The c-Myc-induced PCD and mitoinhibition, which may be attributable to its inhibition of cyclin D1 and induction of p53, may impose a pressure of compensatory proliferation, i.e., regeneration, onto the initiated cells (cancer progenitor cells) that occur sporadically and are resistant to the mitoinhibition. The initiated cells can thus proliferate robustly and progress to a malignancy. This hypothetical thinking, i.e., the concurrent PCD and mitoinhibition induced by c-Myc can promote carcinogenesis, predicts that an optimal balance is achieved between cell death and ensuing regeneration during oncogenic transformation by c-Myc, which can better promote carcinogenesis. In this perspective, we summarize accumulating evidence and challenge the current model that oncoprotein induces carcinogenesis by promoting cellular proliferation and/or inhibiting PCD. Inspired by c-myc oncogene, we surmise that many tumor-suppressive or growth-inhibitory genes may also be able to promote carcinogenesis in a similar way, i.e., by inducing PCD and/or mitoinhibition of normal cells to create a need for compensatory proliferation that drives a robust replication of initiating cells.
Key words: c-Myc, cyclin D1, apoptosis, carcinogenesis, compensatory proliferation
Expression of the c-myc oncogene or its protein product, c-Myc, is elevated in virtually all types of malignant disease.1 Gene amplification also occurs frequently in various cancers but mutations, especially those in the coding region, are rare in most types of cancer, although frequent in some types of lymphoma.1–3 It is a general assumption that the oncogenicity of c-myc requires an elevated expression, but in fact the levels of c-myc in human cancers range from lower than normal to greatly elevated, as pointed out by Chung and Levens.4 A recent study also reports deletion of the c-myc locus in about 5% of breast cancer cases.5 This variation may not be surprising since the c-Myc protein has versatile functions, including the promotion of cell proliferation and programmed cell death (PCD).6,7 It is possible that c-Myc might be elevated initially to promote tumor formation but that it is later decreased or silenced (e.g., by genetic deletion) in order to facilitate the tumor cell progression or to allow the tumor cell to adapt to changes in other genes for a survival purpose,8,9 such as to survive the deficiency of the Apc gene.10–12 In this review, we discuss a possibility that c-Myc-induced PCD may play a positive role in carcinogenesis, a perspective inspired by several classical concepts established from extensive studies on chemical induced carcinogenesis in animals.
C-myc is a Unique Oncogene which Alone can Potently Induce Cell Death and Carcinogenesis in Transgenic Animals
In line with the clinical observations of elevated expression in different cancers, c-myc is the only oncogene, among numerous ones identified, that in its wild type form can induce tumor at a high penetrance, usually 100%, with a relatively short latent time in most transgenic animal models.13,14 Ras family members (H-Ras, N-Ras and K-Ras) may be the only other oncogenes that have similarly potent oncogenicity, but this is not widely tested since most Ras transgenic animals utilize oncogenic mutants (usually at codon 12), not the wild-type, in part because the wild-type form often reverses the transformed phenotype induced by the oncogenic counterparts.15 Other proto-oncogenes (not viral oncogenes) mainly induce proliferating benign lesions, although tumors may develop at a very low penetrance and with long latency in a few transgenic models, such as the MMTV-CCND1,16,17 and LFABP-CCND1 mice.18 The wild-type Neu (erbB2) driven by a mouse mammary tumor virus (MMTV) long terminal repeat in transgenic mice induces mammary tumors at a high frequency,19 but the mechanism involves spontaneous mutations of the Neu transgene, not the wild-type form in most cases.20–22 For most oncogenes at their wild-type form to induce cancer efficiently in transgenic mice, concomitant expression of a second oncogene or deficiency of a tumor suppressor gene is required. Obviously, this “second hit,” i.e., alteration in another gene, can occur spontaneously and efficiently in c-myc transgenic animals, which is not surprising because c-myc induces genomic instability and DNA damage.7,23,24
An intriguing but unanswered question is why c-myc is so different from many other oncogenes in its potency of carcinogenicity. Like other oncoproteins, c-Myc enhances cell proliferation. But unlike others, c-Myc also potently enhances different types of PCD, including senescence24–27 and apoptosis,28–32 in addition to autophagy.33,34 Of the many c-myc transgenic mouse models created to date, very few do not show evident PCD,35 which in some cases may be due to a low expression level of the transgene, since the c-myc driven by another promoter can elicit overt PCD in the same cell types. Because the overarching hypothesis described in this review does not concern a specific type of PCD and also because in many cases c-Myc induced PCD is not typical of any specific kind, as discussed before,36,37 we herein dub all c-Myc induced cell death “PCD” in order to simplify the discussion. Except for the c-myc, none of the canonical oncogenes in its wild type form has been shown to be able to induce prominent PCD in vivo as seen in many c-myc transgene animals, although some oncogenes such as Ras and cyclin D1 (D1) have been shown to cause PCD in cell culture under certain situations as reviewed before by us13 and others.38–40 One may consider E2F1 an exception as it can induce evident PCD in the epidermis of transgenic mice, but its potency is still much weaker than that of c-myc41 and it may serve as a downstream effector of c-Myc in eliciting PCD.42 Restated more clearly, few oncogenes alone can cause cancer as efficiently as c-myc in transgenic models and even fewer, if any, alone can cause tumor with robust apoptosis. The endogenous c-myc has also been shown to be markedly induced to mediate PCD of mammary epithelial cells in Socs3 knockout mice,43 which seems to be the first evidence for an effect of the endogenous c-myc on PCD in animals other than Drosophila. Again, none of other oncoproteins expressed from the endogenous alleles has received such in vivo evidence for its promotion of PCD. For these reasons, we hypothesize that the unique carcinogenicity of the c-myc in transgenic models may be attributable, in part, to its induction of PCD. Although this hypothetical thinking is seemingly counterintuitive, actually one fact that is familiar to pathologists but seldom mentioned is that cancers show a much higher PCD rate than the parental normal tissue or organ,44,45 likely because some cancer cells have accumulated too many genetic changes to survive and some other cells still retain a normal mechanism to avoid being hyperplastic as discussed later. Cancer cells can still form a tumor because their proliferative rate is even higher than the rate of PCD. Therefore, the sentiment that “PCD potential should be inhibited during carcinogenesis” is not always correct, depending on the reference used for the comparison. Cells expressing a c-myc transgene have these features of human cancer cells, i.e., higher in both proliferative and PCD rates.37
Inhibition of c-myc-induced PCD does not Always Promote Carcinogenesis
An early work by Vaux et al. in 1988 sets a milestone for c-myc research with three important findings: (1) c-Myc causes cell death when growth factors are deprived, (2) Bcl-2 can enhance the survival of c-myc expressing cells and (3) Bcl-2 collaborates with c-Myc to immortalize pre-B cells.46 The c-Myc induced demise has later been confirmed by ample studies to be a programmed event and occur in many cell types of different species. Concomitant expression of Bcl-2 has also been shown to enhance c-myc-induced carcinogenesis of lymphocytes,47–49 mammary glands50,51 and other types of cell or tissue. According to Vaux46,52 and others,53,54 the mechanism underlying the Bcl-2 and c-Myc collaboration is that Bcl-2 enhances cell survival whereas c-Myc drives cell proliferation. This notion has been extended to the collaboration between c-myc and other oncogenes; as stated by Naud and Eilers, “suppression of MYC-induced apoptosis is the predominant mechanism through which oncogenes cooperate with MYC during lymphomgenesis.”55 In this pattern of collaboration, inhibition of cell death per se is only very weakly oncogenic, since Bcl-2 transgenic animals develop tumors at a low penetrance with a long latency.56
Intuitively, inhibition of PCD should enhance cancer formation,6,28,57–60 as it should lead to an accumulation of genetic changes and an increase in cell number to form a tumor.61–63 However, there are several lines of evidence opposing this intuition. Tomlin et al. report that co-expression of Bcl-2 does not promote transformation of human B-cell lines by c-myc.64 More surprisingly, Bcl-2 actually inhibits c-myc-induced liver carcinogenesis in L-PK-Bcl-2/L-PK-c-myc double transgenic mice.65 Bcl-2 overexpression also inhibits liver carcinogenesis induced by transforming growth factorα (TGFα), with and without a concomitant treatment with a chemical carcinogen, in a Bcl-2/tgfα double transgenic model.66,67 Moreover, Bcl-2 inhibits chemical-induced mammary carcinogenesis as well.68 All these animal studies suggest that inhibition of apoptosis by Bcl-2 actually prevents cancer formation, which is tentatively explained by a requirement for PCD at certain stages of carcinogenesis65 or by a Bcl-2 caused delay of cell cycle entry69 or progression.70 These results from animal studies dovetail with the clinical observation that Bcl-2 overexpression is associated with a better prognosis of breast cancer71 and probably other cancers as well, which suggests a paradoxical role of apoptosis in human cancers, as discussed by Gurova and Gudkov72 and by Moreno.73
TGFα is known to collaborate with c-Myc in the induction of liver carcinogenesis in a double transgenic model, presumably via inhibition of c-Myc induced PCD.74 Two mutant c-myc alleles, T58A and S71F, are known to lack the PCD-inducing ability but retain a full ability to drive cell proliferation. However, while T58A/tgfα co-expression in the LE6 liver progenitor cells manifests the expected increase in cell proliferation and tumorigenicity when the cells were inoculated to subcutaneous sites of nude mice, S71F/tgfα co-expression actually inhibits proliferation and tumorigenicity, compared with S71F or T58A alone or the wild type c-myc/tgfα co-expression.75 Therefore, inhibition of c-Myc induced PCD is not always associated with enhanced tumorigenicity of liver progenitor cells. On the other hand, co-expression of TGFβ1 or the hepatitis B virus X, both of which are pro-apoptotic genes, has been shown to enhance c-myc induced liver carcinogenesis,76–78 suggesting that counterintuitively, enhancement of PCD may play a positive role in c-myc induced carcinogenesis.
Of the many c-myc transgenic models created to date, there are very few that do not develop a high frequency of tumors79 or do not produce overt tumors at all, either due to a low expression level of c-myc or due to an earlier death of the target cells or the animals.57,80–82 One of these models is the SBM mouse in which the c-myc transgene causes polycystic lesions and certain small renal adenomas that manifest high rates of PCD. No frank cancer is developed because the mice die young from renal failure, about three months of age on average.82 Another is the mouse that expresses an inducible c-myc (pIns-MycERTAM) transgene in the pancreatic β cells. In these mice, the majority of β cells die of PCD within 6–10 days after the c-myc activation and the initial induction of cell proliferation.57 Concomitant expression of Bcl-xL (pIns-MycERTAM/RIP7-Bcl-xL), which is a survival factor in the Bcl-2 family, inhibits c-Myc induced PCD and induces β-cell carcinomas as expected.83 However, concomitant knockout of caspase-3 also inhibits the c-Myc induced PCD of β-cells but does not enhance the tumor formation.84 More surprisingly, concomitant knockout of the p19ARF (pIns-MycERTAM/p19ARF−/−) enhances c-Myc induced PCD but the mice develop β-cell carcinomas.83 Because in this pIns-MycERTAM/p19ARF−/− model increased cell loss is matched by increased cell proliferation,83 it is possible that a certain level of PCD may accelerate carcinogenesis by accelerating cell turnover. Indeed, over inhibition of PCD may actually hinder carcinogenesis, since mammary tumor formation in the MMTV-c-myc transgenic mice is accelerated by the haploid loss of Bax (Bax+/−) but not by the Bax knockout (Bax−/−).85 Moreover, both proliferative and PCD rates are very high in the MMTV-c-myc mammary tumors but very low in the MMTV-Ras mammary tumors, but the latent time for the c-myc tumor (6.3 months of age) is much shorter than that for the Ras tumor (8.8 months),86 again suggesting a positive role of c-Myc-induced PCD in carcinogenesis by accelerating cell turnover.
It needs to be pointed out that many data on gene interactions result from double transgenic mouse models that are usually created by mating one transgenic line with another. In some of these models the two transgenes may be driven by different promoters, such as the WAP-Bcl-2/MMTV-c-myc50 and the pIns-MycERTAM/RIP7-Bcl-xL83 mice. A commonly neglected pitfall in these models is that the two different promoters may activate the two transgenes in different subpopulations of cells in the same organ or at different ages or different physiological statuses, such as the ovarian hormone cycle (which is equivalent to human menstrual stages). As a consequence, the models may involve interaction of different subtypes of cells and/or sequential gene activation. For example, in the MT-tgfα/MMTV-c-myc double transgenic mouse,87 the MMTV-promoter is activated mainly after puberty when the levels of sex hormones are increased, thus probably at a much later age than the activation of the metallothionein-1 (MT) promoter by heavy metals. In contrast, the VavP-c-myc/VavP-Bcl-2,49 and the aforementioned L-PK-Bcl-2/L-PK-c-myc65 double transgenic models utilize the same promoter to drive both transgenes, thus resembling a true situation of gene interaction in the same cells. Many promoters that are used to drive transgenes are not actually studied to the last detail on their targeted subpopulations of the cells and the time point (or time period) of activation. A related issue that may also be neglected easily is that some promoters such as Eµ88 and Mist-1,89 start to activate the transgene as early as prenatally (in utero), and thus the carcinogenic mechanism may be more similar to that of childhood cancers and less relevant to that of sporadic cancers in adulthood. All these issues need to be taken into account when one evaluates a gene-gene interaction in double transgenic models.
Compensatory Proliferation Promotes Carcinogenesis, Especially in a Mitoinhibitory Environment
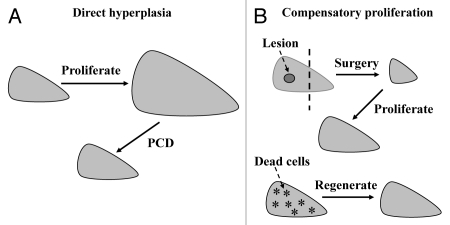
Unlike the situation in cell culture, there are two types of cell proliferation in vivo, i.e., direct hyperplasia and compensatory proliferation. Cell proliferation caused by a direct mitogen in a given cell type or organ results in “hyperplasia”, which is a pathology term to describe a tissue or organ that has extra cells, such as the liver enlargement caused by lead nitrate.90 PCD ensues to eliminate the hyperplastic cells because the cell type or the organ needs to maintain its normal number or physiological size, so-called homeostasis (Fig. 1). Thus, many hyperplastic cells that have acquired spontaneous mutations required for carcinogenesis are also eliminated,90 which may be one of the reasons why many oncogenes or growth factors that directly drive cell replication cannot efficiently induce carcinogenesis in transgenic animals. What still bewilders us is that even in transgenic models most oncogenes such as Ras do not induce evident apoptosis as inferred here and aforementioned. Our conjecture is that in most cases the target organ or tissue may already refrain from transgene-induced proliferation in order to be less hyperplastic, and thus the ensuing PCD is also mild, leading to a mild cell turnover.
Figure 1.
Depiction of direct hyperplasia and compensatory proliferation with the liver as an example. (A) Proliferation of hepatocytes, driven by a direct mitogen such as lead nitrate90 or a growth factor, causes enlargement of the liver. The hyperplastic, i.e., extra, cells will then undergo programmed cell death (PCD) until the organ returns to its normal size. (B) Surgical removal of a lesion in the left part of the liver (top part) or loss of some hepatocytes (stars in the low part) due to reasons such as chemical toxicity, viral infection or expression of a PCD-inducing gene triggers a compensatory proliferation (regeneration) of the remaining hepatocytes to restore the physiological cell number or organ size. No PCD ensues.
When an organ or cell type has cell loss first, due to reasons such as chemical toxicity, viral infection, surgical removal or overexpression of PCD-inducing genes such as the c-myc, it will undergo another type of proliferation, i.e., compensatory proliferation or regeneration, to restore the physiological cell number or organ size (Fig. 1).90,91 Tissue regeneration has been a familiar phenomenon to us for a long time,92,93 but so far little is known about its underlying mechanism in animals other than Drosophila.91,93 Nevertheless, it is conceivable that the more severe the cell loss is, the more potent the driving force for regeneration is. Unlike hyperplasia, compensatory proliferation does not trigger PCD because the nascent cells are needed. Therefore, sporadic mutations occurring during the replication have a higher chance of being inherited by the nascent cells, leading to a more efficient completion of the carcinogenic process.90
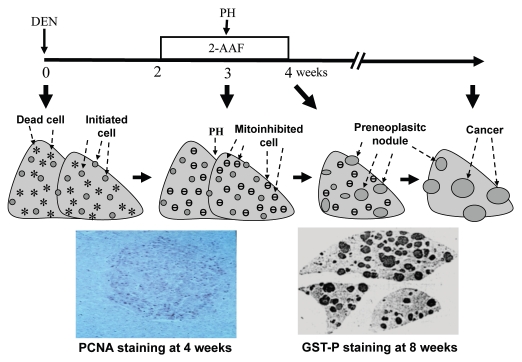
Ample studies of chemical-induced cancer in animals have shown that carcinogenesis is a stepwise process that starts from the sporadic appearance of so-called “initiated cells” in an organ or tissue, followed by a “promotion” period in which the initiated cell or cells replicate in a clonal expansion fashion.94 Many agents or circumstances can promote the proliferation of initiated cells, but the key point is that they act by inhibiting proliferation, i.e., “mitoinhibition”, of the normal cells in the organ or tissue, whereas the initiated cells have found ways to circumvent the mitoinhibition.95,96 This so-called “resistant phenotype” theory was originally proposed by Haddow in 1938,97 and has been extensively tested by Farber and many others, as described in many reviews ever since decades ago98–101 (Fig. 2). Unfortunately, this scriptural principle of carcinogenesis at the histology level is rarely discussed in the studies of molecular carcinogenesis in the past decade. Actually, as stated by Farber and Rubin,102 “virtually every chemical carcinogen is an inhibitor of cell proliferation”. Therefore, it is a widespread misconception that tumor-inducing or -promoting agents should promote cell proliferation, because in many cases their direct effect is mitoinhibition, although it results in proliferation of initiated cells. Because normal cells are mitoinhibited, initiated cells that are resistant to the mitoinhibitory effect become the only cells that can proliferate, and thus replicate robustly when the organ or tissue needs to regenerate to compensate for a physiological or pathological cell loss. In other words, a tumor promoting agent or circumstance causes mitoinhibition of normal cells, which in turn imposes a pressure of compensatory proliferation onto initiated cells, as depicted in Figure 2 with hepatocarcinogenesis as a model system. This effect of normal surrounding cells on the initiated cells has basically not been studied in transgenic or knockout animal models, in part because it is still difficult, if not impossible, to manipulate gene expression specifically in initiated or surrounding normal cells without affecting the other. It is conceivable that some growth-inhibitory or tumor-suppressive genes can enhance carcinogenesis by inhibiting proliferation of normal cells, just like many tumor-promoting agents, as long as initiated cells that occur sporadically have gained a mechanism to resist the mitoinhibition. In other words, many genes may be either oncogenic or tumor-suppressive, and sometimes it depends on which cells we are talking about—the initiated cells or their surrounding normal ones. More often the net result is described, which differs among animal models. Therefore, these genes are generally considered “dual functional,” although it may be a misconception in many cases.
Figure 2.
Use of Farber's “resistant hepatocyte” model of liver carcinogenesis96 as an example to illustrate how mitoinhibition and compensatory proliferation promote carcinogenesis: Rats were injected with a necrotic dose of diethylnitrosamine (DEN) to cause hepatocyte death (stars) and cause some critical genetic damage in some hepatocytes. The liver would regenerate in two weeks, during which the altered genes are passed to nascent hepatocytes and initiated cells are thus created (spots). The rats were then treated with 2-acetylaminofluorene (2-AAF) for two weeks at a low dose that inhibits proliferation of normal hepatocytes, but initiated cells are resistant to this mitoinhibition. When a partial hepatectomy (PH) is performed to remove 2/3 of the liver in the middle of the 2-AAF treatment to stimulate liver regeneration again, initiated cells in the remaining liver will proliferate robustly and form preneoplastic nodules, because normal cells are mitoinhibited (.). As one piece of evidence (the left photo at the bottom part), a liver collected one week after PH was immunohistochemically stained for proliferating cell nuclear antigen (PCNA) to visualize proliferating cells, which are mainly in a colony (i.e., a nodule) of initiated cells, but rarely in the surrounding area that also receives the same regeneration signal from PH.203 Another rat was sacrificed four weeks after cessation of 2-AAF treatment and the remaining three lobes of the liver were sectioned and immunohistochemically stained for the P form of glutathione S transferase (GST-P), which is a marker for the initiated, pre-neoplastic nodules (right, bottom photo). Many nodules will later regress gradually but some will progress to cancers a few months later.
c-Myc Induced PCD may Contribute to Carcinogenesis by Driving Compensatory Proliferation
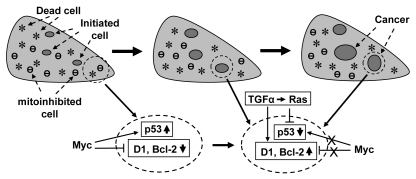
c-Myc induced PCD in transgenic animals is likely a primary event, not secondary to hyperplasia, because c-Myc induced proliferation can be separated from its induction of PCD.38,75,103,104 For instance, knockout of E2F2 enhances c-Myc induced proliferation but does not affect c-Myc induced apoptosis.105 Therefore, the PCD will likely trigger a compensatory proliferation of the target organ or tissue (Fig. 3). Whether c-myc can also cause mitoinhibition is less studied, although growth arrest genes such as p15ink4b,106 p21cip1,107 and Gadd45,108 are known to be suppressed in c-myc overexpressing cells but are elevated in c-myc knockout (c-myc−/−) cells. Nevertheless, c-myc overexpression has been shown to arrest normal fibroblasts at the G2 phase in culture109 and to induce a p53-dependent proliferative arrest of the hepatocytes in controllable transgenic mice.110,111 It is conceivable that growth arrest likely occurs before a cell undergoes PCD or when the c-Myc imposed stress is not strong enough to elicit PCD. Therefore, a c-myc transgenic organ or tissue has three major cell populations, i.e., (1) the dead and dying cells, (2) the mitoinhibitory cells and (3) the proliferating cells. In other words, the organ or tissue manifests a quick cell turnover, which enhances one or several cells to acquire critical genetic changes and thus become initiated cells that may be less mitoinhibitory and have less PCD potential, relative to most other c-myc expressing cells. Therefore, proliferation of initiated cells is driven not only by the c-myc transgene but also by a need for regeneration conferred by PCD and mitoinhibition (Fig. 3), similar to the situation in a chemical-induced carcinogenesis.
Figure 3.
Illustration of our hypothesis on how c-Myc-induced compensatory proliferation and mitoinhibition enhance carcinogenesis in transgenic animals: the c-myc transgene causes cell death (*) and probably also mitoinhibition (θ), which may be mechanistically related to its inhibition of cyclin D1 and other oncogenes (e.g., Bcl-2) as well as to its induction of p53 and other tumor suppressor genes, as shown in an enlarged area in the bottom part. The cell death triggers compensatory proliferation of the organ. Thus, cell proliferation is driven not only by the c-myc transgene per se but also by the cell death, but it still remains unknown which c-myc expressing cells, among many others, decide or are selected to die, to proliferate, or to be mitoinhibited. During the compensatory proliferation, some critical genetic changes occur sporadically in some cells, thus creating initiated cells that may be less mitoinhibitory and have a stronger survival potential, relative to the surrounding cells. The initiated cells proliferate continuously in a clonal expansion fashion and accumulate more and more genetic alternations, developing to preneoplastic lesions and, for some of them, malignant tumors eventually. During this process, some cells may develop mechanisms to escape from the control by c-Myc, resulting in p53 inactivation or D1 induction, as depicted in another enlarged area in the bottom part. At any time of the process, many growth stimuli such as TGFα EGF may collaborate, often via Ras, with c-Myc in the carcinogenesis in part by inducing D1 or inactivating the p53 pathway.
The hypothesis that cell turnover enhances carcinogenesis actually predicts that an optimal balance between cell death and regeneration, not each alone, will better promote c-myc-induced carcinogenesis in transgenic models. A PCD rate that is too low will not be sufficient to drive a compensatory proliferation and thus the initiated cells cannot quickly replicate. On the other hand, if PCD occurs too massively and too quickly, it will prevent accumulation of genetic alterations and thus prevent carcinogenesis, as seen in the aforementioned pIns-MycERTAM mice wherein c-myc activation kills almost all the pancreatic β-cells in only 6–10 days.57
Felsher's group has reported that activation of a controllable c-myc transgene in the liver during adulthood of the mouse induces endoreduplication of hepatocytes without evident cell proliferation or PCD.111 However, the mice still develop liver cancer after a prolonged latency compared with activation of the c-myc in earlier life, and the cancer shows evident PCD.111 This model seems against our hypothesis as neither PCD nor proliferation is essential for the carcinogenesis. Probably, the appearance of endoreduplication functions like proliferation to allow retention of c-Myc-induced genetic alterations in the duplicated DNA, and lack of evident PCD in this case may help retain the altered genes. However, once a tumor is developed and manifests increased cell proliferation, increased PCD ensues. Moreover, the adulthood carcinogenesis can also be accelerated by liver necrosis induced by treatment of the transgenic mice with non-specific small hairpin RNA112 or with hepatotoxin tetrachloride or 5-diethoxycarbonyl-1,4-dihydrocollidine.113 Presumably, the cytotoxicity-induced cell death triggers a compensatory proliferation as the mechanism for the promotion of the carcinogenesis.
c-Myc-induced Epithelial Cancers Retain the Wild-Type p53 for Induction of PCD
PCD usually occurs via mechanisms of decrease in oncoproteins such as Bcl-xL and increase in tumor suppressors such as p53. c-Myc induced PCD seems to involve both of these mechanisms. c-Myc has been shown to induce several tumor suppressor genes, including NOVA,114 RSK4,115 Bim116 and p53.117–120 E2F2 can function as a tumor suppressor105 and c-Myc directly induces it to elicit PCD of T lymphoma cells.121
The MMTV-c-myc mammary tumors, and even cell lines established from these transgenic tumors, retain the wild type p53 and also express relatively high levels of the p53 protein122 (and our unpublished data). Early work independently by Leder's and Dickson's groups has shown that inactivation of p53 does not accelerate c-myc induced mouse mammary carcinogenesis.123,124 Pancreatic tumors from the Ela-myc transgenic mice also retain the wild type p53 (reviewed in ref. 14 and 125 and unpublished data). In the K5-myc transgenic model, c-Myc causes PCD of keratinocytes by DNA damage-triggered p53 induction.41,126 Therefore, it seems that c-myc-induced solid tumors of epithelial origin retain an intact p53 gene and manifest elevated p53 expression, which may be a mechanism for the overt PCD in these tumors (Fig. 3).41
Data from non-epithelial cells such as lymphomas are not consistent. Lymphomas from the CD2-Runx2/CD2-c-myc and the CD2-Runx2/CD2-mycERTM double transgenic mice retain the functional p53, even after the tumors are transplanted into animals, although explanted tumor cells displayed rapid allele loss during culture.127 However, approximately 28% of the lymphomas from the Eµ-myc transgenic mice show deletion or mutations of p53, in addition to about 60% of the tumors showing deletion of ARF or overexpression of Mdm2 that is supposed to lead to inactivation of the p53 pathway as well.128 It is difficult to explain these incongruous data in lymphomas, in part because the c-myc is driven by different promoters (CD2 and Eµ) in these transgenic mice and thus may be activated at different time points or different subpopulations of lymphocytes. Moreover, the relationship between c-Myc and p53 also depends on the functional status of the c-myc per se and other genes such as Bim.116,117 Therefore, the actual relationship between c-Myc and the p53 status in spontaneous malignancies remains to be explored further.
c-Myc may Inhibit Cyclin D1 and Other Oncogenes to Induce PCD
As reviewed in more detail previously in reference 13, several studies have shown that c-Myc strikingly inhibits cyclin D1 (D1) expression in fibroblasts129–131 and mammary epithelial cells,132 which may occur via forming a complex with ZO-2133 or TCEAL7134 protein to bind to the D1 promoter and repress D1 transcription. D1 mRNA135 and protein levels136,137 are higher in c-myc−/− rat embryonic fibroblasts (REF) and mouse embryonic fibroblasts (MEF), and the D1-CDK4/6 activities are also 12-fold higher, compared with the c-myc+/+ counterparts. In astrocytoma cells, downregulation of the c-myc with antisense increases D1 protein level but decreases CDK4 protein level.138 Altogether, these data show a reciprocal relationship between c-Myc and D1, not only when a c-myc is ectopically expressed but also when the level of the endogenous c-myc is decreased. Therefore, c-Myc inhibits D1 transcription and it may occur physiologically. It was suggested that inhibition of D1 by c-Myc occurred only in transformed and Rb-deficient MEF,130 but we found that the inhibition occurred also in the Rb-wild type mouse pancreatic cancer cells.125,139–141 In MMTV-c-myc transgenic mammary tumors, D1 is expressed only in certain focal areas that have lost the expression of the c-myc transgene, not in the areas showing high c-Myc levels.36,42,87 These results, together with the report that c-Myc and D1 proteins are reciprocally expressed in colorectal adenocarcinomas,142 suggest that suppression of D1 by c-Myc may occur in vivo as well. However, there are exceptions that are still unexplainable to us, as the D1 level is higher in K5-c-myc transgenic dermal keratinocytes than in the non-c-myc counterparts.143 D1 has also been shown to be induced by c-Myc in the liver and, together with an induction of p53, to contribute to c-Myc-induced PCD.120 Probably, c-Myc may also recruit D1 to cause PCD in certain situations wherein D1 causes PCD.13,144
c-Myc has also been shown to inhibit the mRNA expression of oncogenes other than D1, including the Neu (erbB2),145 vascular endothelial growth factor (VEGF),146 IGF2,147 and most components of the NFκB complex.148–151 Inhibition of Neu by c-Myc reverses Neu-induced transformed morphology,145 which is a surprise because c-Myc is supposed to transform, not to reverse a transformed status. C-Myc can, indirectly, inhibit Bcl-xL and Bcl-2 expression as well.57,152–154 We have also shown that c-Myc inhibits the DNA binding activity of NFκB in mouse pancreatic cancer cells.140 It is reasonable to infer that inhibition of these oncogenes, some of which are known survival factors, may be part of the mechanism for c-Myc-induced PCD (Fig. 3). Indeed, inhibition of NFκB activity has been shown to sensitize murine hepatocytes to c-Myc-induced PCD.74 Expression of the eukaryotic translation initiation factor 4E was shown to repress c-Myc-induced apoptosis of REFs by inducing D1, and expression of D1 attenuated c-Myc-sensitized apoptosis of REFs induced by several cytostatic agents.155 We also observed that ectopic c-myc could abolish D1 expression in pancreatic cancer cells and cause apoptosis, whereas restoration of D1 expression inhibited the apoptosis.140 Therefore, D1 may have a novel function, i.e., serving as a survival factor, in certain situations.
D1 and p53 may Escape from c-Myc's Control Sometime during Carcinogenesis
Although inhibition of D1 is an initial effect of c-Myc, in a chronic situation cells may find ways to activate D1 expression to gain survival ability (Fig. 3). One way is to silence the c-myc transgene, as seen in some focal areas in the MMTV-c-myc transgenic mammary tumors.42 Actually, in some human solid tumors, a spontaneous decrease in the level of the endogenous c-Myc to gain survival has been observed in the areas distant from the blood vessels,9 although it is unclear whether this decrease is companied by an increase in D1. A second mechanism may be oncogenic activation of Ras via such as mutation, as seen in c-myc transgenic mammary tumors,156,157 lung cancers158 and lymphomas,159 since Ras proteins are known to induce D1 expression.13 Actually, Ras mutation and silencing of the c-myc transgene may be related as they are co-localized in the epithelial-mesenchymal-transition areas of MMTV-c-myc mammary tumors.160 Genetic changes in the D1 (CCND1) gene per se or in other D1-regulating genes (besides Ras), leading to ecoptic expression of D1 as seen in the Eµ-myc transgenic lymphomas,161 may be a third mechanism but this still waits for confirmation. In several cell lines we developed from the MMTV-c-myc transgenic mammary tumors and the Ela-myc transgenic pancreatic tumors, D1 is readily detectable as well141 (and our unpublished data). It is likely that those cells that express a higher D1 level have advantages for growth and survival than the others and thus are selected out during the cell line development.
Also for a better survival, some c-myc expressing cells may sooner or later develop mechanisms to block the activation of p53 by c-Myc and eventually inactivate the p53 pathway by p53 mutation, ARF deletion or Mdm2 overexpression as seen in many Eµ-myc transgenic lymphomas.128 In c-myc induced lymphomas, loss of p53 also confers the tumor cells independence of c-myc.8,162,163 However, these changes may occur at a much lower frequency and a much later stage in c-myc induced epithelial cancers, relative to lymphomas. Since loss of p53 or increase in D1 has been shown to enhance cell survival (reviewed in ref. 164 and 165) and to be associated with chemoresistance and radioresistance,139,140,166–168 these changes may render a stronger survival ability to the cells (Fig. 3) but may not be part of the mechanism for the establishment of initiated cells, at least not in epithelial cells, as discussed above.
c-myc Induced Carcinogenesis does not Require Several Survival Factors
Although ectopic Bcl-2 enhances c-myc induced lymphomagenesis in double transgenic mice,47–49 the endogenous Bcl-2 gene is not required for it because c-Myc can still efficiently induce lymphomas in Bcl-2 knockout mice.169 This is understandable since c-Myc inhibits Bcl-2 to a very low level.152,153 D1 and c-Myc seem to have a similar relationship: D1 is inhibited by c-Myc and the endogenous D1 is not required for c-myc induced mammary carcinogenesis in MMTV-c-myc/D1−/− mice,170 but ectopic D1 enhances c-myc induced lymphomagenesis in the Eµ-D1/Eµ-myc double transgenic mice.161 Also similarly, several NFκB components are inhibited by c-Myc and are dispensable for c-myc induced lymphomagenesis.148–151 The fact that c-myc induced carcinogenesis does not require the endogenous alleles of these survival factors supports our overarching hypothesis that inhibition of PCD is not required by, but may promote under certain situations, c-myc induced carcinogenesis, all depending on the balance between cell death and compensatory proliferation. Thus, although suppression of the Nfκb2 gene by c-Myc accelerates lymphomagenesis,150 we anticipate that enhanced NFκB activity may still promote c-myc induced carcinogenesis in some cell types. Moreover, since c-Myc also inhibits Neu expression,145 we speculate that c-myc induced carcinogenesis in certain tissues or organs may not require the endogenous Neu but may be enhanced by overexpressed Neu. Although Neu is not classified as a canonical survival factor, it is reported to enhance cancer cell survival in some situations.171
Do c-Myc Expressing Cells Commit Suicide or Homicide?
In the target organ of a c-myc transgenic animal, millions of cells simultaneously express the c-myc transgene. Since c-Myc induces either PCD or cell proliferation, an intriguing but unanswered question is how the c-myc expressing cells decide, or are selected, to proliferate or to die. In mammalian cells, it seems that a low level of c-Myc is sufficient to drive cell proliferation, but malignant transformation may require increased expression whereas induction of PCD may require an even higher level of overexpression.58,172–174 Moreover, once cells are transformed and develop to tumors, maintenance of their survival may also require a threshold level of c-Myc in many cases,8,173–176 a phenomenon called oncogene addiction.177–179 However, studies on Drosophila Myc (dMyc) suggest that the decision may be made under the influence of cell-cell interaction, because dMyc can cause not only cell-autonomous apoptosis but also cell competition,180,181 in which those cells with a higher dMyc level out-compete those with a lower level and will survive, while those cells with a lower dMyc level will die of apoptosis.181–189 When a S2 Drosophila cell clone that bears an inducible MT-dMyc construct was co-cultured with its empty vector clone, the so-called “cell competition” can be observed soon after the induction of the dMyc by metal.190 The cell-cell interaction seems to be mediated by soluble factors in the culture medium because it does not require a direct contact of the two cell types.190
Although dMyc is known to be functionally equivalent to c-Myc in mammals,186,187,191 currently there is still insufficient evidence for whether the cell competition also occurs in mammals. Nevertheless, it has been shown that when the c-myc is conditionally deleted from some intestinal cells or hepatocytes of mouse, a normal intestine or liver can be rapidly regenerated from a proportion of the wild type cells that out-compete the c-myc deleted regions.192–195 It remains possible that those dying or dead cells seen in c-myc transgenic mice have a relatively lower, although still overexpressing, level of c-myc compared with non-PCD cells and thus die of cell competition. A problem is that although the cell-competition theory also says that c-Myc can cause apoptosis, the cells with a higher level of c-Myc are actually the killers, not the ones that will die, which is obviously different from the concept of “c-Myc induced apoptosis” in human or mouse cells that is defined as a suicidal event, not as a homicide. Unfortunately, although numerous mechanistic studies on mammalian cells have identified the p53 or other genes as a downstream effector to elicit the killing effect of c-Myc, few, if any, of these studies really trace the whole procedure, from the beginning to the end, of the c-Myc action in each individual c-myc expressing cell. Usually, results from combined cells are collected instead, and thus cannot really tell us whether a cell with a relatively higher c-Myc level kills itself or kills others, although the data seem to be some inkling that the death is a suicide. Hence, the conflicting data obtained from Drosophila seem to mean that after all these years of efforts, we come back to the square one again and start to ask who dies (the high c-Myc cell or the low c-Myc cell) and dies of what (suicide or homicide). It is imperative to determine whether c-Myc also elicits cell competition in mice and, if yes, in what situation it commits cell-autonomous apoptosis or cell competition.
If the above described cell-competition also occurs in humans, it provides us a straightforward explanation for the clinical observation of elevated c-Myc levels in human cancers: the elevated c-Myc renders the cells a survival advantage over the surrounding normal cells, and the death of the loser (i.e., the normal) cells may provide a need for a compensatory proliferation to drive the replication of the winner cells. Hence, high c-Myc level cells become dominant and develop to a tumor.73,196,197 This inference implies that a higher c-Myc level may be a bad omen for the patients. However, if it is those with higher c-Myc levels who will die of PCD, a high c-Myc level may be auspicious.61 Unfortunately, the dMyc-triggered cell-competition is known to be influenced by other factors such as the strength of ribosome biogenesis184 or the presence of growth factors such as cyclin D (Drosophila has only one form of cyclin D).181 In a sporadic cancer of humans, the fate of a cancer cell is also influenced by factors other than the c-Myc level, such as activation of other oncogenes or inactivation of tumor suppressor genes. For example, high c-Myc level cells may commit senescence when the Werner gene is concomitantly lost.27 On the other hand, presence of CDK2 will allow c-Myc to suppress senescence25,26,198–201 whereas loss of CDK2 prevents c-myc expressing cells from apoptosis119 although it does not affect c-myc induced tumorigenesis.202 Therefore, the real situation is actually more complex.
No matter whether c-Myc caused death is a suicide or a homicide, it depends not only on the c-Myc level per se but also on the differentiation status of the cells. In a tissue or organ that has a better differentiation status and a lower proliferation potential, it may be more difficult for c-Myc to induce carcinogenesis but easier for it to induce cell death, which may be the case in the c-myc transgenic pancreatic β-cells,57 cardiomyocytes,80 and neural cells.81 In this situation, inhibition of the c-Myc-induced massive cell death is much more needed for an induction of carcinogenesis,57 as seen in the pIns-MycERTAM/RIP7-Bcl-xL double transgenic mice.83 This supposition anticipates that c-myc overexpression in the earlier life of an animal, when cells have not yet reached the terminal differentiation status and have a stronger intrinsic proliferation potential, may be more efficient in the induction of carcinogenesis, relative to its action at an older age. It is also anticipated that the more potent physiological proliferation ability a cell has, the less PCD is required for c-myc induced carcinogenesis. Sporadic cancer (not childhood cancer that is already initiated at an embryonic stage) cannot occur in a cell type that has reached its terminal differentiation and lost the ability to regenerate, such as the heart muscle, nerve or retina. Unfortunately, different organs and tissues of an adult human body have a total of 60 billion cells dying of PCD every day and thus need to regenerate the same number of cells,6 which creates risks for cancer formation in these organs or tissues.
Summary and Perspective
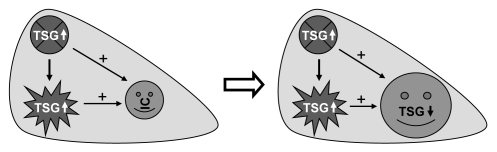
The c-myc is distinguished from other oncogenes by its ability to efficiently induce cancer and massive PCD in transgenic animals. There is also some inkling that c-myc may cause mitoinhibition. The PCD and mitoinhibition first facilitate creation, and then drive proliferation and progression, of the initiated cells that are refractory to PCD, less mitoinhibitory, and thus capable of proliferating to compensate for the cell loss of the organ or tissue. Mechanistically, c-Myc induced PCD and mitoinhibition may involve activation of p53 and inhibition of D1. However, some premalignant or malignant cells may later find ways to escape from these c-Myc's controls, resulting in inactivation of the p53 pathway and/or induction of D1 to gain survival abilities. Most studies on the oncogenicity of c-myc have so far focused on the lineage from a normal status to a malignancy whereas the interaction between initiated cells and their surrounding normal cells is much understudied. Technical constraints may be one of the reasons, since cell proliferation in culture is not for a compensation purpose, whereas transgenic or knockout approach does not allow us to manipulate gene expression specifically in the initiated cells or the surrounding normal cells without affecting the other. Studies on the interaction of these two cell populations may help us to understand why many genes are dual-functional, i.e., both oncogenic and tumor-suppressive. Since most tumor-promoting agents or circumstances promote cancer formation by inhibiting proliferation of normal cells, it is conceivable that many growth-inhibitory or tumor-suppressive genes may also promote carcinogenesis by inhibiting normal cells' proliferation and/or inducing their demise (Fig. 4). This hypothesis deserves further exploration, probably by using chemical carcinogens to induce sporadic formation of initiated cells in controllable transgenic or knockout mice, followed by manipulation of the gene expression.
Figure 4.
Illustration of an extended hypothesis on how increased expression of tumor suppressor gene (TSG) may promote carcinogenesis: Overexpression of TSGs or growth inhibitory genes in a given organ such as the liver may induce programmed cell death (PCD; the irregular star) or inhibit cell proliferation (the crossed spot). Some growth arrested cells may later die of PCD as well. Because the cell death triggers compensatory proliferation (regeneration) of the organ but the normal cells are growth arrested, the initiated cells (IC in the smiley phase) that occur sporadically to be resistant to the mitoinhibition will proliferate in a clonal expansion fashion in order to restore the physiological cell number. Continuous proliferation of the initiated cells allows accumulation of genetic alterations, leading to the development of preneoplastic lesions and eventually malignant tumors. The reason for the resistance of the initiated cells to the mitoinhibition varies, including a relatively lower expression level or inactivation of the TSG, but in many cases decreased or lost function of the TGS may occur later as a step of the process towards the malignancy. In this way, a TSG or growth-inhibitory gene may play a positive role in carcinogenesis, although its expression level in the whole organ may be “lower” or “higher” than the normal control, depending on the ratio of the normal cells to the initiated cells in the organ at the time of measurement (“+” indicates promotion of proliferation).
Acknowledgements
We wish to thank Dr. Fred Bogott at Austin Medical Center, Austin, MN, for his excellent English editing and valuable discussions. This work is supported by NIH grant RO1 CA100864 to D.J.L.
References
- 1.Nesbit CE, Tersak JM, Prochownik EV. MYC oncogenes and human neoplastic disease. Oncogene. 1999;18:3004–3016. doi: 10.1038/sj.onc.1202746. [DOI] [PubMed] [Google Scholar]
- 2.Graves JA, Rothermund K, Wang T, Qian W, Van HB, Prochownik EV. Point mutations in c-Myc uncouple neoplastic transformation from multiple other phenotypes in rat fibroblasts. PLoS One. 2010;5:13717. doi: 10.1371/journal.pone.0013717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Boxer LM, Dang CV. Translocations involving c-myc and c-myc function. Oncogene. 2001;20:5595–5610. doi: 10.1038/sj.onc.1204595. [DOI] [PubMed] [Google Scholar]
- 4.Chung HJ, Levens D. c-myc expression: keep the noise down! Mol Cells. 2005;20:157–166. [PubMed] [Google Scholar]
- 5.Jensen LB, Bartlett JM, Witton CJ, Kirkegaard T, Brown S, Muller S, et al. Frequent amplifications and deletions of G1/S-phase transition genes, CCND1 and MYC in early breast cancers: a potential role in G1/S escape. Cancer Biomark. 2009;5:41–49. doi: 10.3233/CBM-2009-0570. [DOI] [PubMed] [Google Scholar]
- 6.Cotter TG. Apoptosis and cancer: the genesis of a research field. Nat Rev Cancer. 2009;9:501–507. doi: 10.1038/nrc2663. [DOI] [PubMed] [Google Scholar]
- 7.Meyer N, Penn LZ. Reflecting on 25 years with MYC. Nat Rev Cancer. 2008;8:976–990. doi: 10.1038/nrc2231. [DOI] [PubMed] [Google Scholar]
- 8.Felsher DW. myc inactivation elicits oncogene addiction through both tumor cell-intrinsic and host-dependent mechanisms. Genes Cancer. 2010;1:597–604. doi: 10.1177/1947601910377798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Okuyama H, Endo H, Akashika T, Kato K, Inoue M. Downregulation of c-MYC protein levels contributes to cancer cell survival under dual deficiency of oxygen and glucose. Cancer Res. 2010;70:10213–10223. doi: 10.1158/0008-5472.CAN-10-2720. [DOI] [PubMed] [Google Scholar]
- 10.Sansom OJ, Meniel VS, Muncan V, Phesse TJ, Wilkins JA, Reed KR, et al. Myc deletion rescues Apc deficiency in the small intestine. Nature. 2007;446:676–679. doi: 10.1038/nature05674. [DOI] [PubMed] [Google Scholar]
- 11.Wilkins JA, Sansom OJ. C-Myc is a critical mediator of the phenotypes of Apc loss in the intestine. Cancer Res. 2008;68:4963–4966. doi: 10.1158/0008-5472.CAN-07-5558. [DOI] [PubMed] [Google Scholar]
- 12.Athineos D, Sansom OJ. Myc heterozygosity attenuates the phenotypes of APC deficiency in the small intestine. Oncogene. 2010;29:2585–2590. doi: 10.1038/onc.2010.5. [DOI] [PubMed] [Google Scholar]
- 13.Liao D, Thakur A, Wu J, Biliran H, Sarkar FH. Perspectives on c-Myc, cyclin d1 and their interaction in cancer formation, progression and response to chemotherapy. Crit Rev Oncog. 2007;13:93–158. doi: 10.1615/critrevoncog.v13.i2.10. [DOI] [PubMed] [Google Scholar]
- 14.Liao JD, Adsay NV, Khannani F, Grignon D, Thakur A, Sarkar FH. Histological complexities of pancreatic lesions from transgenic mouse models are consistent with biological and morphological heterogeneity of human pancreatic cancer. Histol Histopathol. 2007;22:661–676. doi: 10.14670/hh-22.661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Singh A, Sowjanya AP, Ramakrishna G. The wild-type Ras: road ahead. FASEB J. 2005;19:161–169. doi: 10.1096/fj.04-2584hyp. [DOI] [PubMed] [Google Scholar]
- 16.Lin DI, Lessie MD, Gladden AB, Bassing CH, Wagner KU, Diehl JA. Disruption of cyclin D1 nuclear export and proteolysis accelerates mammary carcinogenesis. Oncogene. 2008;27:1231–1242. doi: 10.1038/sj.onc.1210738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang TC, Cardiff RD, Zukerberg L, Lees E, Arnold A, Schmidt EV. Mammary hyperplasia and carcinoma in MMTV-cyclin D1 transgenic mice. Nature. 1994;369:669–671. doi: 10.1038/369669a0. [DOI] [PubMed] [Google Scholar]
- 18.Deane NG, Parker MA, Aramandla R, Diehl L, Lee WJ, Washington MK, et al. Hepatocellular carcinoma results from chronic cyclin D1 overexpression in transgenic mice. Cancer Res. 2001;61:5389–5395. [PubMed] [Google Scholar]
- 19.Guy CT, Webster MA, Schaller M, Parsons TJ, Cardiff RD, Muller WJ. Expression of the neu protooncogene in the mammary epithelium of transgenic mice induces metastatic disease. Proc Natl Acad Sci USA. 1992;89:10578–10582. doi: 10.1073/pnas.89.22.10578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Siegel PM, Dankort DL, Hardy WR, Muller WJ. Novel activating mutations in the neu proto-oncogene involved in induction of mammary tumors. Mol Cell Biol. 1994;14:7068–7077. doi: 10.1128/mcb.14.11.7068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Siegel PM, Muller WJ. Mutations affecting conserved cysteine residues within the extracellular domain of Neu promote receptor dimerization and activation. Proc Natl Acad Sci USA. 1996;93:8878–8883. doi: 10.1073/pnas.93.17.8878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Siegel PM, Ryan ED, Cardiff RD, Muller WJ. Elevated expression of activated forms of Neu/ErbB-2 and ErbB-3 are involved in the induction of mammary tumors in transgenic mice: implications for human breast cancer. EMBO J. 1999;18:2149–2164. doi: 10.1093/emboj/18.8.2149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Herold S, Herkert B, Eilers M. Facilitating replication under stress: an oncogenic function of MYC? Nat Rev Cancer. 2009;9:441–444. doi: 10.1038/nrc2640. [DOI] [PubMed] [Google Scholar]
- 24.Prochownik EV. c-Myc: linking transformation and genomic instability. Curr Mol Med. 2008;8:446–458. doi: 10.2174/156652408785747988. [DOI] [PubMed] [Google Scholar]
- 25.van RJ, Felsher DW. Myc and a Cdk2 senescence switch. Nat Cell Biol. 2010;12:7–9. doi: 10.1038/ncb0110-7. [DOI] [PubMed] [Google Scholar]
- 26.Campaner S, Doni M, Hydbring P, Verrecchia A, Bianchi L, Sardella D, et al. Cdk2 suppresses cellular senescence induced by the c-myc oncogene. Nat Cell Biol. 2010;12:54–59. doi: 10.1038/ncb2004. [DOI] [PubMed] [Google Scholar]
- 27.Grandori C, Robinson KL, Galloway DA, Swisshelm K. Functional link between Myc and the Werner gene in tumorigenesis. Cell Cycle. 2004;3:22–25. [PubMed] [Google Scholar]
- 28.Evan G, Littlewood T. A matter of life and cell death. Science. 1998;281:1317–1322. doi: 10.1126/science.281.5381.1317. [DOI] [PubMed] [Google Scholar]
- 29.Evan GI, Vousden KH. Proliferation, cell cycle and apoptosis in cancer. Nature. 2001;411:342–348. doi: 10.1038/35077213. [DOI] [PubMed] [Google Scholar]
- 30.Ewen ME, Lamb J. The activities of cyclin D1 that drive tumorigenesis. Trends Mol Med. 2004;10:158–162. doi: 10.1016/j.molmed.2004.02.005. [DOI] [PubMed] [Google Scholar]
- 31.Fiers W, Beyaert R, Declercq W, Vandenabeele P. More than one way to die: apoptosis, necrosis and reactive oxygen damage. Oncogene. 1999;18:7719–7730. doi: 10.1038/sj.onc.1203249. [DOI] [PubMed] [Google Scholar]
- 32.Hoffman B, Liebermann DA. Apoptotic signaling by c-MYC. Oncogene. 2008;27:6462–6472. doi: 10.1038/onc.2008.312. [DOI] [PubMed] [Google Scholar]
- 33.Tsuneoka M, Umata T, Kimura H, Koda Y, Nakajima M, Kosai K, et al. c-myc induces autophagy in rat 3Y1 fibroblast cells. Cell Struct Funct. 2003;28:195–204. doi: 10.1247/csf.28.195. [DOI] [PubMed] [Google Scholar]
- 34.Dang CV. Antimalarial therapy prevents Myc-induced lymphoma. J Clin Invest. 2008;118:15–17. doi: 10.1172/JCI34503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pelengaris S, Rudolph B, Littlewood T. Action of Myc in vivo—proliferation and apoptosis. Curr Opin Genet Dev. 2000;10:100–105. doi: 10.1016/s0959-437x(99)00046-5. [DOI] [PubMed] [Google Scholar]
- 36.Liao DJ, Dickson RB. Cell death in MMTV-c-myc transgenic mouse mammary tumors may not be typical apoptosis. Lab Invest. 2003;83:1437–1449. doi: 10.1097/01.lab.0000090153.13977.ae. [DOI] [PubMed] [Google Scholar]
- 37.Liao DJ. The scavenger cell hypothesis of apoptosis: apoptosis redefined as a process by which a cell in living tissue is destroyed by phagocytosis. Med Hypotheses. 2005;65:23–28. doi: 10.1016/j.mehy.2005.01.028. [DOI] [PubMed] [Google Scholar]
- 38.Lowe SW, Cepero E, Evan G. Intrinsic tumour suppression. Nature. 2004;432:307–315. doi: 10.1038/nature03098. [DOI] [PubMed] [Google Scholar]
- 39.Han EK, Ng SC, Arber N, Begemann M, Weinstein IB. Roles of cyclin D1 and related genes in growth inhibition, senescence and apoptosis. Apoptosis. 1999;4:213–219. doi: 10.1023/a:1009618824145. [DOI] [PubMed] [Google Scholar]
- 40.Cagnol S, Chambard JC. ERK and cell death: mechanisms of ERK-induced cell death—apoptosis, autophagy and senescence. FEBS J. 2010;277:2–21. doi: 10.1111/j.1742-4658.2009.07366.x. [DOI] [PubMed] [Google Scholar]
- 41.Rounbehler RJ, Schneider-Broussard R, Conti CJ, Johnson DG. Myc lacks E2F1's ability to suppress skin carcinogenesis. Oncogene. 2001;20:5341–5349. doi: 10.1038/sj.onc.1204691. [DOI] [PubMed] [Google Scholar]
- 42.Liao DJ, Natarajan G, Deming SL, Jamerson MH, Johnson M, Chepko G, Dickson RB. Cell cycle basis for the onset and progression of c-Myc-induced, TGFalpha-enhanced mouse mammary gland carcinogenesis. Oncogene. 2000;19:1307–1317. doi: 10.1038/sj.onc.1203430. [DOI] [PubMed] [Google Scholar]
- 43.Sutherland KD, Vaillant F, Alexander WS, Wintermantel TM, Forrest NC, Holroyd SL, et al. c-myc as a mediator of accelerated apoptosis and involution in mammary glands lacking Socs3. EMBO J. 2006;25:5805–5815. doi: 10.1038/sj.emboj.7601455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schulte-Hermann R, Bursch W, Kraupp-Grasl B, Oberhammer F, Wagner A. Programmed cell death and its protective role with particular reference to apoptosis. Toxicol Lett. 1992;64:569–574. doi: 10.1016/0378-4274(92)90233-a. [DOI] [PubMed] [Google Scholar]
- 45.Schulte-Hermann R, Bursch W, Kraupp-Grasl B, Oberhammer F, Wagner A, Jirtle R. Cell proliferation and apoptosis in normal liver and preneoplastic foci. Environ Health Perspect. 1993;101:87–90. doi: 10.1289/ehp.93101s587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Vaux DL, Cory S, Adams JM. Bcl-2 gene promotes haemopoietic cell survival and cooperates with c-myc to immortalize pre-B cells. Nature. 1988;335:440–442. doi: 10.1038/335440a0. [DOI] [PubMed] [Google Scholar]
- 47.Marin MC, Hsu B, Stephens LC, Brisbay S, McDonnell TJ. The functional basis of c-myc and bcl-2 complementation during multistep lymphomagenesis in vivo. Exp Cell Res. 1995;217:240–247. doi: 10.1006/excr.1995.1083. [DOI] [PubMed] [Google Scholar]
- 48.Strasser A, Harris AW, Bath ML, Cory S. Novel primitive lymphoid tumours induced in transgenic mice by cooperation between myc and bcl-2. Nature. 1990;348:331–333. doi: 10.1038/348331a0. [DOI] [PubMed] [Google Scholar]
- 49.Smith DP, Bath ML, Metcalf D, Harris AW, Cory S. MYC levels govern hematopoietic tumor type and latency in transgenic mice. Blood. 2006;108:653–661. doi: 10.1182/blood-2006-01-0172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Jager R, Herzer U, Schenkel J, Weiher H. Overexpression of Bcl-2 inhibits alveolar cell apoptosis during involution and accelerates c-myc-induced tumorigenesis of the mammary gland in transgenic mice. Oncogene. 1997;15:1787–1795. doi: 10.1038/sj.onc.1201353. [DOI] [PubMed] [Google Scholar]
- 51.Jager R. Targeting the death machinery in mammary epithelial cells: Implications for breast cancer from transgenic and tissue culture experiments. Crit Rev Oncol Hematol. 2007;63:231–240. doi: 10.1016/j.critrevonc.2007.05.006. [DOI] [PubMed] [Google Scholar]
- 52.Vaux DL. Early work on the function of Bcl-2, an interview with David Vaux. Cell Death Differ. 2004;11:28–32. doi: 10.1038/sj.cdd.4401439. [DOI] [PubMed] [Google Scholar]
- 53.Wagner AJ, Small MB, Hay N. Myc-mediated apoptosis is blocked by ectopic expression of Bcl-2. Mol Cell Biol. 1993;13:2432–2440. doi: 10.1128/mcb.13.4.2432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Cory S, Vaux DL, Strasser A, Harris AW, Adams JM. Insights from Bcl-2 and Myc: malignancy involves abrogation of apoptosis as well as sustained proliferation. Cancer Res. 1999;59:1685–1692. [PubMed] [Google Scholar]
- 55.Naud JF, Eilers M. PIM1 and MYC: a changing relationship? Nat Cell Biol. 2007;9:873–875. doi: 10.1038/ncb0807-873. [DOI] [PubMed] [Google Scholar]
- 56.Gerl R, Vaux DL. Apoptosis in the development and treatment of cancer. Carcinogenesis. 2005;26:263–270. doi: 10.1093/carcin/bgh283. [DOI] [PubMed] [Google Scholar]
- 57.Pelengaris S, Khan M, Evan GI. Suppression of Myc-induced apoptosis in beta cells exposes multiple oncogenic properties of Myc and triggers carcinogenic progression. Cell. 2002;109:321–334. doi: 10.1016/s0092-8674(02)00738-9. [DOI] [PubMed] [Google Scholar]
- 58.Soucek L, Evan GI. The ups and downs of Myc biology. Curr Opin Genet Dev. 2010;20:91–95. doi: 10.1016/j.gde.2009.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Pelengaris S, Khan M, Evan G. c-MYC: more than just a matter of life and death. Nat Rev Cancer. 2002;2:764–776. doi: 10.1038/nrc904. [DOI] [PubMed] [Google Scholar]
- 60.Lowe SW, Lin AW. Apoptosis in cancer. Carcinogenesis. 2000;21:485–495. doi: 10.1093/carcin/21.3.485. [DOI] [PubMed] [Google Scholar]
- 61.Wodarz D, Komarova N. Can loss of apoptosis protect against cancer? Trends Genet. 2007;23:232–237. doi: 10.1016/j.tig.2007.03.005. [DOI] [PubMed] [Google Scholar]
- 62.Pettigrew CA, Cotter TG. Deregulation of cell death (apoptosis): implications for tumor development. Discov Med. 2009;8:61–63. [PubMed] [Google Scholar]
- 63.Jaattela M. Multiple cell death pathways as regulators of tumour initiation and progression. Oncogene. 2004;23:2746–2756. doi: 10.1038/sj.onc.1207513. [DOI] [PubMed] [Google Scholar]
- 64.Tomlin JL, Guinn BA, Penn LZ, Berinstein NL. Bcl-2 and c-Myc co-operate in the Epstein-Barr virus-immortalized human B-cell line GM607 but do not confer tumorigenicity. Leuk Lymphoma. 2005;46:581–592. doi: 10.1080/10428190400019867. [DOI] [PubMed] [Google Scholar]
- 65.de La CA, Mignon A, Fabre M, Gilbert E, Porteu A, Van DT, et al. Paradoxical inhibition of c-myc-induced carcinogenesis by Bcl-2 in transgenic mice. Cancer Res. 1999;59:5017–5022. [PubMed] [Google Scholar]
- 66.Pierce RH, Vail ME, Ralph L, Campbell JS, Fausto N. Bcl-2 expression inhibits liver carcinogenesis and delays the development of proliferating foci. Am J Pathol. 2002;160:1555–1560. doi: 10.1016/S0002-9440(10)61101-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Vail ME, Pierce RH, Fausto N. Bcl-2 delays and alters hepatic carcinogenesis induced by transforming growth factoralpha. Cancer Res. 2001;61:594–601. [PubMed] [Google Scholar]
- 68.Murphy KL, Kittrell FS, Gay JP, Jager R, Medina D, Rosen JM. Bcl-2 expression delays mammary tumor development in dimethylbenz(a)anthracene-treated transgenic mice. Oncogene. 1999;18:6597–6604. doi: 10.1038/sj.onc.1203099. [DOI] [PubMed] [Google Scholar]
- 69.Greider C, Chattopadhyay A, Parkhurst C, Yang E. Bcl-x(L) and Bcl2 delay Myc-induced cell cycle entry through elevation of p27 and inhibition of G1 cyclin-dependent kinases. Oncogene. 2002;21:7765–7775. doi: 10.1038/sj.onc.1205928. [DOI] [PubMed] [Google Scholar]
- 70.Vail ME, Chaisson ML, Thompson J, Fausto N. Bcl-2 expression delays hepatocyte cell cycle progression during liver regeneration. Oncogene. 2002;21:1548–1555. doi: 10.1038/sj.onc.1205212. [DOI] [PubMed] [Google Scholar]
- 71.Joensuu H, Pylkkanen L, Toikkanen S. Bcl-2 protein expression and long-term survival in breast cancer. Am J Pathol. 1994;145:1191–1198. [PMC free article] [PubMed] [Google Scholar]
- 72.Gurova KV, Gudkov AV. Paradoxical role of apoptosis in tumor progression. J Cell Biochem. 2003;88:128–137. doi: 10.1002/jcb.10382. [DOI] [PubMed] [Google Scholar]
- 73.Moreno E. Is cell competition relevant to cancer? Nat Rev Cancer. 2008;8:141–147. doi: 10.1038/nrc2252. [DOI] [PubMed] [Google Scholar]
- 74.Cavin LG, Wang F, Factor VM, Kaur S, Venkatraman M, Thorgeirsson SS, et al. Transforming growth factoralpha inhibits the intrinsic pathway of c-Myc-induced apoptosis through activation of nuclear factorkappaB in murine hepatocellular carcinomas. Mol Cancer Res. 2005;3:403–412. doi: 10.1158/1541-7786.MCR-04-0186. [DOI] [PubMed] [Google Scholar]
- 75.Cheung RS, Brooling JT, Johnson MM, Riehle KJ, Campbell JS, Fausto N. Interactions between MYC and transforming growth factoralpha alter the growth and tumorigenicity of liver progenitor cells. Carcinogenesis. 2007;28:2624–2631. doi: 10.1093/carcin/bgm184. [DOI] [PubMed] [Google Scholar]
- 76.Factor VM, Kao CY, Santoni-Rugiu E, Woitach JT, Jensen MR, Thorgeirsson SS. Constitutive expression of mature transforming growth factor beta1 in the liver accelerates hepatocarcinogenesis in transgenic mice. Cancer Res. 1997;57:2089–2095. [PubMed] [Google Scholar]
- 77.Kim H, Lee H, Yun Y. X-gene product of hepatitis B virus induces apoptosis in liver cells. J Biol Chem. 1998;273:381–385. doi: 10.1074/jbc.273.1.381. [DOI] [PubMed] [Google Scholar]
- 78.Terradillos O, Billet O, Renard CA, Levy R, Molina T, Briand P, et al. The hepatitis B virus X gene potentiates c-myc-induced liver oncogenesis in transgenic mice. Oncogene. 1997;14:395–404. doi: 10.1038/sj.onc.1200850. [DOI] [PubMed] [Google Scholar]
- 79.Cheung WC, Kim JS, Linden M, Peng L, Van NB, Polakiewicz RD, et al. Novel targeted deregulation of c-Myc cooperates with Bcl-X(L) to cause plasma cell neoplasms in mice. J Clin Invest. 2004;113:1763–1773. doi: 10.1172/JCI20369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Lee HG, Chen Q, Wolfram JA, Richardson SL, Liner A, Siedlak SL, et al. Cell cycle re-entry and mitochondrial defects in myc-mediated hypertrophic cardiomyopathy and heart failure. PLoS One. 2009;4:7172. doi: 10.1371/journal.pone.0007172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Lee HG, Casadesus G, Nunomura A, Zhu X, Castellani RJ, Richardson SL, et al. The neuronal expression of MYC causes a neurodegenerative phenotype in a novel transgenic mouse. Am J Pathol. 2009;174:891–897. doi: 10.2353/ajpath.2009.080583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Trudel M, Lanoix J, Barisoni L, Blouin MJ, Desforges M, L'Italien C, et al. C-myc-induced apoptosis in polycystic kidney disease is Bcl-2 and p53 independent. J Exp Med. 1997;186:1873–1884. doi: 10.1084/jem.186.11.1873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Finch A, Prescott J, Shchors K, Hunt A, Soucek L, Dansen TB, et al. Bcl-xL gain of function and p19 ARF loss of function cooperate oncogenically with Myc in vivo by distinct mechanisms. Cancer Cell. 2006;10:113–120. doi: 10.1016/j.ccr.2006.06.017. [DOI] [PubMed] [Google Scholar]
- 84.Radziszewska A, Schroer SA, Choi D, Tajmir P, Radulovich N, Ho JC, et al. Absence of caspase-3 protects pancreatic {beta}-cells from c-Myc-induced apoptosis without leading to tumor formation. J Biol Chem. 2009;284:10947–10956. doi: 10.1074/jbc.M806960200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Jamerson MH, Johnson MD, Korsmeyer SJ, Furth PA, Dickson RB. Bax regulates c-Myc-induced mammary tumour apoptosis but not proliferation in MMTV-c-myc transgenic mice. Br J Cancer. 2004;91:1372–1379. doi: 10.1038/sj.bjc.6602137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Hundley JE, Koester SK, Troyer DA, Hilsenbeck SG, Barrington RE, Windle JJ. Differential regulation of cell cycle characteristics and apoptosis in MMTV-myc and MMTV-ras mouse mammary tumors. Cancer Res. 1997;57:600–603. [PubMed] [Google Scholar]
- 87.Liao DJ, Dickson RB. c-Myc in breast cancer. Endocr Relat Cancer. 2000;7:143–164. doi: 10.1677/erc.0.0070143. [DOI] [PubMed] [Google Scholar]
- 88.Verbeek S, van Lohuizen M, van der Valk, Domen J, Kraal G, Berns A. Mice bearing the E mu-myc and E mu-pim-1 transgenes develop pre-B-cell leukemia prenatally. Mol Cell Biol. 1991;11:1176–1179. doi: 10.1128/mcb.11.2.1176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Tuveson DA, Zhu L, Gopinathan A, Willis NA, Kachatrian L, Grochow R, et al. Mist1-KrasG12D knock-in mice develop mixed differentiation metastatic exocrine pancreatic carcinoma and hepatocellular carcinoma. Cancer Res. 2006;66:242–247. doi: 10.1158/0008-5472.CAN-05-2305. [DOI] [PubMed] [Google Scholar]
- 90.Columbano A, Shinozuka H. Liver regeneration versus direct hyperplasia. FASEB J. 1996;10:1118–1128. doi: 10.1096/fasebj.10.10.8751714. [DOI] [PubMed] [Google Scholar]
- 91.Fan Y, Bergmann A. Apoptosis-induced compensatory proliferation. The Cell is dead. Long live the Cell! Trends Cell Biol. 2008;18:467–473. doi: 10.1016/j.tcb.2008.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Brockes JP, Kumar A. Comparative aspects of animal regeneration. Annu Rev Cell Dev Biol. 2008;24:525–549. doi: 10.1146/annurev.cellbio.24.110707.175336. [DOI] [PubMed] [Google Scholar]
- 93.Bergmann A, Steller H. Apoptosis, stem cells and tissue regeneration. Sci Signal. 2010;3:8. doi: 10.1126/scisignal.3145re8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Laconi E, Doratiotto S, Vineis P. The microenvironments of multistage carcinogenesis. Semin Cancer Biol. 2008;18:322–329. doi: 10.1016/j.semcancer.2008.03.019. [DOI] [PubMed] [Google Scholar]
- 95.Farber E. Cell proliferation as a major risk factor for cancer: a concept of doubtful validity. Cancer Res. 1995;55:3759–3762. [PubMed] [Google Scholar]
- 96.Solt DB, Farber E. A new principle for the analysis of chemical carcinogenesis. Nature. 1976;263:702–703. [Google Scholar]
- 97.Haddow A. Cellular inhibition and the origin of cancer. Acta Unio Int Contra Cancrum. 1938;3:342–353. [Google Scholar]
- 98.Laconi E, Pani P, Farber E. The resistance phenotype in the development and treatment of cancer. Lancet Oncol. 2000;1:235–241. doi: 10.1016/s1470-2045(00)00154-6. [DOI] [PubMed] [Google Scholar]
- 99.Laconi S, Pani P, Pillai S, Pasciu D, Sarma DS, Laconi E. A growth-constrained environment drives tumor progression in vivo. Proc Natl Acad Sci USA. 2001;98:7806–7811. doi: 10.1073/pnas.131210498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Marongiu F, Doratiotto S, Montisci S, Pani P, Laconi E. Liver repopulation and carcinogenesis: two sides of the same coin? Am J Pathol. 2008;172:857–864. doi: 10.2353/ajpath.2008.070910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Kohle C, Schwarz M, Bock KW. Promotion of hepatocarcinogenesis in humans and animal models. Arch Toxicol. 2008;82:623–631. doi: 10.1007/s00204-007-0273-7. [DOI] [PubMed] [Google Scholar]
- 102.Farber E, Rubin H. Cellular adaptation in the origin and development of cancer. Cancer Res. 1991;51:2751–2761. [PubMed] [Google Scholar]
- 103.Chang DW, Claassen GF, Hann SR, Cole MD. The c-Myc transactivation domain is a direct modulator of apoptotic versus proliferative signals. Mol Cell Biol. 2000;20:4309–4319. doi: 10.1128/mcb.20.12.4309-4319.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Dang CV, O'Donnell KA, Juopperi T. The great MYC escape in tumorigenesis. Cancer Cell. 2005;8:177–178. doi: 10.1016/j.ccr.2005.08.005. [DOI] [PubMed] [Google Scholar]
- 105.Pusapati RV, Weaks RL, Rounbehler RJ, McArthur MJ, Johnson DG. E2F2 suppresses Myc-induced proliferation and tumorigenesis. Mol Carcinog. 2009 doi: 10.1002/mc.20584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Staller P, Peukert K, Kiermaier A, Seoane J, Lukas J, Karsunky H, et al. Repression of p15INK4b expression by Myc through association with Miz-1. Nat Cell Biol. 2001;3:392–399. doi: 10.1038/35070076. [DOI] [PubMed] [Google Scholar]
- 107.Coller HA, Grandori C, Tamayo P, Colbert T, Lander ES, Eisenman RN, et al. Expression analysis with oligonucleotide microarrays reveals that MYC regulates genes involved in growth, cell cycle, signaling and adhesion. Proc Natl Acad Sci USA. 2000;97:3260–3265. doi: 10.1073/pnas.97.7.3260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Bush A, Mateyak M, Dugan K, Obaya A, Adachi S, Sedivy J, et al. c-myc null cells misregulate cad and gadd45 but not other proposed c-Myc targets. Genes Dev. 1998;12:3797–3802. doi: 10.1101/gad.12.24.3797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Felsher DW, Zetterberg A, Zhu J, Tlsty T, Bishop JM. Overexpression of MYC causes p53-dependent G2 arrest of normal fibroblasts. Proc Natl Acad Sci USA. 2000;97:10544–10548. doi: 10.1073/pnas.190327097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Arvanitis C, Felsher DW. Conditional transgenic models define how MYC initiates and maintains tumorigenesis. Semin Cancer Biol. 2006;16:313–317. doi: 10.1016/j.semcancer.2006.07.012. [DOI] [PubMed] [Google Scholar]
- 111.Beer S, Zetterberg A, Ihrie RA, McTaggart RA, Yang Q, Bradon N, et al. Developmental context determines latency of MYC-induced tumorigenesis. PLoS Biol. 2004;2:332. doi: 10.1371/journal.pbio.0020332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Beer S, Bellovin DI, Lee JS, Komatsubara K, Wang LS, Koh H, et al. Low-level shRNA cytotoxicity can contribute to MYC-induced hepatocellular carcinoma in adult mice. Mol Ther. 2010;18:161–170. doi: 10.1038/mt.2009.222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Beer S, Komatsubara K, Bellovin DI, Kurobe M, Sylvester K, Felsher DW. Hepatotoxin-induced changes in the adult murine liver promote MYC-induced tumorigenesis. PLoS One. 2008;3:2493. doi: 10.1371/journal.pone.0002493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Nikiforov MA, Riblett M, Tang WH, Gratchouck V, Zhuang D, Fernandez Y, et al. Tumor cell-selective regulation of NOXA by c-MYC in response to proteasome inhibition. Proc Natl Acad Sci USA. 2007;104:19488–19493. doi: 10.1073/pnas.0708380104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Thakur A, Sun Y, Bollig A, Wu J, Biliran H, Banerjee S, et al. Anti-invasive and antimetastatic activities of ribosomal protein S6 kinase 4 in breast cancer cells. Clin Cancer Res. 2008;14:4427–4436. doi: 10.1158/1078-0432.CCR-08-0458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Egle A, Harris AW, Bouillet P, Cory S. Bim is a suppressor of Myc-induced mouse B cell leukemia. Proc Natl Acad Sci USA. 2004;101:6164–6169. doi: 10.1073/pnas.0401471101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Hemann MT, Bric A, Teruya-Feldstein J, Herbst A, Nilsson JA, Cordon-Cardo C, et al. Evasion of the p53 tumour surveillance network by tumour-derived MYC mutants. Nature. 2005;436:807–811. doi: 10.1038/nature03845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Hermeking H, Eick D. Mediation of c-Myc-induced apoptosis by p53. Science. 1994;265:2091–2093. doi: 10.1126/science.8091232. [DOI] [PubMed] [Google Scholar]
- 119.Deb-Basu D, Aleem E, Kaldis P, Felsher DW. CDK2 is required by MYC to induce apoptosis. Cell Cycle. 2006;5:1342–1347. doi: 10.4161/cc.5.12.2859. [DOI] [PubMed] [Google Scholar]
- 120.Yang H, Li TW, Ko KS, Xia M, Lu SC. Switch from Mnt-Max to Myc-Max induces p53 and cyclin D1 expression and apoptosis during cholestasis in mouse and human hepatocytes. Hepatology. 2009;49:860–870. doi: 10.1002/hep.22720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Opavsky R, Tsai SY, Guimond M, Arora A, Opavska J, Becknell B, et al. Specific tumor suppressor function for E2F2 in Myc-induced T cell lymphomagenesis. Proc Natl Acad Sci USA. 2007;104:15400–15405. doi: 10.1073/pnas.0706307104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Nass SJ, Li M, Amundadottir LT, Furth PA, Dickson RB. Role for Bcl-xL in the regulation of apoptosis by EGF and TGFbeta1 in c-myc overexpressing mammary epithelial cells. Biochem Biophys Res Commun. 1996;227:248–256. doi: 10.1006/bbrc.1996.1497. [DOI] [PubMed] [Google Scholar]
- 123.Elson A, Deng C, Campos-Torres J, Donehower LA, Leder P. The MMTV/c-myc transgene and p53 null alleles collaborate to induce T-cell lymphomas, but not mammary carcinomas in transgenic mice. Oncogene. 1995;11:181–190. [PubMed] [Google Scholar]
- 124.McCormack SJ, Weaver Z, Deming S, Natarajan G, Torri J, Johnson MD, et al. Myc/p53 interactions in transgenic mouse mammary development, tumorigenesis and chromosomal instability. Oncogene. 1998;16:2755–2766. doi: 10.1038/sj.onc.1201804. [DOI] [PubMed] [Google Scholar]
- 125.Liao DJ, Wang Y, Wu J, Adsay NV, Grignon D, Khanani F, et al. Characterization of pancreatic lesions from MT-tgfalpha, Ela-myc and MT-tgfalpha/Ela-myc single and double transgenic mice. J Carcinog. 2006;5:1–19. doi: 10.1186/1477-3163-5-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Hong S, Pusapati RV, Powers JT, Johnson DG. Oncogenes and the DNA damage response: Myc and E2F1 engage the ATM signaling pathway to activate p53 and induce apoptosis. Cell Cycle. 2006;5:801–803. doi: 10.4161/cc.5.8.2638. [DOI] [PubMed] [Google Scholar]
- 127.Blyth K, Vaillant F, Hanlon L, Mackay N, Bell M, Jenkins A, et al. Runx2 and MYC collaborate in lymphoma development by suppressing apoptotic and growth arrest pathways in vivo. Cancer Res. 2006;66:2195–2201. doi: 10.1158/0008-5472.CAN-05-3558. [DOI] [PubMed] [Google Scholar]
- 128.Eischen CM, Weber JD, Roussel MF, Sherr CJ, Cleveland JL. Disruption of the ARF-Mdm2-p53 tumor suppressor pathway in Myc-induced lymphoma-genesis. Genes Dev. 1999;13:2658–2669. doi: 10.1101/gad.13.20.2658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Jansen-Durr P, Meichle A, Steiner P, Pagano M, Finke K, Botz J, et al. Differential modulation of cyclin gene expression by MYC. Proc Natl Acad Sci USA. 1993;90:3685–3689. doi: 10.1073/pnas.90.8.3685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Marhin WW, Hei YJ, Chen S, Jiang Z, Gallie BL, Phillips RA, et al. Loss of Rb and Myc activation co-operate to suppress cyclin D1 and contribute to transformation. Oncogene. 1996;12:43–52. [PubMed] [Google Scholar]
- 131.Philipp A, Schneider A, Vasrik I, Finke K, Xiong Y, Beach D, et al. Repression of cyclin D1: a novel function of MYC. Mol Cell Biol. 1994;14:4032–4043. doi: 10.1128/mcb.14.6.4032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Cowling VH. Enhanced mRNA cap methylation increases cyclin D1 expression and promotes cell transformation. Oncogene. 2010;29:930–936. doi: 10.1038/onc.2009.368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Huerta M, Munoz R, Tapia R, Soto-Reyes E, Ramirez L, Recillas-Targa F, et al. Cyclin D1 is transcriptionally downregulated by ZO-2 via an E box and the transcription factor c-Myc. Mol Biol Cell. 2007;18:4826–4836. doi: 10.1091/mbc.E07-02-0109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Chien J, Narita K, Rattan R, Giri S, Shridhar R, Staub J, et al. A role for candidate tumor-suppressor gene TCEAL7 in the regulation of c-Myc activity, cyclin D1 levels and cellular transformation. Oncogene. 2008;27:7223–7234. doi: 10.1038/onc.2008.360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Guo QM, Malek RL, Kim S, Chiao C, He M, Ruffy M, et al. Identification of c-myc responsive genes using rat cDNA microarray. Cancer Res. 2000;60:5922–5928. [PubMed] [Google Scholar]
- 136.Mateyak MK, Obaya AJ, Sedivy JM. c-Myc regulates cyclin D-Cdk4 and -Cdk6 activity but affects cell cycle progression at multiple independent points. Mol Cell Biol. 1999;19:4672–4683. doi: 10.1128/mcb.19.7.4672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Hermeking H, Rago C, Schuhmacher M, Li Q, Barrett JF, Obaya AJ, et al. Identification of CDK4 as a target of c-MYC. Proc Natl Acad Sci USA. 2000;97:2229–2234. doi: 10.1073/pnas.050586197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Amendola D, De SM, Marchese R, Verga FC, Stigliano A, Carico E, et al. Myc downregulation affects cyclin D1/cdk4 activity and induces apoptosis via Smac/Diablo pathway in an astrocytoma cell line. Cell Prolif. 2009;42:94–109. doi: 10.1111/j.1365-2184.2008.00576.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Biliran H, Jr, Wang Y, Banerjee S, Xu H, Heng H, Thakur A, et al. Overexpression of cyclin D1 promotes tumor cell growth and confers resistance to cisplatin-mediated apoptosis in an elastase-myc transgeneexpressing pancreatic tumor cell line. Clin Cancer Res. 2005;11:6075–6086. doi: 10.1158/1078-0432.CCR-04-2419. [DOI] [PubMed] [Google Scholar]
- 140.Biliran H, Jr, Banerjee S, Thakur A, Sarkar FH, Bollig A, Ahmed F, et al. c-Myc-induced chemosensitization is mediated by suppression of cyclin D1 expression and nuclear factorkappa B activity in pancreatic cancer cells. Clin Cancer Res. 2007;13:2811–2821. doi: 10.1158/1078-0432.CCR-06-1844. [DOI] [PubMed] [Google Scholar]
- 141.Wu J, Wu SH, Bollig A, Thakur A, Liao DJ. Identification of the cyclin D1b mRNA variant in mouse. Mol Biol Rep. 2009;36:953–957. doi: 10.1007/s11033-008-9267-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Wang HL, Wang J, Xiao SY, Haydon R, Stoiber D, He TC, et al. Elevated protein expression of cyclin D1 and Fra-1 but decreased expression of c-Myc in human colorectal adenocarcinomas overexpressing beta-catenin. Int J Cancer. 2002;101:301–310. doi: 10.1002/ijc.10630. [DOI] [PubMed] [Google Scholar]
- 143.Miliani de Marval PL, Macias E, Rounbehler R, Sicinski P, Kiyokawa H, Johnson DG, et al. Lack of cyclin-dependent kinase 4 inhibits c-myc tumorigenic activities in epithelial tissues. Mol Cell Biol. 2004;24:7538–7547. doi: 10.1128/MCB.24.17.7538-7547.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Li Z, Jiao X, Wang C, Shirley LA, Elsaleh H, Dahl O, et al. Alternative cyclin D1 splice forms differentially regulate the DNA damage response. Cancer Res. 2010;70:8802–8811. doi: 10.1158/0008-5472.CAN-10-0312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Suen TC, Hung MC. c-myc reverses neu-induced transformed morphology by transcriptional repression. Mol Cell Biol. 1991;11:354–362. doi: 10.1128/mcb.11.1.354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Barr LF, Campbell SE, Diette GB, Gabrielson EW, Kim S, Shim H, et al. c-Myc suppresses the tumorigenicity of lung cancer cells and downregulates vascular endothelial growth factor expression. Cancer Res. 2000;60:143–149. [PubMed] [Google Scholar]
- 147.Barsyte-Lovejoy D, Lau SK, Boutros PC, Khosravi F, Jurisica I, Andrulis IL, et al. The c-Myc oncogene directly induces the H19 noncoding RNA by allelespecific binding to potentiate tumorigenesis. Cancer Res. 2006;66:5330–5337. doi: 10.1158/0008-5472.CAN-06-0037. [DOI] [PubMed] [Google Scholar]
- 148.Keller U, Nilsson JA, Maclean KH, Old JB, Cleveland JL. NFκB 1 is dispensable for Myc-induced lymphomagenesis. Oncogene. 2005;24:6231–6240. doi: 10.1038/sj.onc.1208779. [DOI] [PubMed] [Google Scholar]
- 149.Schlee M, Holzel M, Bernard S, Mailhammer R, Schuhmacher M, Reschke J, et al. C-myc activation impairs the NFkappaB and the interferon response: implications for the pathogenesis of Burkitt's lymphoma. Int J Cancer. 2007;120:1387–1395. doi: 10.1002/ijc.22372. [DOI] [PubMed] [Google Scholar]
- 150.Keller U, Huber J, Nilsson JA, Fallahi M, Hall MA, Peschel C, et al. Myc suppression of Nfkb2 accelerates lymphomagenesis. BMC Cancer. 2010;10:348. doi: 10.1186/1471-2407-10-348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Klapproth K, Sander S, Marinkovic D, Baumann B, Wirth T. The IKK2/NF-{kappa}B pathway suppresses MYC-induced lymphomagenesis. Blood. 2009;114:2448–2458. doi: 10.1182/blood-2008-09-181008. [DOI] [PubMed] [Google Scholar]
- 152.Eischen CM, Woo D, Roussel MF, Cleveland JL. Apoptosis triggered by Myc-induced suppression of Bcl-X(L) or Bcl-2 is bypassed during lymphomagenesis. Mol Cell Biol. 2001;21:5063–5070. doi: 10.1128/MCB.21.15.5063-5070.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Eischen CM, Packham G, Nip J, Fee BE, Hiebert SW, Zambetti GP, et al. Bcl-2 is an apoptotic target suppressed by both c-Myc and E2F-1. Oncogene. 2001;20:6983–6993. doi: 10.1038/sj.onc.1204892. [DOI] [PubMed] [Google Scholar]
- 154.Maclean KH, Keller UB, Rodriguez-Galindo C, Nilsson JA, Cleveland JL. c-Myc augments gamma irradiation-induced apoptosis by suppressing Bcl-XL. Mol Cell Biol. 2003;23:7256–7270. doi: 10.1128/MCB.23.20.7256-7270.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Tan A, Bitterman P, Sonenberg N, Peterson M, Polunovsky V. Inhibition of Myc-dependent apoptosis by eukaryotic translation initiation factor 4E requires cyclin D1. Oncogene. 2000;19:1437–1447. doi: 10.1038/sj.onc.1203446. [DOI] [PubMed] [Google Scholar]
- 156.D'Cruz CM, Gunther EJ, Boxer RB, Hartman JL, Sintasath L, Moody SE, et al. c-MYC induces mammary tumorigenesis by means of a preferred pathway involving spontaneous Kras2 mutations. Nat Med. 2001;7:235–239. doi: 10.1038/84691. [DOI] [PubMed] [Google Scholar]
- 157.Jang JW, Boxer RB, Chodosh LA. Isoform-specific ras activation and oncogene dependence during MYC- and Wnt-induced mammary tumorigenesis. Mol Cell Biol. 2006;26:8109–8121. doi: 10.1128/MCB.00404-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Rapp UR, Korn C, Ceteci F, Karreman C, Luetkenhaus K, Serafin V, et al. MYC is a metastasis gene for non-small-cell lung cancer. PLoS One. 2009;4:6029. doi: 10.1371/journal.pone.0006029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Alexander WS, Bernard O, Cory S, Adams JM. Lymphomagenesis in Emu-myc transgenic mice can involve ras mutations. Oncogene. 1989;4:575–581. [PubMed] [Google Scholar]
- 160.Andrechek ER, Cardiff RD, Chang JT, Gatza ML, Acharya CR, Potti A, et al. Genetic heterogeneity of Myc-induced mammary tumors reflecting diverse phenotypes including metastatic potential. Proc Natl Acad Sci USA. 2009;106:16387–16392. doi: 10.1073/pnas.0901250106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Bodrug SE, Warner BJ, Bath ML, Lindeman GJ, Harris AW, Adams JM. Cyclin D1 transgene impedes lymphocyte maturation and collaborates in lymphomagenesis with the myc gene. EMBO J. 1994;13:2124–2130. doi: 10.1002/j.1460-2075.1994.tb06488.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Giuriato S, Ryeom S, Fan AC, Bachireddy P, Lynch RC, Rioth MJ, et al. Sustained regression of tumors upon MYC inactivation requires p53 or thrombospondin-1 to reverse the angiogenic switch. Proc Natl Acad Sci USA. 2006;103:16266–16271. doi: 10.1073/pnas.0608017103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Wu CH, van RJ, Yetil A, Fan AC, Bachireddy P, Felsher DW. Cellular senescence is an important mechanism of tumor regression upon c-Myc inactivation. Proc Natl Acad Sci USA. 2007;104:13028–13033. doi: 10.1073/pnas.0701953104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Gan L, Liu P, Lu H, Chen S, Yang J, McCarthy JB, et al. Cyclin D1 promotes anchorage-independent cell survival by inhibiting FOXO-mediated anoikis. Cell Death Differ. 2009;16:1408–1417. doi: 10.1038/cdd.2009.86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Roue G, Pichereau V, Lincet H, Colomer D, Sola B. Cyclin D1 mediates resistance to apoptosis through upregulation of molecular chaperones and consequent redistribution of cell death regulators. Oncogene. 2008;27:4909–4920. doi: 10.1038/onc.2008.126. [DOI] [PubMed] [Google Scholar]
- 166.Noel EE, Yeste-Velasco M, Mao X, Perry J, Kudahetti SC, Li NF, et al. The association of CCND1 overexpression and cisplatin resistance in testicular germ cell tumors and other cancers. Am J Pathol. 2010;176:2607–2615. doi: 10.2353/ajpath.2010.090780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Shimura T, Kakuda S, Ochiai Y, Nakagawa H, Kuwahara Y, Takai Y, et al. Acquired radioresistance of human tumor cells by DNA-PK/AKT/GSK3beta-mediated cyclin D1 overexpression. Oncogene. 2010;29:4826–4837. doi: 10.1038/onc.2010.238. [DOI] [PubMed] [Google Scholar]
- 168.Smalley KS, Lioni M, Dalla PM, Xiao M, Desai B, Egyhazi S, et al. Increased cyclin D1 expression can mediate BRAF inhibitor resistance in BRAF V600E-mutated melanomas. Mol Cancer Ther. 2008;7:2876–2883. doi: 10.1158/1535-7163.MCT-08-0431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Kelly PN, Puthalakath H, Adams JM, Strasser A. Endogenous bcl-2 is not required for the development of Emu-myc-induced B-cell lymphoma. Blood. 2007;109:4907–4913. doi: 10.1182/blood-2006-10-051847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Yu Q, Geng Y, Sicinski P. Specific protection against breast cancers by cyclin D1 ablation. Nature. 2001;411:1017–1021. doi: 10.1038/35082500. [DOI] [PubMed] [Google Scholar]
- 171.Freudenberg JA, Wang Q, Katsumata M, Drebin J, Nagatomo I, Greene MI. The role of HER2 in early breast cancer metastasis and the origins of resistance to HER2-targeted therapies. Exp Mol Pathol. 2009;87:1–11. doi: 10.1016/j.yexmp.2009.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Freie BW, Eisenman RN. Ratcheting Myc. Cancer Cell. 2008;14:425–426. doi: 10.1016/j.ccr.2008.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Murphy DJ, Junttila MR, Pouyet L, Karnezis A, Shchors K, Bui DA, et al. Distinct thresholds govern Myc's biological output in vivo. Cancer Cell. 2008;14:447–457. doi: 10.1016/j.ccr.2008.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Shachaf CM, Gentles AJ, Elchuri S, Sahoo D, Soen Y, Sharpe O, et al. Genomic and proteomic analysis reveals a threshold level of MYC required for tumor maintenance. Cancer Res. 2008;68:5132–5142. doi: 10.1158/0008-5472.CAN-07-6192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Felsher DW. Tumor dormancy: death and resurrection of cancer as seen through transgenic mouse models. Cell Cycle. 2006;5:1808–1811. doi: 10.4161/cc.5.16.3111. [DOI] [PubMed] [Google Scholar]
- 176.Felsher DW. Tumor dormancy and oncogene addiction. APMIS. 2008;116:629–637. doi: 10.1111/j.1600-0463.2008.01037.x. [DOI] [PubMed] [Google Scholar]
- 177.Weinstein IB, Joe A. Oncogene addiction. Cancer Res. 2008;68:3077–3080. doi: 10.1158/0008-5472.CAN-07-3293. [DOI] [PubMed] [Google Scholar]
- 178.Weinstein IB, Joe AK. Mechanisms of disease: Oncogene addiction—a rationale for molecular targeting in cancer therapy. Nat Clin Pract Oncol. 2006;3:448–457. doi: 10.1038/ncponc0558. [DOI] [PubMed] [Google Scholar]
- 179.Weinstein IB. Cancer. Addiction to oncogenes—the Achilles heal of cancer. Science. 2002;297:63–64. doi: 10.1126/science.1073096. [DOI] [PubMed] [Google Scholar]
- 180.Montero L, Muller N, Gallant P. Induction of apoptosis by Drosophila Myc. Genesis. 2008;46:104–111. doi: 10.1002/dvg.20373. [DOI] [PubMed] [Google Scholar]
- 181.de la Cova C, Abril M, Bellosta P, Gallant P, Johnston LA. Drosophila myc regulates organ size by inducing cell competition. Cell. 2004;117:107–116. doi: 10.1016/s0092-8674(04)00214-4. [DOI] [PubMed] [Google Scholar]
- 182.Donaldson TD, Duronio RJ. Cancer cell biology: Myc wins the competition. Curr Biol. 2004;14:425–427. doi: 10.1016/j.cub.2004.05.035. [DOI] [PubMed] [Google Scholar]
- 183.Gallant P. Myc, cell competition and compensatory proliferation. Cancer Res. 2005;65:6485–6487. doi: 10.1158/0008-5472.CAN-05-1101. [DOI] [PubMed] [Google Scholar]
- 184.Moreno E, Basler K. dMyc transforms cells into supercompetitors. Cell. 2004;117:117–129. doi: 10.1016/s0092-8674(04)00262-4. [DOI] [PubMed] [Google Scholar]
- 185.Gallant P. Drosophila Myc. Adv Cancer Res. 2009;103:111–144. doi: 10.1016/S0065-230X(09)03005-X. [DOI] [PubMed] [Google Scholar]
- 186.Siddall NA, Lin JI, Hime GR, Quinn LM. Myc—what we have learned from flies. Curr Drug Targets. 2009;10:590–601. doi: 10.2174/138945009788680400. [DOI] [PubMed] [Google Scholar]
- 187.Bellosta P, Gallant P. Myc Function in Drosophila. Genes Cancer. 2010;1:542–546. doi: 10.1177/1947601910377490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Gallant P, Steiger D. Myc's secret life without Max. Cell Cycle. 2009;8:3848–3853. doi: 10.4161/cc.8.23.10088. [DOI] [PubMed] [Google Scholar]
- 189.Portela M, Casas-Tinto S, Rhiner C, Lopez-Gay JM, Dominguez O, Soldini D, et al. Drosophila SPARC is a self-protective signal expressed by loser cells during cell competition. Dev Cell. 2010;19:562–573. doi: 10.1016/j.devcel.2010.09.004. [DOI] [PubMed] [Google Scholar]
- 190.Senoo-Matsuda N, Johnston LA. Soluble factors mediate competitive and cooperative interactions between cells expressing different levels of Drosophila Myc. Proc Natl Acad Sci USA. 2007;104:18543–18548. doi: 10.1073/pnas.0709021104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Benassayag C, Montero L, Colombie N, Gallant P, Cribbs D, Morello D. Human c-Myc isoforms differentially regulate cell growth and apoptosis in Drosophila melanogaster. Mol Cell Biol. 2005;25:9897–9909. doi: 10.1128/MCB.25.22.9897-9909.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Muncan V, Sansom OJ, Tertoolen L, Phesse TJ, Begthel H, Sancho E, et al. Rapid loss of intestinal crypts upon conditional deletion of the Wnt/Tcf-4 target gene c-Myc. Mol Cell Biol. 2006;26:8418–8426. doi: 10.1128/MCB.00821-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Bettess MD, Dubois N, Murphy MJ, Dubey C, Roger C, Robine S, et al. c-Myc is required for the formation of intestinal crypts but dispensable for homeostasis of the adult intestinal epithelium. Mol Cell Biol. 2005;25:7868–7878. doi: 10.1128/MCB.25.17.7868-7878.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Li F, Xiang Y, Potter J, Dinavahi R, Dang CV, Lee LA. Conditional deletion of c-myc does not impair liver regeneration. Cancer Res. 2006;66:5608–5612. doi: 10.1158/0008-5472.CAN-05-4242. [DOI] [PubMed] [Google Scholar]
- 195.Levens D. You don't muck with MYC. Genes Cancer. 2010;1:547–554. doi: 10.1177/1947601910377492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Baker NE, Li W. Cell competition and its possible relation to cancer. Cancer Res. 2008;68:5505–5507. doi: 10.1158/0008-5472.CAN-07-6348. [DOI] [PubMed] [Google Scholar]
- 197.Rhiner C, Moreno E. Super competition as a possible mechanism to pioneer precancerous fields. Carcinogenesis. 2009;30:723–728. doi: 10.1093/carcin/bgp003. [DOI] [PubMed] [Google Scholar]
- 198.Hydbring P, Larsson LG. Tipping the balance: Cdk2 enables Myc to suppress senescence. Cancer Res. 2010;70:6687–6691. doi: 10.1158/0008-5472.CAN-10-1383. [DOI] [PubMed] [Google Scholar]
- 199.Hydbring P, Larsson LG. Cdk2: a key regulator of the senescence control function of Myc. Aging (Albany NY) 2010;2:244–250. doi: 10.18632/aging.100140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Hydbring P, Bahram F, Su Y, Tronnersjo S, Hogstrand K, Von Der LN, et al. Phosphorylation by Cdk2 is required for Myc to repress Ras-induced senescence in cotransformation. Proc Natl Acad Sci USA. 2010;107:58–63. doi: 10.1073/pnas.0900121106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Campaner S, Doni M, Verrecchia A, Faga G, Bianchi L, Amati B. Myc, Cdk2 and cellular senescence: Old players, new game. Cell Cycle. 2010;9:3655–3661. [PubMed] [Google Scholar]
- 202.Macias E, Kim Y, Miliani de Marval PL, Klein-Szanto A, Rodriguez-Puebla ML. Cdk2 deficiency decreases ras/CDK4-dependent malignant progression, but not myc-induced tumorigenesis. Cancer Res. 2007;67:9713–9720. doi: 10.1158/0008-5472.CAN-07-2119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Liao DZ, Porsch-Hallstrom I, Gustafsson JA, Blanck A. Persistent sex differences in growth control of early rat liver lesions are programmed during promotion in the resistant hepatocyte model. Hepatology. 1996;23:835–839. doi: 10.1002/hep.510230426. [DOI] [PubMed] [Google Scholar]