Abstract
The cadherin family is classified into classical cadherins, desmosomal cadherins and protocadherins (PCDHs). Genomic structures distinguish between PCDHs and other cadherins, and between clustered and non-clustered PCDHs. The phylogenetic analysis with full sequences of non-clustered PCDHs enabled them to be further classified into three subgroups: δ1 (PCDH1, PCDH7, PCDH9, PCDH11 and PCDH20), δ2 (PCDH8, PCDH10, PCDH12, PCDH17, PCDH18 and PCDH19) and ε (PCDH15, PCDH16, PCDH21 and MUCDHL). ε-PCDH members except PCDH21 have either higher or lower numbers of cadherin repeats than those of other PCDHs. Non-clustered PCDHs are expressed predominantly in the nervous system and have spatiotemporally diverse expression patterns. Especially, the region-specific expressions of non-clustered PCDHs have been observed in cortical area of early postnatal stage and in caudate putaman and/or hippocampal formation of mature brains, suggesting that non-clustered PCDHs play roles in the circuit formation and maintenance. The non-clustered PCDHs appear to have homophilic/heterophilic cell-cell adhesion properties, and each member has diverse cell signaling partnership distinct from those of other members (PCDH7/TAF1; PCDH8/TAO2δ; PCDH10/Nap1; PCDH11/δ-catenin; PCDH18/mDab1). Furthermore, each PCDH has several isoforms with differential cytoplasmic sequences, suggesting that one PCDH isoform could activate intracellular signaling differential from other isoforms. These facts suggest that non-clustered PCDHs play roles as a mediator of a regulator of other molecules as well as cell-cell adhesion. Furthermore, some non-clustered PCDHs have been considered to be involved in neuronal diseases such as autism-spectrum disorders, schizophrenia and female-limited epilepsy and cognitive impairment, suggesting that they play multiple, tightly regulated roles in normal brain function. In addition, some non-clustered PCDHs have been suggested as candidate tumor suppressor genes in several tissues. Although molecular adhesive and regulatory properties of some PCDHs began to be unveiled, the endeavor to understand the molecular mechanism of non-clustered PCDH is still in its infancy and requires future study.
Key words: non-clustered protocadherins, gene structure, cell adhesion, intracellular signaling, neural disease, cancer
Introduction
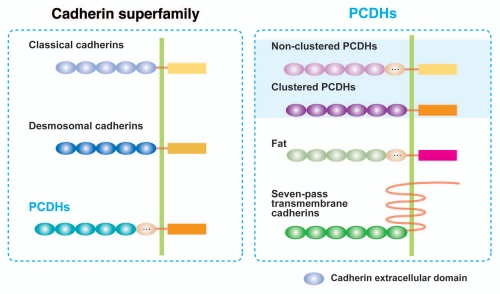
Cadherin is a calcium-dependent adhesion protein that constitutes a large family of cell adhesion molecules. Cadherins have been identified by the presence of extracellular cadherin repeats of about 110 amino acid residues, and can be classified into several subfamilies based on shared properties and sequence similarity (Fig. 1): the classical cadherins, desmosomal cadherins and protocadherins (PCDHs).1,2 The PCDH family can be divided largely into two groups, based on their genomic structure: clustered PCDHs and non-clustered PCDHs.3–5 The term “PCDH,” however, sometimes includes Fats and seven-pass transmembrane cadherins (Flamingo/CELSER) in the broad sense.6–9 Here, the term “PCDH” is used in a restricted sense, including only clustered and non-clustered PCDHs. PCDHs are expressed predominantly in the nervous system,10,11 and constitute the largest subgroup (about 80 members) of the cadherin superfamily.12,13
Figure 1.
Classification of cadherin superfamily including PCDHs. All cadherin superfamily members have calcium-binding cadherin repeats. The number of cadherin repeats varies from one subfamily to another. On average, a single cadherin repeat contains 110 amino acids in length. PCDHs are largely classified into the clustered PCDHs and non-clustered PCDHs, yet also include fats and seven-pass transmembrane cadherins in some cases. The clustered PCDHs are consisted of the PCDHα, β and γ family, which is clustered in a small genome locus. Non-clustered PCDHs are scattered in several genome loci.
In this review, we will focus on recent findings of non-clustered PCDHs, and attempt to provide further insights into the molecular mechanisms and disease-relationship of non-clustered PCDH members on which the findings have been accumulated over the past few years.
Classification and Genomic Structures of Non-Clustered PCDHs
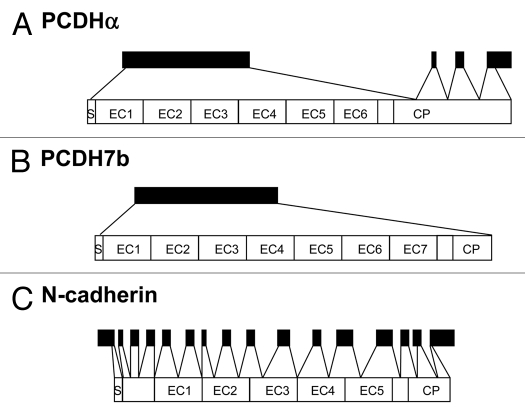
Clustered PCDHs (PCDHα, δ and γ family) are encoded as a large cluster in the genome,4,14–16 while non-clustered PCDH genes are scattered in the genome.13 Non-clustered PCDHs which have so far been found are summarized in Table 1. Most nonclustered PCDHs typically have six or seven cadherin repeats, while PCDH15, PCDH16 and MUCDHL has 11, 27 and 4 cadherin repeats, respectively. Human non-clustered PCDH genes are often located at three chromosomal loci: 4q28–31, 5q31–33 and 13q21. A striking difference in the genomic organization of classical cadherin genes and PCDH genes is the presence of unusually large exons in PCDH genes.9 The ectodomain of each member of the PCDH gene is encoded by a single large exon (Fig. 2A and B), while the classical cadherin extracellular domain is encoded by multiple exons (Fig. 2C).17 Typically, this PCDH large exon encodes the entire extracellular portion as well as the transmembrane domain and a short cytoplasmic part, thus giving rise to a complete PCDH molecule. If additional exons for an extension of the cytoplasmic domain are absent, the corresponding PCDH would be a single-exon gene such as the β subfamily of the clustered PCDHs18,19 and PCDH7b of nonclustered PCDHs (Fig. 2A and B). Large exons are also found in Fat and Flamingo cadherins, thus sometimes being classified into PCDH subgroup: however, these exons encode only some parts of the extracellular domains.9 On the other hand, there are a few exceptions in non-clustered PCDH members: The extracellular domains of PCDH11, PCDH15, PCDH16 and µ-PCDH are encoded by multiple exons.5
Table 1.
Features of non-clustered protocadherin family
| Gene symbol | Name | Other designation | # EC | # Known isoform | Locus (human) | Related diseases |
| PCDH1 | Protocadherin 1 | Cadherin-like protein 1, protocadherin 42 (PCDH42, pc42), Axial protocadherin (AXPC) | 7 | 2 | 5q31.3 | Asthma83 |
| PCDH7 | BH-protocadherin | Protocadherin7, BHPCDH, BH-pc, Neural fold protocadherin (NFPC) | 7 | 4 | 4p15 | Non-small-cell lung cancer84 |
| PCDH8 | Protocadherin 8 | Arcadlin, Paraxial protocadherin (PAPC) | 6 | 2 | 13q21.1 | Cocaine abuse85/tumor suppressor (breast cancer70/mantle cell lympoma86) |
| PCDH9 | Protocadherin 9 | Cadherin superfamily protin VR4-11 | 7 | 3 | 13q21.32 | Autism spectrum disorder55/auditory neuropathy87/tumor suppressor (glioblastoma72) |
| PCDH10 | Protocadherin 10 | OL-protocadherin (OL-PCDH, OLpcad) | 6 | 2 | 4q28.3 | Autism54/tumor suppressor (gastric,73,74 cervical,77,78 and other cancers73,75,76,79,80) |
| PCDH11 | Protocadherin 11X-linked | Protocadherin11X (PCDH11X), protocadherinX (PCDHX), protocadherin-S | 6 | 8 | Xq21.3 | Late-onset Alzheimer's disease57,58 |
| Protocadherin 11Y-linked | Protocadherin11Y (PCDH11Y), protocadherinY (PCDHY), protocadherin22 (PCDH22) | 6 | 3 | Yp11.2 | Prostate cancer47,88 | |
| PCDH12 (PCDH14) | Protocadherin 12 | Vascular endothelial cadherin 2 (VE-cadherin-2, VECAD2), vascular cadherin2, protocadherin 14 | 6 | 1 | 5q31 | |
| PCDH15 | Protocadherin-related 15 | Usher syndrome 1F (USH1F), deafness autosomal recessive 23 (DFNB23) | 11 | 12 | 10p21.1 | Usher syndrome63–66,89/hyperlipidemia90 |
| PCDH16 (DCHS1) | Dachsous 1 (Drosophila) | Dachsous-like, fibroblast cadherin 1, fibroblast cadherin FIB1, protocadherin 16 (PCDH16), CDH25, FIB1 | 27 | 1 | 11p15.4 | |
| PCDH17 | Protocadherin 17 | Protocadherin68 (PCDH68, PCH68) | 6 | 1 | 13q21.1 | Schizophrenia56/tumor suppressor (esophageal carcinoma81) |
| PCDH18 | Protocadherin 18 | Protocadheinr 68-like protein (PCDH68L) | 6 | 1 | 4q31 | |
| PCDH19 | Protocadherin 19 | Epilepsy female-restricted with mental retardation (EFMR) | 6 | 2 | Xq13.3 | |
| Female-limited epilepsy and mental retardation59,60/Dravet syndrome61 | PCDH20 (PCDH13) | Protocadherin 20 | Protocadherin 13 (PCDH13) | 6 | 1 | 13q21 |
| Huntington disease91/non-small-cell lung cancer82 | PCDH21 (CDHR1) | Protocadherin 21 | MT-protocadherin, photoreceptor cadherin (PRCAD), cadherin-related family member1 (CDHR1) | 6 | 1 | 10q23.1 Retinal dystrophy67–69 |
| MUCDHL (CDHR5) | Mucin and cadherin-like protein | µ-protocadherin (MU-PCDH), MUCDHL, MUPCD | 4 | 3 | 11p15.5 |
The number of extracelluar cadherin repeats is predicted by SMART program. Non-clustered protocadherins typically have six or seven cadherin repeats, and the ectodomain is encoded by a single large exon. However, the cadherin domains of PCDH11, PCDH15, PCDH16, MUCDHL are encoded by multiple exons. δ1-PCDHs are indicated with red background, ε2-PCDHs are indicated with yellow and ε-PCDHs are indicated with green. The numbers of isoforms and related diseases have been updated on July 25, 2010. The largest numbers of isoforms are present in human, rat and mouse species, based on the information of GeneID at Pubmed. Only those of PCDH7 and PCDH11X are based on the submitted sequences (PCDH7a, AY69613; PCDH7b, AY690614; PCDH7c, AY690615; PCDH7c1, AY690616) and a published paper.92
Figure 2.
Comparison of the genomic organization of α-PCDHs (A), mouse PCDH7b (B) and N-cadherin gene (C).
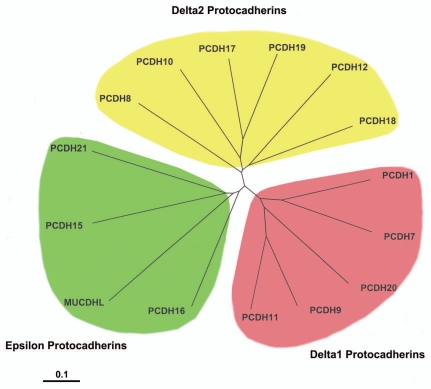
At present, non-clustered PCDHs have been divided into three groups: PCDHδ1 (PCDH1, PCDH7, PCDH9 and PCDH11), PCDHδ2 (PCDH8, PCDH10, PCDH17, PCDH18 and PCDH19) and solitary PCDHs (PCDH12, PCDH15, PCDH20 and PCDH21) in the phylogenic tree.5 All PCDHδ members contain highly conserved motifs [Conserved motif (CM) 1, 27 amino acids; CM2, 17 amino acids] in their cytoplasmic domains.5,20,21 Members of PCDHδ are further divided into PCDHδ1 with protein phosphatase-1α (PP1α) binding domain (RRVTF, CM3) and PCDHδ2 without PP1α binding domain.5,20 We performed phylogenetic analysis again in order to include all known non-clustered PCDHs which are summarized in Table 1, and all analyses, using either the whole protein sequences, extracellular protein sequences or intracellular protein sequences, showed similar results irrespective of the species. In the present study, therefore, we present the analysis based on the sequence of each human nonclustered PCDH to curtail the complexity (Fig. 3 and Sup. Fig. 1A and B). If PCDHs have isoforms, we presented one isoform. In our analyses, non-clustered PCDHs were classified into three groups (Fig. 3 and Sup. Fig. 1A and B): δ1, δ2 and ε subgroups. We obtained similar but a slightly different result from that by Redies et al.5,20 We classified solitary non-clustered PCDHs into one group, which we refer to as ε group. In our analyses, PCDH12 and PCDH20 are classified into δ2 and δ1 subgroup, respectively, whereas PCDH15, PCDH16, PCDH21 and MUCDHL are classified into ε group. One characteristic feature of ε-PCDHs is that the members except PCDH21 have either higher or lower numbers of cadherin repeats compared to other δ-PCDH members (Table 1): 11 repeats in PCDH15, 27 repeats in PCDH16 and four repeats in MUCDHL. Although PCDH12 and PCDH20 are classified into δ-PCDHs, we could not find the CM1, CM2 and CM3 motifs in the cytoplasmic domains of PCDH12 and PCDH20.
Figure 3.
Phylogenetic tree of human non-clustered PCDHs on the basis of their total protein sequences. Multiple alignments and phylogenetic analysis were performed using the ClustalW2 program (www.ebi.ac.uk/Tools/clustalw2/), and the tree was visualized using the Treeview program (http://taxonomy.zoology.gla.ac.uk/rod/). The scale bar represents the substitution rate of amino acid per ten. All sequences used were obtained from the National Center for Biotechnology Information (NCBI). Accession numbers are as follows: PCDH1 (NM_002587), PCDH7 (NM_002589), PCDH8 (NM_002590), PCDH9 (NM_020403), PCDH10 (NM_032961), PCDH11 (NM_014522), PCDH12 (NM_016580), PCDH15 (NM_033056), PCDH16 (NM_003737), PCDH17 (NM_001040429), PCDH18 (NM_019035), PCDH19 (NM_020766), PCDH20 (NM_022843), PCDH21 (NM_033100) and MUCDHL (NM_021924).
Spatial and Temporal Expression of Non-Clustered PCDHs in the CNS
Each classical cadherin tends to be expressed at the highest levels in various types of tissue during development: E-cadherin in epithelia, N-cadherin in neural tissue and muscle, R-cadherin in forebrain and bone, and P-cadherin in the basal layer of epidermis.2 However, PCDHs appear to be expressed mainly in the central nervous system (CNS).10,11,20,22
Expression patterns of non-clustered PCDHs in the CNS system have been studied well at protein and/or mRNA levels, although some non-clustered PCDHs such as PCDH1 and PCDH19 are expressed in non-neuronal tissue.23,24 Expression of PCDH10/OL-PC protein is most extensively studied. PCDH10 protein is expressed in certain local circuits of functional systems such as the olfactory system, nigrostriatal projection, olivocerebellar projection and visual system.25,26 These results are consistent with the finding that PCDH10-deficient mice have defects in axon pathfindings of striatal neurons and thalamocortical projections.27 PCDH19 protein is also expressed in retinofugal projections.28
Studies on mRNA expressions have been carried out more systemically. Some non-clustered PCDHs show the region-specificity in the basal ganglia25,29 with gradients (PCDH8, PCDH9, PCDH10, PCDH17 and PCDH19) and/or the matrix/striosome-based expression patterns (PCDH1, PCDH8, PCDH9, PCDH10, PCDH11 and PCDH17),11,29 suggesting the circuit correlation of their expressions. PCDH7, PCDH9, PCDH11 and PCDH17 also revealed characteristic expression correlated to thalamo-cortical circuits at early postnatal stages,11 and most non-clustered PCDHs showed topographical preferences along the septotemporal axis of adult hippocampus.30 Furthermore, most of non-clustered PCDH is constitutively expressed in the CNS; however, PCDH8/arcadlin is inducible, and PCDH19 and PCDH20 are reducible in the hippocampus and cerebral cortex by elevated activity, such as epileptic seizure.30–32 These diverse and circuit-correlated expression patterns of non-clustered PCDHs in the CNS suggest that non-clustered PCDHs play roles in the wiring of neural circuit formation and maintenance through their adhesive and regulatory mechanism.
Molecular Function of Non-Clustered PCDHs
Adhesive properties play important roles in morphogenesis during the developmental to adult stage. The formation of germ layers and tissues, cell rearrangement and migration, cell sorting, neurite outgrowth, axon pathfinding and synaptic formation in neurons depend on the cell adhesion ability. The function of classical cadherin is mediated by strong cell-cell adhesion through homophilic interactions, whereas the PCDHs appear to have more varied physiological functions as a mediator of cell-cell adhesion or a regulator of other molecules. Recently, the molecular functions of non-clustered PCDHs have been clarified. We next discuss the role of non-clustered PCDHs as a mediator of cell-cell adhesion and/or a regulator of other molecules.
Mediator of cell-cell adhesion.
Adhesion properties and cytoplasmic partners of non-clustered PCDHs are still poorly understood. Most of the cadherin superfamily proteins show calcium-dependent homophilic adhesion activities.33,34 Although several non-clustered PCDHs (PCDH1, PCDH7, PCDH8, PCDH10, PCDH18 and PCDH19) exhibit homophilic binding ability, some of these (PCDH8, PCDH10 and PCDH19) show only weak binding ability.26,32,35–39 Nevertheless, the cell-cell adhesion is strengthened when the cytoplasmic tail of PCDH1/axial protocadherin (AXPC) or PCDH8/paraxial protocadherin (PAPC) is removed36,37,39 or the cytoplasmic tail of PCDH19 is replaced with that of E-cadherin,39 suggesting that the extracellular domain of non-clustered PCDHs is able to form cell-cell adhesive interactions, and that the cytoplasmic domain may not efficiently stabilize those interactions to facilitate adhesion or may regulate negatively their extracellular adhesions.
PCDH1/AXPC, PCDH7/neural fold protocadherin (NFPC) and PCDH8/PAPC exhibited substantial adhesive activity in vivo. A Xenopus PCDH1-homolog AXPC and a PCDH8/arcadlin ortholog PAPC are complementally expressed in paraxial mesoderm, and mediate cell sorting and cell movements during embryonic gastrulation.36,37 In addition, PCDH7/NFPC has been shown to regulate differentiation of the embryonic ectoderm, 40 neural tube formation,41 cell morphology,38 and axonal elongation in retinal ganglion cells.42 As for the mechanism for strong adhesive activity of PCDH7, its interacting protein may be involved. Template-activating factor1 (TAF1) interacts with the cytoplasmic region of PCDH7, and may regulate the adhesive activity of PCDH7 (Fig. 5A).40 Thus, the homophilic interaction of some PCDHs may mediate cell-cell adhesion as classical cadherins.
Figure 5.
Schematic overview of intracellular signaling proteins bound to the cytoplasmic domain of each PCDH. (A) PCDH7c (PCDH7c and 7c1, but not PCDH7a or 7b) has CM1, CM2 and CM3 motifs, and CM3 could be bound by PP1α. This interaction inactivates PP1α.29,93 In addition, all PCDH7 isoforms interact with histone-regulating protein TAF1. The PCDH7-TAF1 interaction is involved in retinal axon initiation and elongation in developing retinal ganglion cells.42 (B) All PCDH11Y isoforms (11Ya, Yb and Yc) have the binding sequences to δ-catenins, however, only PCDH11Yc (but not 11Ya or 11Yb) has CM1, CM2 and CM3 motifs. PCDH11Y is biochemically associated with δ-catenin, and this interaction might affect wnt signaling and tumorgenesis.47 (C) PCDH8/arcadlin binds to a serine-threonine kinase TAO2δ. This interaction causes N-cadherin endocytosis at synaptic membrane in a p38 MAPK-dependent manner.31 (D) PCDH10/OL-PC interacts with Nap1, and this interaction recruits WAVE1, a Nap1 binding protein, to cell-cell contact sites. The formation of PCDH10/Nap1/WAVE complex regulates actin assembly, and subsequently promotes the migration of cells.50 (E) PCDH18 is associated with mDab1, and this interaction might be involved in the correct formation of cortical nerve cell layers.52
On the other hand, heterophilic cell adhesion activity has been reported between PCDHα4 (one of clustered PCDHs) and β1 integrin in an in vitro cell aggregation assay with HEK293T cells.43 Integrins recognize the RGD motif that is essential for integrin-dependent cell adhesion. This RGD motif is found in fibronectin, vitronectin, fibrinogen, von Willebrand factor and many other large glycoproteins.44 Interestingly, this RGD motif has also been seen in the extracellular domain (EC1 or EC2) of certain non-clustered PCDHs (PCDH17, PCDH19 and MUCDHL) (Fig. 4). This suggests a possibility that non-clustered PCDHs may also have heterophilic adhesion activity, acting as membrane-associated ligands or receptors for integrins. In addition, PAPC, a putative mammalian PCDH8/arcadlin homolog, participates in early cell sorting by regulating the adhesive activity of a classical C-cadherin.45 This suggests that PCDH8/PAPC may have heterophilic interaction with classical cadherins. PCDH8/arcadlin shows also a lateral (cis) interaction with N-cadherin in the same plane of plasma membrane, and regulates the endocytosis of N-cadherin.31 Recently, the heterophilic intereaction between PCDH15 and classical cadherin (cadherin 23) has been reported.46 Thus, PCDHs may mediate homophilic, heterophilic or both cell adhesions in vivo.
Figure 4.
The extracellular domain sequences of PCDH17, PCDH19 and MUCDHL. The RGD motif is found in the extracellular domains of PCDH17, PCDH19 and MUCDHL. The RGD motif is an essential residue for integrin-dependent cell adhesion activity. Other species are: H, human; M, mouse; R, rat; C, chick; DR, Danio rerio. PCDH17 (H, NM_014459; M, XM_127786; R, XM_224389; C, XM_417021; DR, XM_684743), PCDH19 (H, NM_001105243; M, NM_001105245; C, NM_001098607; DR, NP_001120991), MUCDHL (H, NM_021924; M, NM_028069; R, NM_138525).
Regulator of various “effector” molecules.
Recently, nonclustered PCDHs have been clarified as a regulator of other molecules. PCDHs lack a β-catenin binding cytoplasmic site present in classical cadherins. The cytoplasmic domains of non-clustered PCDHs are different from each other, and their homology ranges from low to moderate.2,3 Therefore, non-clustered PCDHs could act as a regulator via interaction with a variety of intracellular binding partners.
δ-PCDHs have conserved cytoplasmic motifs (CM1, CM2, CM3 and CM4), whose binding molecules remain largely elusive; Only CM3 region is known to interact with PP1α.20 PCDH7 (NFPC) has four isoforms (7a, 7b, 7c and 7c1), and PCDH7c and 7c1 have CM1, CM2 and CM3 motifs (Fig. 5A). PCDH7c1 is an 8 amino acid-deleted 7c from the region between CM2 and CM3. All PCDH7 isoforms interact with template-activating factor1 (TAF1).40 PCDH11Y has three isoforms (11Ya, Yb and Yc), and only PCDH11Yc has CM1, CM2 and CM3 motifs (Fig. 5B). All isoforms of PCDH11Y bind to β-catenin, and this interaction may regulate wnt signaling and tumorgenesis.47
The intracellular domain of PCDH8/arcadlin interacts with thousand and one amino acid protein kinase 2β (TAO2β) which activates p38 MAPK pathway, and subsequently promotes endocytosis of N-cadherin (Fig. 5C).31 Because N-cadherin regulates spine dynamics and maintains the shape and density of spines,48,49 this PCDH8-TAO2-p38 MAPK pathway may transfer epileptic activity into dendritic spine morphology via N-cadherin endocytosis.
PCDH10/OL-PC interacts with Nck-associated protein 1 (Nap1)/WAVE1 (Fig. 5D),50 and PCDH10/Nap1/WAVE1 complex affects actin assembly and subsequently regulates cell migration. 51 However, it is not known how PCDH10/Nap1/WAVE1 complex controls actin assembly.
PCDH18 interacts with mouse Disabled homolog 1 (mDab1) (Fig. 5E),52 which functions downstream of Reelin and mediates neural circuit formation.53
Finally, each PCDH has several isoforms that are differentiated from their cytoplasmic domains. This suggests that PCDH isoforms could play diverse roles as intracellular signaling regulators.
In summary, weak homophilic or heterophilic interaction and diverse intracellular sequences of non-clustered PCDHs suggest that they may function as a regulator of cell-cell adhesive, and/or intracellular effect molecules rather than only physical glues between cells.
Non-Clustered PCDHs and Disease
Abnormalities in non-clustered PCDHs may be responsible for the pathogenesis of several neurological disorders and carcinogenesis. Especially, the relationship between δ-PCDHs and cognitive dysfunction has been well investigated, and as described below, some epsilon PCDHs are related to sensory impairment. Also, the emergence or silencing of non-clustered PCDHs on chromosome 13q21 influences oncogenesis.
Delta PCDH and cognitive dysfunction.
Several lines of evidence indicate that the dysfunction of non-clustered PCDHs is associated with some cognitive dysfunction. For instance, the homozygous deletion within a protocadherin cluster (between PCDH10 and PCDH18 loci on 4q28.3) proximal to PCDH10 has been shown to be associated significantly with the pathophysiology of cognitive impairment such as autism,54 and recurrent and overlapping copy number variations, including PCDH9 loci, have been identified in autism patients.55 Another delta protocadherin PCDH17 is involved in the pathogenesis of schizophrenia.56
On the other hand, a genome-wide association study showed that SNP on Xq21.3 in PCDH11X is associated strongly with late-onset Alzheimer's disease susceptibility,57 although recent studies show non-statistical association between PCDH11X polymorphisms and late-onset Alzheimer's disease susceptibility.58 Nonsense mutation of PCDH19 has been found in seven families of mental retardation limited to females, characterized by seizure onset in infancy or early childhood and cognitive dysfunction.59,60 Furthermore, the dysfunction of PCDH19 causes Dravet syndrome-like epileptic encephalopathy, which is marked by seizures, developmental and language delays, behavioral disturbances and cognitive regression.61 The fact that some PCDHs regulate synaptic function and morphology30,31,54 leads us to speculate that delta PCDHs are important for normal function of neural circuitry as well as wiring development of neural circuitry, and the disruption of delta PCDH may cause abnormal neural circuitry and subsequent cognitive impairment.
Epsilon PCDH and retinal pigmentosa.
Usher syndrome type 1F (USH1F) is characterized by a loss of vision due to retinitis pigmentosa (RP), a genetic disease with progressive dysfunction and degeneration of the rod and cone photoreceptors, and bilateral sensorineural deafness.62 PCDH15 is expressed in inner ear hair cell stereocilia and retinal photoreceptors,63,64 and may play a pivotal role in the morphogenesis and cohesion of stereocilia bundles and retinal photoreceptor cell maintenance or function. The mutation, splicing abnormality, frameshift, nonsense or large deletions of PCDH15 gene have been shown to cause USH1F,63,65,66 indicating that the dysfunction of PCDH15 plays a pathogenetic role in the RP and hearing loss associated with USH1F. Moreover, null mutations in PCDH21, which is known as a photoreceptor-specific gene,67,68 cause the RP.69 These results suggest that the abnormality of epsilon PCDHs might disrupt photoreceptors and induce visual dysfunction.
Non-clustered PCDHs on chromosome 13q21 as tumor suppressors.
Recently, some delta PCDHs (PCDH8, PCDH9, PCDH10, PCDH17 and PCDH20) have been reported as candidate tumor suppressor genes. The expressions of PCDH8 in breast70 and hematologic cancers,71 PCDH9 in glioblastoma,72 PCDH10 in gastric,73,74 colorectal,73 nasopharyngeal, esophageal,75 breast,76 cervical,77,78 lung, hepatocellular,75 tesicular79 and hematologic cancers,80 PCDH17 in esophageal squamous cell carcinoma81 and PCDH20 in non-small-cell lung cancers82 are reduced or silenced through gene inactivation such as promoter hypermethylation and/or somatic mutation, and re-expression of PCDH8,70 or PCDH10,74 suppresses tumor cell proliferation and inhibits cell migration. Notably, PCDH8, PCDH9, PCDH17 and PCDH20 genes are located around 13q21.1 and closely positioned within 16 megabases. These results suggest that PCDHs on chromosome 13q21 (Table 1) might be broadly involved in tumor suppression in a variety of tumors. Also, PCDHs on chromosome 13q21 might be regulated by common genetic or epigenetic factors and further involved in a variety of cellular and brain function together.
Conclusions
At present, non-clustered PCDHs are considered to play critical roles in brain development, including normal brain function and oncogenesis. Although the involvements of non-clustered PCDHs in the pathogenesis of some neural diseases and tumor are relatively well established, the endeavors to understand the molecular functions of non-clustered PCDH are still in its infancy and more detailed functional analyses are required at cellular and molecular levels in the future studies.
Acknowledgements
This work was supported by the Korea Science and Engineering Foundation grant (2009K001284 to H.K.) funded by the Korean government, KAKENHI (20591426 to S.Y.; 19590247 to H.T.; 20650052 to K.Y.) and a grant from the Naito Foundation (to K.Y.). A part of this work was technically supported by the core facility service of 21C Frontier Brain Research Center.
Abbreviations
- AXPC
axial protocadherin
- CM
conserved motif
- CNS
central nervous system
- ECS
electrical convulsive shock
- Nap1
Nck-associated protein 1
- NFPC
neural fold protocadherin
- PCDH
protocadherin
- PAPC
paraxial protocadherin
- RP
retinitis pigmentosa
- USH1F
Usher syndrome type 1F
Supplementary Material
References
- 1.Nollet F, Kools P, van Roy F. Phylogenetic analysis of the cadherin superfamily allows identification of six major subfamilies besides several solitary members. J Mol Biol. 2000;299:551–572. doi: 10.1006/jmbi.2000.3777. [DOI] [PubMed] [Google Scholar]
- 2.Halbleib JM, Nelson WJ. Cadherins in development: cell adhesion, sorting and tissue morphogenesis. Genes Dev. 2006;20:3199–3214. doi: 10.1101/gad.1486806. [DOI] [PubMed] [Google Scholar]
- 3.Morishita H, Yagi T. Protocadherin family: diversity, structure and function. Curr Opin Cell Biol. 2007;19:584–592. doi: 10.1016/j.ceb.2007.09.006. [DOI] [PubMed] [Google Scholar]
- 4.Wu Q, Maniatis T. A striking organization of a large family of human neural cadherin-like cell adhesion genes. Cell. 1999;97:779–790. doi: 10.1016/s0092-8674(00)80789-8. [DOI] [PubMed] [Google Scholar]
- 5.Redies C, Vanhalst K, Roy F. delta-Protocadherins: unique structures and functions. Cell Mol Life Sci. 2005;62:2840–2852. doi: 10.1007/s00018-005-5320-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tissir F, Bar I, Jossin Y, De Backer O, Goffinet AM. Protocadherin Celsr3 is crucial in axonal tract development. Nat Neurosci. 2005;8:451–457. doi: 10.1038/nn1428. [DOI] [PubMed] [Google Scholar]
- 7.Ciani L, Patel A, Allen ND, ffrench-Constant C. Mice lacking the giant protocadherin mFAT1 exhibit renal slit junction abnormalities and a partially penetrant cyclopia and anophthalmia phenotype. Mol Cell Biol. 2003;23:3575–3582. doi: 10.1128/MCB.23.10.3575-3582.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chae J, Kim MJ, Goo JH, Collier S, Gubb D, Charlton J, et al. The Drosophila tissue polarity gene starry night encodes a member of the protocadherin family. Development. 1999;126:5421–5429. doi: 10.1242/dev.126.23.5421. [DOI] [PubMed] [Google Scholar]
- 9.Wu Q, Maniatis T. Large exons encoding multiple ectodomains are a characteristic feature of protocadherin genes. Proc Natl Acad Sci USA. 2000;97:3124–3129. doi: 10.1073/pnas.060027397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sano K, Tanihara H, Heimark RL, Obata S, Davidson M, St. John T, et al. Protocadherins: a large family of cadherin-related molecules in central nervous system. EMBO J. 1993;12:2249–2256. doi: 10.1002/j.1460-2075.1993.tb05878.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kim SY, Chung HS, Sun W, Kim H. Spatiotemporal expression pattern of non-clustered protocadherin family members in the developing rat brain. Neuroscience. 2007;147:996–1021. doi: 10.1016/j.neuroscience.2007.03.052. [DOI] [PubMed] [Google Scholar]
- 12.Tepass U, Truong K, Godt D, Ikura M, Peifer M. Cadherins in embryonic and neural morphogenesis. Nat Rev Mol Cell Biol. 2000;1:91–100. doi: 10.1038/35040042. [DOI] [PubMed] [Google Scholar]
- 13.Frank M, Kemler R. Protocadherins. Curr Opin Cell Biol. 2002;14:557–562. doi: 10.1016/s0955-0674(02)00365-4. [DOI] [PubMed] [Google Scholar]
- 14.Suzuki ST. Recent progress in protocadherin research. Exp Cell Res. 2000;261:13–18. doi: 10.1006/excr.2000.5039. [DOI] [PubMed] [Google Scholar]
- 15.Junghans D, Haas IG, Kemler R. Mammalian cadherins and protocadherins: about cell death, synapses and processing. Curr Opin Cell Biol. 2005;17:446–452. doi: 10.1016/j.ceb.2005.08.008. [DOI] [PubMed] [Google Scholar]
- 16.Hirayama T, Yagi T. The role and expression of the protocadherin-alpha clusters in the CNS. Curr Opin Neurobiol. 2006;16:336–342. doi: 10.1016/j.conb.2006.05.003. [DOI] [PubMed] [Google Scholar]
- 17.Wallis J, Fox MF, Walsh FS. Structure of the human N-cadherin gene: YAC analysis and fine chromosomal mapping to 18q11.2. Genomics. 1994;22:172–179. doi: 10.1006/geno.1994.1358. [DOI] [PubMed] [Google Scholar]
- 18.Vanhalst K, Kools P, Vanden Eynde E, van Roy F. The human and murine protocadherin-beta one-exon gene families show high evolutionary conservation, despite the difference in gene number. FEBS Lett. 2001;495:120–125. doi: 10.1016/s0014-5793(01)02372-9. [DOI] [PubMed] [Google Scholar]
- 19.Wu Q, Zhang T, Cheng JF, Kim Y, Grimwood J, Schmutz J, et al. Comparative DNA sequence analysis of mouse and human protocadherin gene clusters. Genome Res. 2001;11:389–404. doi: 10.1101/gr.167301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Vanhalst K, Kools P, Staes K, van Roy F, Redies C. delta-Protocadherins: a gene family expressed differentially in the mouse brain. Cell Mol Life Sci. 2005;62:1247–1259. doi: 10.1007/s00018-005-5021-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wolverton T, Lalande M. Identification and characterization of three members of a novel subclass of protocadherins. Genomics. 2001;76:66–72. doi: 10.1006/geno.2001.6592. [DOI] [PubMed] [Google Scholar]
- 22.Zou C, Huang W, Ying G, Wu Q. Sequence analysis and expression mapping of the rat clustered protocadherin gene repertoires. Neuroscience. 2007;144:579–603. doi: 10.1016/j.neuroscience.2006.10.011. [DOI] [PubMed] [Google Scholar]
- 23.Gaitan Y, Bouchard M. Expression of the delta-protocadherin gene Pcdh19 in the developing mouse embryo. Gene Expr Patterns. 2006;6:893–899. doi: 10.1016/j.modgep.2006.03.001. [DOI] [PubMed] [Google Scholar]
- 24.Redies C, Heyder J, Kohoutek T, Staes K, Van Roy F. Expression of protocadherin-1 (Pcdh1) during mouse development. Dev Dyn. 2008;237:2496–2505. doi: 10.1002/dvdy.21650. [DOI] [PubMed] [Google Scholar]
- 25.Aoki E, Kimura R, Suzuki ST, Hirano S. Distribution of OL-protocadherin protein in correlation with specific neural compartments and local circuits in the postnatal mouse brain. Neuroscience. 2003;117:593–614. doi: 10.1016/s0306-4522(02)00944-2. [DOI] [PubMed] [Google Scholar]
- 26.Hirano S, Yan Q, Suzuki ST. Expression of a novel protocadherin, OL-protocadherin, in a subset of functional systems of the developing mouse brain. J Neurosci. 1999;19:995–1005. doi: 10.1523/JNEUROSCI.19-03-00995.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Uemura M, Nakao S, Suzuki ST, Takeichi M, Hirano S. OL-protocadherin is essential for growth of striatal axons and thalamocortical projections. Nat Neurosci. 2007;10:1151–1159. doi: 10.1038/nn1960. [DOI] [PubMed] [Google Scholar]
- 28.Tai K, Kubota M, Shiono K, Tokutsu H, Suzuki ST. Adhesion properties and retinofugal expression of chicken protocadherin-19. Brain Res. 2010;1344:13–24. doi: 10.1016/j.brainres.2010.04.065. [DOI] [PubMed] [Google Scholar]
- 29.Hertel N, Krishna K, Nuernberger M, Redies C. A cadherin-based code for the divisions of the mouse basal ganglia. J Comp Neurol. 2008;508:511–528. doi: 10.1002/cne.21696. [DOI] [PubMed] [Google Scholar]
- 30.Kim SY, Mo JW, Han S, Choi SY, Han SB, Moon BH, et al. The expression of non-clustered protocadherins in adult rat hippocampal formation and the connecting brain regions. Neuroscience. 2010;170:189–199. doi: 10.1016/j.neuroscience.2010.05.027. [DOI] [PubMed] [Google Scholar]
- 31.Yasuda S, Tanaka H, Sugiura H, Okamura K, Sakaguchi T, Tran U, et al. Activity-induced protocadherin arcadlin regulates dendritic spine number by triggering N-cadherin endocytosis via TAO2beta and p38 MAP kinases. Neuron. 2007;56:456–471. doi: 10.1016/j.neuron.2007.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yamagata K, Andreasson KI, Sugiura H, Maru E, Dominique M, Irie Y, et al. Arcadlin is a neural activity-regulated cadherin involved in long term potentiation. J Biol Chem. 1999;274:19473–19479. doi: 10.1074/jbc.274.27.19473. [DOI] [PubMed] [Google Scholar]
- 33.Takeichi M. Morphogenetic roles of classic cadherins. Curr Opin Cell Biol. 1995;7:619–627. doi: 10.1016/0955-0674(95)80102-2. [DOI] [PubMed] [Google Scholar]
- 34.Shapiro L, Colman DR. The diversity of cadherins and implications for a synaptic adhesive code in the CNS. Neuron. 1999;23:427–430. doi: 10.1016/s0896-6273(00)80796-5. [DOI] [PubMed] [Google Scholar]
- 35.Aamar E, Dawid IB. Protocadherin-18a has a role in cell adhesion, behavior and migration in zebrafish development. Dev Biol. 2008;318:335–346. doi: 10.1016/j.ydbio.2008.03.040. [DOI] [PubMed] [Google Scholar]
- 36.Kim SH, Yamamoto A, Bouwmeester T, Agius E, Robertis EM. The role of paraxial protocadherin in selective adhesion and cell movements of the mesoderm during Xenopus gastrulation. Development. 1998;125:4681–4690. doi: 10.1242/dev.125.23.4681. [DOI] [PubMed] [Google Scholar]
- 37.Kuroda H, Inui M, Sugimoto K, Hayata T, Asashima M. Axial protocadherin is a mediator of prenotochord cell sorting in Xenopus. Dev Biol. 2002;244:267–277. doi: 10.1006/dbio.2002.0589. [DOI] [PubMed] [Google Scholar]
- 38.Yoshida K. Fibroblast cell shape and adhesion in vitro is altered by overexpression of the 7a and 7b isoforms of protocadherin 7, but not the 7c isoform. Cell Mol Biol Lett. 2003;8:735–741. [PubMed] [Google Scholar]
- 39.Tai K, Kubota M, Shiono K, Tokutsu H, Suzuki ST. Adhesion properties and retinofugal expression of chicken protocadherin-19. Brain Res. 2010;1344:13–24. doi: 10.1016/j.brainres.2010.04.065. [DOI] [PubMed] [Google Scholar]
- 40.Heggem MA, Bradley RS. The cytoplasmic domain of Xenopus NF-protocadherin interacts with TAF1/set. Dev Cell. 2003;4:419–429. doi: 10.1016/s1534-5807(03)00036-4. [DOI] [PubMed] [Google Scholar]
- 41.Rashid D, Newell K, Shama L, Bradley R. A requirement for NF-protocadherin and TAF1/Set in cell adhesion and neural tube formation. Dev Biol. 2006;291:170–181. doi: 10.1016/j.ydbio.2005.12.027. [DOI] [PubMed] [Google Scholar]
- 42.Piper M, Dwivedy A, Leung L, Bradley RS, Holt CE. NF-protocadherin and TAF1 regulate retinal axon initiation and elongation in vivo. J Neurosci. 2008;28:100–105. doi: 10.1523/JNEUROSCI.4490-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mutoh T, Hamada S, Senzaki K, Murata Y, Yagi T. Cadherin-related neuronal receptor 1 (CNR1) has cell adhesion activity with beta1 integrin mediated through the RGD site of CNR1. Exp Cell Res. 2004;294:494–508. doi: 10.1016/j.yexcr.2003.11.019. [DOI] [PubMed] [Google Scholar]
- 44.Takagi J. Structural basis for ligand recognition by RGD (Arg-Gly-Asp)-dependent integrins. Biochem Soc Trans. 2004;32:403–406. doi: 10.1042/BST0320403. [DOI] [PubMed] [Google Scholar]
- 45.Chen X, Gumbiner BM. Paraxial protocadherin mediates cell sorting and tissue morphogenesis by regulating C-cadherin adhesion activity. J Cell Biol. 2006;174:301–313. doi: 10.1083/jcb.200602062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kazmierczak P, Sakaguchi H, Tokita J, Wilson-Kubalek EM, Milligan RA, Muller U, et al. Cadherin 23 and protocadherin 15 interact to form tip-link filaments in sensory hair cells. Nature. 2007;449:87–91. doi: 10.1038/nature06091. [DOI] [PubMed] [Google Scholar]
- 47.Chen MW, Vacherot F, De La Taille A, Gil-Diez-De-Medina S, Shen R, Friedman RA, et al. The emergence of protocadherin-PC expression during the acquisition of apoptosis-resistance by prostate cancer cells. Oncogene. 2002;21:7861–17871. doi: 10.1038/sj.onc.1205991. [DOI] [PubMed] [Google Scholar]
- 48.Togashi H, Abe K, Mizoguchi A, Takaoka K, Chisaka O, Takeichi M. Cadherin regulates dendritic spine morphogenesis. Neuron. 2002;35:77–89. doi: 10.1016/s0896-6273(02)00748-1. [DOI] [PubMed] [Google Scholar]
- 49.Okamura K, Tanaka H, Yagita Y, Saeki Y, Taguchi A, Hiraoka Y, et al. Cadherin activity is required for activity-induced spine remodeling. J Cell Biol. 2004;167:961–972. doi: 10.1083/jcb.200406030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Nakao S, Platek A, Hirano S, Takeichi M. Contact-dependent promotion of cell migration by the OL-protocadherin-Nap1 interaction. J Cell Biol. 2008;182:395–410. doi: 10.1083/jcb.200802069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Grove EA. Turning neurons into a nervous system. Development. 2008;135:2203–2206. doi: 10.1242/dev.020511. [DOI] [PubMed] [Google Scholar]
- 52.Homayouni R, Rice DS, Curran T. Disabled-1 interacts with a novel developmentally regulated protocadherin. Biochem Biophys Res Commun. 2001;289:539–547. doi: 10.1006/bbrc.2001.5998. [DOI] [PubMed] [Google Scholar]
- 53.Borrell V, Pujadas L, Simo S, Dura D, Sole M, Cooper JA, et al. Reelin and mDab1 regulate the development of hippocampal connections. Mol Cell Neurosci. 2007;36:158–173. doi: 10.1016/j.mcn.2007.06.006. [DOI] [PubMed] [Google Scholar]
- 54.Morrow EM, Yoo SY, Flavell SW, Kim TK, Lin Y, Hill RS, et al. Identifying autism loci and genes by tracing recent shared ancestry. Science. 2008;321:218–223. doi: 10.1126/science.1157657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Marshall CR, Noor A, Vincent JB, Lionel AC, Feuk L, Skaug J, et al. Structural variation of chromosomes in autism spectrum disorder. Am J Hum Genet. 2008;82:477–488. doi: 10.1016/j.ajhg.2007.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Dean B, Keriakous D, Scarr E, Thomas EA. Gene expression profiling in Brodmann's area 46 from subjects with schizophrenia. Aust N Z J Psychiatry. 2007;41:308–320. doi: 10.1080/00048670701213245. [DOI] [PubMed] [Google Scholar]
- 57.Carrasquillo MM, Zou F, Pankratz VS, Wilcox SL, Ma L, Walker LP, et al. Genetic variation in PCDH11X is associated with susceptibility to late-onset Alzheimer's disease. Nat Genet. 2009;41:192–198. doi: 10.1038/ng.305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Beecham GW, Naj AC, Gilbert JR, Haines JL, Buxbaum JD, Pericak-Vance MA. PCDH11X variation is not associated with late-onset Alzheimer disease susceptibility. Psychiatr Genet. 2010;20(6):321–324. doi: 10.1097/YPG.0b013e32833b635d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Dibbens LM, Tarpey PS, Hynes K, Bayly MA, Scheffer IE, Smith R, et al. X-linked protocadherin 19 mutations cause female-limited epilepsy and cognitive impairment. Nat Genet. 2008;40:776–781. doi: 10.1038/ng.149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hynes K, Tarpey P, Dibbens LM, Bayly MA, Berkovic SF, Smith R, et al. Epilepsy and mental retardation limited to females with PCDH19 mutations can present de novo or in single generation families. J Med Genet. 47:211–216. doi: 10.1136/jmg.2009.068817. [DOI] [PubMed] [Google Scholar]
- 61.Depienne C, Bouteiller D, Keren B, Cheuret E, Poirier K, Trouillard O, et al. Sporadic infantile epileptic encephalopathy caused by mutations in PCDH19 resembles Dravet syndrome but mainly affects females. PLoS Genet. 2009;5:1000381. doi: 10.1371/journal.pgen.1000381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Smith RJ, Berlin CI, Hejtmancik JF, Keats BJ, Kimberling WJ, Lewis RA, et al. Clinical diagnosis of Syndrome Consortium. Am J Med Genet. 1994;50:32–38. doi: 10.1002/ajmg.1320500107. [DOI] [PubMed] [Google Scholar]
- 63.Ahmed ZM, Riazuddin S, Aye S, Ali RA, Venselaar H, Anwar S, et al. Gene structure and mutant alleles of PCDH15: nonsyndromic deafness DFNB23 and type 1 Usher syndrome. Hum Genet. 2008;124:215–223. doi: 10.1007/s00439-008-0543-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Alagramam KN, Yuan H, Kuehn MH, Murcia CL, Wayne S, Srisailpathy CR, et al. Mutations in the novel protocadherin PCDH15 cause Usher syndrome type 1F. Hum Mol Genet. 2001;10:1709–1718. doi: 10.1093/hmg/10.16.1709. [DOI] [PubMed] [Google Scholar]
- 65.Ben-Yosef T, Ness SL, Madeo AC, Bar-Lev A, Wolfman JH, Ahmed ZM, et al. A mutation of PCDH15 among Ashkenazi Jews with the type 1 Usher syndrome. N Engl J Med. 2003;348:1664–1670. doi: 10.1056/NEJMoa021502. [DOI] [PubMed] [Google Scholar]
- 66.Doucette L, Merner ND, Cooke S, Ives E, Galutira D, Walsh V, et al. Profound, prelingual nonsyndromic deafness maps to chromosome 10q21 and is caused by a novel missense mutation in the Usher syndrome type IF gene PCDH15. Eur J Hum Genet. 2009;17:554–564. doi: 10.1038/ejhg.2008.231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Rattner A, Smallwood PM, Williams J, Cooke C, Savchenko A, Lyubarsky A, et al. A photoreceptor-specific cadherin is essential for the structural integrity of the outer segment and for photoreceptor survival. Neuron. 2001;32:775–786. doi: 10.1016/s0896-6273(01)00531-1. [DOI] [PubMed] [Google Scholar]
- 68.Rattner A, Chen J, Nathans J. Proteolytic shedding of the extracellular domain of photoreceptor cadherin. Implications for outer segment assembly. J Biol Chem. 2004;279:42202–42210. doi: 10.1074/jbc.M407928200. [DOI] [PubMed] [Google Scholar]
- 69.Henderson RH, Li Z, Abd El Aziz MM, Mackay DS, Eljinini MA, Zeidan M, et al. Biallelic mutation of protocadherin-21 (PCDH21) causes retinal degeneration in humans. Mol Vis. 2010;16:46–52. [PMC free article] [PubMed] [Google Scholar]
- 70.Yu JS, Koujak S, Nagase S, Li CM, Su T, Wang X, et al. PCDH8, the human homolog of PAPC, is a candidate tumor suppressor of breast cancer. Oncogene. 2008;27:4657–4665. doi: 10.1038/onc.2008.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Leshchenko VV, Kuo PY, Shaknovich R, Yang DT, Gellen T, Petrich A, et al. Genomewide DNA methylation analysis reveals novel targets for drug development in mantle cell lymphoma. Blood. 2009;116:1025–1034. doi: 10.1182/blood-2009-12-257485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.de Tayrac M, Etcheverry A, Aubry M, Saikali S, Hamlat A, Quillien V, et al. Integrative genome-wide analysis reveals a robust genomic glioblastoma signature associated with copy number driving changes in gene expression. Genes Chromosomes Cancer. 2009;48:55–68. doi: 10.1002/gcc.20618. [DOI] [PubMed] [Google Scholar]
- 73.Yu B, Yang H, Zhang C, Wu Q, Shao Y, Zhang J, et al. High-resolution melting analysis of PCDH10 methylation levels in gastric, colorectal and pancreatic cancers. Neoplasma. 2010;57:247–252. doi: 10.4149/neo_2010_03_247. [DOI] [PubMed] [Google Scholar]
- 74.Yu J, Cheng YY, Tao Q, Cheung KF, Lam CN, Geng H, et al. Methylation of protocadherin 10, a novel tumor suppressor, is associated with poor prognosis in patients with gastric cancer. Gastroenterology. 2009;136:640–651. doi: 10.1053/j.gastro.2008.10.050. [DOI] [PubMed] [Google Scholar]
- 75.Ying J, Li H, Seng TJ, Langford C, Srivastava G, Tsao SW, et al. Functional epigenetics identifies a protocadherin PCDH10 as a candidate tumor suppressor for nasopharyngeal, esophageal and multiple other carcinomas with frequent methylation. Oncogene. 2006;25:1070–1080. doi: 10.1038/sj.onc.1209154. [DOI] [PubMed] [Google Scholar]
- 76.Miyamoto K, Fukutomi T, Akashi-Tanaka S, Hasegawa T, Asahara T, Sugimura T, et al. Identification of 20 genes aberrantly methylated in human breast cancers. Int J Cancer. 2005;116:407–414. doi: 10.1002/ijc.21054. [DOI] [PubMed] [Google Scholar]
- 77.Wang KH, Liu HW, Lin SR, Ding DC, Chu TY. Field methylation silencing of the protocadherin 10 gene in cervical carcinogenesis as a potential specific diagnostic test from cervical scrapings. Cancer Sci. 2009;100:2175–2180. doi: 10.1111/j.1349-7006.2009.01285.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Narayan G, Scotto L, Neelakantan V, Kottoor SH, Wong AH, Loke SL, et al. Protocadherin PCDH10, involved in tumor progression, is a frequent and early target of promoter hypermethylation in cervical cancer. Genes Chromosomes Cancer. 2009;48:983–992. doi: 10.1002/gcc.20703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Cheung HH, Lee TL, Davis AJ, Taft DH, Rennert OM, Chan WY. Genome-wide DNA methylation profiling reveals novel epigenetically regulated genes and non-coding RNAs in human testicular cancer. Br J Cancer. 2010;102:419–427. doi: 10.1038/sj.bjc.6605505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Ying J, Gao Z, Li H, Srivastava G, Murray PG, Goh HK, et al. Frequent epigenetic silencing of protocadherin 10 by methylation in multiple haematologic malignancies. Br J Haematol. 2007;136:829–832. doi: 10.1111/j.1365-2141.2007.06512.x. [DOI] [PubMed] [Google Scholar]
- 81.Haruki S, Imoto I, Kozaki K, Matsui T, Kawachi H, Komatsu S, et al. Frequent silencing of protocadherin 17, a candidate tumour suppressor for esophageal squamous cell carcinoma. Carcinogenesis. 2010;31:1027–1036. doi: 10.1093/carcin/bgq053. [DOI] [PubMed] [Google Scholar]
- 82.Imoto I, Izumi H, Yokoi S, Hosoda H, Shibata T, Hosoda F, et al. Frequent silencing of the candidate tumor suppressor PCDH20 by epigenetic mechanism in non-small-cell lung cancers. Cancer Res. 2006;66:4617–4626. doi: 10.1158/0008-5472.CAN-05-4437. [DOI] [PubMed] [Google Scholar]
- 83.Koppelman GH, Meyers DA, Howard TD, Zheng SL, Hawkins GA, Ampleford EJ, et al. Identification of PCDH1 as a novel susceptibility gene for bronchial hyperresponsiveness. Am J Respir Crit Care Med. 2009;180:929–935. doi: 10.1164/rccm.200810-1621OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Huang YT, Heist RS, Chirieac LR, Lin X, Skaug V, Zienolddiny S, et al. Genome-wide analysis of survival in early-stage non-small-cell lung cancer. J Clin Oncol. 2009;27:2660–2667. doi: 10.1200/JCO.2008.18.7906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Mash DC, ffrench-Mullen J, Adi N, Qin Y, Buck A, Pablo J. Gene expression in human hippocampus from cocaine abusers identifies genes which regulate extracellular matrix remodeling. PLoS One. 2007;2:1187. doi: 10.1371/journal.pone.0001187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Leshchenko VV, Kuo PY, Shaknovich R, Yang DT, Gellen T, Petrich A, et al. Genomewide DNA methylation analysis reveals novel targets for drug development in mantle cell lymphoma. Blood. 2009;116:1025–1034. doi: 10.1182/blood-2009-12-257485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Grati FR, Lesperance MM, De Toffol S, Chinetti S, Selicorni A, Emery S, et al. Pure monosomy and pure trisomy of 13q21.2–31.1 consequent to a familial insertional translocation: exclusion of PCDH9 as the responsible gene for autosomal dominant auditory neuropathy (AUNA1) Am J Med Genet A. 2009;149:906–913. doi: 10.1002/ajmg.a.32754. [DOI] [PubMed] [Google Scholar]
- 88.Terry S, Queires L, Gil-Diez-de-Medina S, Chen MW, de la Taille A, Allory Y, et al. Protocadherin-PC promotes androgen-independent prostate cancer cell growth. Prostate. 2006;66:1100–1113. doi: 10.1002/pros.20446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Ahmed ZM, Riazuddin S, Ahmad J, Bernstein SL, Guo Y, Sabar MF, et al. PCDH15 is expressed in the neurosensory epithelium of the eye and ear and mutant alleles are responsible for both USH1F and DFNB23. Hum Mol Genet. 2003;12:3215–3223. doi: 10.1093/hmg/ddg358. [DOI] [PubMed] [Google Scholar]
- 90.Huertas-Vazquez A, Plaisier CL, Geng R, Haas BE, Lee J, Greevenbroek MM, et al. A nonsynonymous SNP within PCDH15 is associated with lipid traits in familial combined hyperlipidemia. Hum Genet. 2010;127:83–89. doi: 10.1007/s00439-009-0749-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Becanovic K, Pouladi MA, Lim RS, Kuhn A, Pavlidis P, Luthi-Carter R, et al. Transcriptional changes in Huntington disease identified using genome-wide expression profiling and cross-platform analysis. Hum Mol Genet. 19:1438–1452. doi: 10.1093/hmg/ddq018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Kung A, Jae-Won H, Dae-Soo K, Hong-Seok H, Yun-Ji K, Ja-Rang L, et al. Quantitative analysis of alternative transcripts of human PCDH11X/Y genes. Am J Med Genet B Neuropsychiatr Genet. 2010;153:736–744. doi: 10.1002/ajmg.b.31041. [DOI] [PubMed] [Google Scholar]
- 93.Yoshida K, Watanabe M, Kato H, Dutta A, Sugano S. BH-protocadherin-c, a member of the cadherin superfamily, interacts with protein phosphatase 1alpha through its intracellular domain. FEBS Lett. 1999;460:93–98. doi: 10.1016/s0014-5793(99)01309-5. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.