Abstract
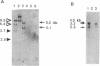
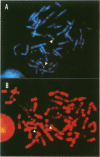
A complementary DNA encoding DNA-dependent ATPase Q1 possessing DNA helicase activity, which is the major DNA-dependent ATPase in human cell extracts, was cloned from a cDNA library of human KB cells. The predicted amino acid sequence has seven consecutive motifs conserved in the RNA and DNA helicase super family and DNA helicase Q1 belongs to DEXH helicase family. A homology search indicated that helicase Q1 had 47% homology in the seven conserved regions with Escherichia coli RecQ protein. Three RNA bands of 4.0, 3.3, and 2.2 kilobases were detected in HeLa cells by Northern blotting. Analysis of the genomic DNA indicated the presence of a homologous gene in mouse cells. The DNA helicase Q1 gene was localized on the short arm of human chromosome 12 at 12p12.
Full text
PDF






Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Aboussekhra A., Chanet R., Zgaga Z., Cassier-Chauvat C., Heude M., Fabre F. RADH, a gene of Saccharomyces cerevisiae encoding a putative DNA helicase involved in DNA repair. Characteristics of radH mutants and sequence of the gene. Nucleic Acids Res. 1989 Sep 25;17(18):7211–7219. doi: 10.1093/nar/17.18.7211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adams M. D., Kelley J. M., Gocayne J. D., Dubnick M., Polymeropoulos M. H., Xiao H., Merril C. R., Wu A., Olde B., Moreno R. F. Complementary DNA sequencing: expressed sequence tags and human genome project. Science. 1991 Jun 21;252(5013):1651–1656. doi: 10.1126/science.2047873. [DOI] [PubMed] [Google Scholar]
- Backendorf C., Spaink H., Barbeiro A. P., van de Putte P. Structure of the uvrB gene of Escherichia coli. Homology with other DNA repair enzymes and characterization of the uvrB5 mutation. Nucleic Acids Res. 1986 Apr 11;14(7):2877–2890. doi: 10.1093/nar/14.7.2877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cleaver J. E. It was a very good year for DNA repair. Cell. 1994 Jan 14;76(1):1–4. doi: 10.1016/0092-8674(94)90165-1. [DOI] [PubMed] [Google Scholar]
- Feinberg A. P., Vogelstein B. A technique for radiolabeling DNA restriction endonuclease fragments to high specific activity. Anal Biochem. 1983 Jul 1;132(1):6–13. doi: 10.1016/0003-2697(83)90418-9. [DOI] [PubMed] [Google Scholar]
- Finch P. W., Emmerson P. T. The nucleotide sequence of the uvrD gene of E. coli. Nucleic Acids Res. 1984 Jul 25;12(14):5789–5799. doi: 10.1093/nar/12.14.5789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finch P. W., Storey A., Chapman K. E., Brown K., Hickson I. D., Emmerson P. T. Complete nucleotide sequence of the Escherichia coli recB gene. Nucleic Acids Res. 1986 Nov 11;14(21):8573–8582. doi: 10.1093/nar/14.21.8573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flejter W. L., McDaniel L. D., Johns D., Friedberg E. C., Schultz R. A. Correction of xeroderma pigmentosum complementation group D mutant cell phenotypes by chromosome and gene transfer: involvement of the human ERCC2 DNA repair gene. Proc Natl Acad Sci U S A. 1992 Jan 1;89(1):261–265. doi: 10.1073/pnas.89.1.261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foury F., Lahaye A. Cloning and sequencing of the PIF gene involved in repair and recombination of yeast mitochondrial DNA. EMBO J. 1987 May;6(5):1441–1449. doi: 10.1002/j.1460-2075.1987.tb02385.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilchrist C. A., Denhardt D. T. Escherichia coli rep gene: sequence of the gene, the encoded helicase, and its homology with uvrD. Nucleic Acids Res. 1987 Jan 26;15(2):465–475. doi: 10.1093/nar/15.2.465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorbalenya A. E., Koonin E. V., Donchenko A. P., Blinov V. M. Two related superfamilies of putative helicases involved in replication, recombination, repair and expression of DNA and RNA genomes. Nucleic Acids Res. 1989 Jun 26;17(12):4713–4730. doi: 10.1093/nar/17.12.4713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gustafson G., Armour S. L. The complete nucleotide sequence of RNA beta from the type strain of barley stripe mosaic virus. Nucleic Acids Res. 1986 May 12;14(9):3895–3909. doi: 10.1093/nar/14.9.3895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodgman T. C. A new superfamily of replicative proteins. Nature. 1988 May 5;333(6168):22–23. doi: 10.1038/333022b0. [DOI] [PubMed] [Google Scholar]
- Hoshino S., Miyazawa H., Enomoto T., Hanaoka F., Kikuchi Y., Kikuchi A., Ui M. A human homologue of the yeast GST1 gene codes for a GTP-binding protein and is expressed in a proliferation-dependent manner in mammalian cells. EMBO J. 1989 Dec 1;8(12):3807–3814. doi: 10.1002/j.1460-2075.1989.tb08558.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Human gene mapping 11. London Conference (1991). Eleventh International Workshop on Human Gene Mapping. London, UK, August 18-22, 1991. Cytogenet Cell Genet. 1991;58(3-4):986–2156. doi: 10.1159/000133716. [DOI] [PubMed] [Google Scholar]
- Irino N., Nakayama K., Nakayama H. The recQ gene of Escherichia coli K12: primary structure and evidence for SOS regulation. Mol Gen Genet. 1986 Nov;205(2):298–304. doi: 10.1007/BF00430442. [DOI] [PubMed] [Google Scholar]
- Kawai J., Ando A., Sato T., Nakatsuji T., Tsuji K., Inoko H. Analysis of gene structure and antigen determinants of DR2 antigens using DR gene transfer into mouse L cells. J Immunol. 1989 Jan 1;142(1):312–317. [PubMed] [Google Scholar]
- Koonin E. V., Choi G. H., Nuss D. L., Shapira R., Carrington J. C. Evidence for common ancestry of a chestnut blight hypovirulence-associated double-stranded RNA and a group of positive-strand RNA plant viruses. Proc Natl Acad Sci U S A. 1991 Dec 1;88(23):10647–10651. doi: 10.1073/pnas.88.23.10647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Legerski R., Peterson C. Expression cloning of a human DNA repair gene involved in xeroderma pigmentosum group C. Nature. 1992 Sep 3;359(6390):70–73. doi: 10.1038/359070a0. [DOI] [PubMed] [Google Scholar]
- Linder P., Lasko P. F., Ashburner M., Leroy P., Nielsen P. J., Nishi K., Schnier J., Slonimski P. P. Birth of the D-E-A-D box. Nature. 1989 Jan 12;337(6203):121–122. doi: 10.1038/337121a0. [DOI] [PubMed] [Google Scholar]
- Lloyd R. G., Sharples G. J. Molecular organization and nucleotide sequence of the recG locus of Escherichia coli K-12. J Bacteriol. 1991 Nov;173(21):6837–6843. doi: 10.1128/jb.173.21.6837-6843.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lovett S. T., Clark A. J. Genetic analysis of regulation of the RecF pathway of recombination in Escherichia coli K-12. J Bacteriol. 1983 Mar;153(3):1471–1478. doi: 10.1128/jb.153.3.1471-1478.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masutani C., Sugasawa K., Yanagisawa J., Sonoyama T., Ui M., Enomoto T., Takio K., Tanaka K., van der Spek P. J., Bootsma D. Purification and cloning of a nucleotide excision repair complex involving the xeroderma pigmentosum group C protein and a human homologue of yeast RAD23. EMBO J. 1994 Apr 15;13(8):1831–1843. doi: 10.1002/j.1460-2075.1994.tb06452.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naumovski L., Chu G., Berg P., Friedberg E. C. RAD3 gene of Saccharomyces cerevisiae: nucleotide sequence of wild-type and mutant alleles, transcript mapping, and aspects of gene regulation. Mol Cell Biol. 1985 Jan;5(1):17–26. doi: 10.1128/mcb.5.1.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naumovski L., Friedberg E. C. Analysis of the essential and excision repair functions of the RAD3 gene of Saccharomyces cerevisiae by mutagenesis. Mol Cell Biol. 1986 Apr;6(4):1218–1227. doi: 10.1128/mcb.6.4.1218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen P. J., McMaster G. K., Trachsel H. Cloning of eukaryotic protein synthesis initiation factor genes: isolation and characterization of cDNA clones encoding factor eIF-4A. Nucleic Acids Res. 1985 Oct 11;13(19):6867–6880. doi: 10.1093/nar/13.19.6867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oakley B. R., Kirsch D. R., Morris N. R. A simplified ultrasensitive silver stain for detecting proteins in polyacrylamide gels. Anal Biochem. 1980 Jul 1;105(2):361–363. doi: 10.1016/0003-2697(80)90470-4. [DOI] [PubMed] [Google Scholar]
- Pai E. F., Krengel U., Petsko G. A., Goody R. S., Kabsch W., Wittinghofer A. Refined crystal structure of the triphosphate conformation of H-ras p21 at 1.35 A resolution: implications for the mechanism of GTP hydrolysis. EMBO J. 1990 Aug;9(8):2351–2359. doi: 10.1002/j.1460-2075.1990.tb07409.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pause A., Sonenberg N. Mutational analysis of a DEAD box RNA helicase: the mammalian translation initiation factor eIF-4A. EMBO J. 1992 Jul;11(7):2643–2654. doi: 10.1002/j.1460-2075.1992.tb05330.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reynolds P., Higgins D. R., Prakash L., Prakash S. The nucleotide sequence of the RAD3 gene of Saccharomyces cerevisiae: a potential adenine nucleotide binding amino acid sequence and a nonessential acidic carboxyl terminal region. Nucleic Acids Res. 1985 Apr 11;13(7):2357–2372. doi: 10.1093/nar/13.7.2357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaeffer L., Roy R., Humbert S., Moncollin V., Vermeulen W., Hoeijmakers J. H., Chambon P., Egly J. M. DNA repair helicase: a component of BTF2 (TFIIH) basic transcription factor. Science. 1993 Apr 2;260(5104):58–63. doi: 10.1126/science.8465201. [DOI] [PubMed] [Google Scholar]
- Seki M., Enomoto T., Watanabe Y., Tawaragi Y., Kawasaki K., Hanaoka F., Yamada M. Purification and characterization of a deoxyribonucleic acid dependent adenosinetriphosphatase from mouse FM3A cells: effects of ribonucleoside triphosphates on the interaction of the enzyme with single-stranded DNA. Biochemistry. 1986 Jun 3;25(11):3239–3245. doi: 10.1021/bi00359a024. [DOI] [PubMed] [Google Scholar]
- Seki M., Enomoto T., Yanagisawa J., Hanaoka F., Ui M. Further characterization of DNA helicase activity of mouse DNA-dependent adenosinetriphosphatase B (DNA helicase B). Biochemistry. 1988 Mar 8;27(5):1766–1771. doi: 10.1021/bi00405a057. [DOI] [PubMed] [Google Scholar]
- Seki M., Yanagisawa J., Kohda T., Sonoyama T., Ui M., Enomoto T. Purification of two DNA-dependent adenosinetriphosphatases having DNA helicase activity from HeLa cells and comparison of the properties of the two enzymes. J Biochem. 1994 Mar;115(3):523–531. doi: 10.1093/oxfordjournals.jbchem.a124369. [DOI] [PubMed] [Google Scholar]
- Smith G. R. Homologous recombination in E. coli: multiple pathways for multiple reasons. Cell. 1989 Sep 8;58(5):807–809. doi: 10.1016/0092-8674(89)90929-x. [DOI] [PubMed] [Google Scholar]
- Sung P., Bailly V., Weber C., Thompson L. H., Prakash L., Prakash S. Human xeroderma pigmentosum group D gene encodes a DNA helicase. Nature. 1993 Oct 28;365(6449):852–855. doi: 10.1038/365852a0. [DOI] [PubMed] [Google Scholar]
- Sung P., Higgins D., Prakash L., Prakash S. Mutation of lysine-48 to arginine in the yeast RAD3 protein abolishes its ATPase and DNA helicase activities but not the ability to bind ATP. EMBO J. 1988 Oct;7(10):3263–3269. doi: 10.1002/j.1460-2075.1988.tb03193.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi E., Hori T., O'Connell P., Leppert M., White R. R-banding and nonisotopic in situ hybridization: precise localization of the human type II collagen gene (COL2A1). Hum Genet. 1990 Nov;86(1):14–16. doi: 10.1007/BF00205165. [DOI] [PubMed] [Google Scholar]
- Takahashi E., Yamauchi M., Tsuji H., Hitomi A., Meuth M., Hori T. Chromosome mapping of the human cytidine-5'-triphosphate synthetase (CTPS) gene to band 1p34.1-p34.3 by fluorescence in situ hybridization. Hum Genet. 1991 Nov;88(1):119–121. doi: 10.1007/BF00204942. [DOI] [PubMed] [Google Scholar]
- Umezu K., Nakayama K., Nakayama H. Escherichia coli RecQ protein is a DNA helicase. Proc Natl Acad Sci U S A. 1990 Jul;87(14):5363–5367. doi: 10.1073/pnas.87.14.5363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vermeulen W., Stefanini M., Giliani S., Hoeijmakers J. H., Bootsma D. Xeroderma pigmentosum complementation group H falls into complementation group D. Mutat Res. 1991 Sep;255(2):201–208. doi: 10.1016/0921-8777(91)90054-s. [DOI] [PubMed] [Google Scholar]
- Walker J. E., Saraste M., Runswick M. J., Gay N. J. Distantly related sequences in the alpha- and beta-subunits of ATP synthase, myosin, kinases and other ATP-requiring enzymes and a common nucleotide binding fold. EMBO J. 1982;1(8):945–951. doi: 10.1002/j.1460-2075.1982.tb01276.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber C. A., Salazar E. P., Stewart S. A., Thompson L. H. ERCC2: cDNA cloning and molecular characterization of a human nucleotide excision repair gene with high homology to yeast RAD3. EMBO J. 1990 May;9(5):1437–1447. doi: 10.1002/j.1460-2075.1990.tb08260.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weeda G., van Ham R. C., Vermeulen W., Bootsma D., van der Eb A. J., Hoeijmakers J. H. A presumed DNA helicase encoded by ERCC-3 is involved in the human repair disorders xeroderma pigmentosum and Cockayne's syndrome. Cell. 1990 Aug 24;62(4):777–791. doi: 10.1016/0092-8674(90)90122-u. [DOI] [PubMed] [Google Scholar]
- Yanagisawa J., Seki M., Ui M., Enomoto T. Alteration of a DNA-dependent ATPase activity in xeroderma pigmentosum complementation group C cells. J Biol Chem. 1992 Feb 25;267(6):3585–3588. [PubMed] [Google Scholar]