Abstract
Objective
To assess the influence of rs5848 polymorphism in serum progranulin (PGRN) level in a cohort of subjects with Alzheimer and related dementias from a tertiary referral clinic.
Background
Mutations in the GRN gene cause autosomal dominant frontotemporal dementia (FTD) with TDP-43 pathology (FTLD-TDP) through haploinsufficiency. It has recently been shown that homozygous carriers of the T-allele of rs5848 have an elevated risk developing FTD, and this polymorphism may play a role in the pathogenesis of other dementia by modifying progranulin level. We hypothesize that genotype of rs5848 may influence serum PGRN level in AD, FTD, and other dementias.
Methods
Blood samples were obtained from patients with cognitive impairment and dementia referred to a tertiary dementia clinic, as well as samples from a cohort of healthy controls. Serum PGRN level was measured using an ELISA assay, and rs5848 genotype was determined by a TaqMan assay.
Results
We found that rs5848 SNP significantly influenced serum PGRN level, with TT genotype having the lowest levels, CC the highest. This relationship is observed in each of the subgroups. We also confirmed that GRN mutation carriers had significantly lower serum PGRN levels than all other groups.
Conclusions
The rs5848 polymorphism significantly influences serum PGRN with TT carriers having a lower level of serum PGRN then CT and CC carriers. This is consistent with the finding that miR-659 binding to the high risk T allele of rs5848 may augment translational inhibition of GRN and alter risk of FTD and possibly other dementias.
Keywords: Frontotemporal Dementia, Progranulin, PGRN, GRN, rs5848, genetic polymorphism, biomarker
Background
The progranulin protein (PGRN) is an 88 kDa secreted growth factor with multiple physiological functions, including wound healing, tumour growth and embryonic brain development. Full-length PGRN undergoes proteolytic cleavage by elastase to generate granulin peptides (GRN), that have potent pro-inflammatory effects. PGRN is constitutively expressed in a number of cells, including skin, gastrointestinal tract, the reproductive system and immune cells. In the brain, PGRN is expressed by specific neuronal populations and microglial cells.
Since the discovery that mutations in the progranulin gene (GRN) cause familial frontotemporal lobar degeneration (FTLD), over 60 different mutations have been identified in over 200 families world-wide. Most pathogenic GRN mutations lead to a frameshift or premature stop codon and result in abnormal mRNA transcripts that undergo nonsense-mediated mRNA decay and are not expressed. Based on this haploinsufficiency mechanism, it is hypothesized that serum PGRN levels will be lower in GRN-mutation carriers. Several studies have now shown that cerebral spinal fluid (CSF), serum and plasma PGRN levels are significantly lower in GRN mutations carriers than non-carriers and that measurement of PGRN levels predicts a pathogenic GRN mutation with almost 100% sensitivity and specificity.
In addition to causing FTLD via null mutations, a common genetic variation in GRN has recently been shown to influence the risk of sporadic FTLD-TDP. A case-control study found that homozygosity for the T allele of the common rs5848 polymorphism in the 3′-untranslated region of GRN was more frequent in patients with pathologically proven FTLD-TDP (odds ratio of 3.2). The rs5848 polymorphism is predicted to be a binding site for the microRNA miR-659, and more efficient binding to the T allele results in increased translational suppression of GRN. Consistent with this was the finding that homozygosity for the T allele was associated with ~30% reduction in PGRN expression in FTLD-TDP brain tissue (compared with CC), despite normal levels of GRN mRNA. These findings suggest that rs5848 influences the risk of sporadic FTLD-TDP by affecting PGRN expression, although this has not been confirmed in two other clinical FTLD patients cohorts.
The role of PGRN in other neurodegenerative disorders is also unclear. Some recent studies have found that GRN expression is increased in activated microglia as well as in peripheral blood in Alzheimer disease (AD). In another study, no significant differences were found in relative peripheral blood mononuclear cells and CSF PGRN expression in AD patients compared to controls, but patients with TT genotype had a lower GRN mRNA expression level in brain samples compared to the CC genotype.
To further examined the role of PGRN serum level in the risk of developing other dementias and its relationship to the rs5848 polymorphisms, we compared the serum PGRN levels and examined the rs5848 genotype frequency in a cohort of subjects assessed at a tertiary dementia referral clinic.
Methods
Subjects
Study subjects were recruited from the University of British Columbia hospital clinic for Alzheimer and related disorders (UBCH-CARD) with approval of our institutional Clinical Research Ethics Board. Clinical diagnosis was made by specialty-trained neurologists and geriatricians according to current criteria. Blood samples were obtained from 100 patients, including 57 with a clinical diagnosis of AD, 13 with amnestic mild cognitive impairment (MCI), 12 with a combination of frontotemporal dementia and amyotrophic lateral sclerosis (FTD-ALS), 6 with dementia with Lewy bodies, 5 with vascular dementia, 3 with Parkinson disease with dementia, 2 with corticobasal syndrome, and 2 with progressive supranuclear palsy. Samples were also obtained from spouses of patients as well as healthy community subjects (n = 36), all of whom scored within the normal range in the Montreal Cognitive Assessment. A small set of samples (n = 6) from known carriers of pathogenic GRN mutations from 3 different families (C31LfsX34, R418X, and Q130SfsX124 mutations) was also included to confirm the sensitivity of this assay.
Biochemical and genetic analyses
Serum levels of PGRN were measured using a commercial ELISA assay (Human Progranulin ELISA kit, AdipoGen, Inc, South Korea) according to the manufacturer’s instructions. GRN carrier status was determined by direct sequencing of exonic and flanking intronic regions using previously published primers in both directions using the Big Dye Terminator v3.1 Cycle Sequencing kit (Applied Biosystems, Foster City, CA).
DNA was extracted from whole blood using an automated DNA extraction machine (AutogenflexStar, Autogen Inc, Hollisten, MA). Genotyping of SNP rs5848 was performed with a TaqMan chemistry-based allelic discrimination assay, with Assay by Design probes (Applied Biosystems) and an ABI 7900 PCR system, followed by analysis with Sequence Detection System 2.2.1 software (Applied Biosystems).
Statistical Analysis
PGRN levels among different diagnostic groups were compared by ANOVA with post-hoc Student-Neuman-Keuls test to determine group allocations. We also compared serum and plasma PGRN level in 12 healthy controls using paired sample t-test. Multi-variable linear regression was used to examine the effect of age, sex, APOE and rs5848 genotype on serum PGRN levels among non-GRN mutation carriers.
Results
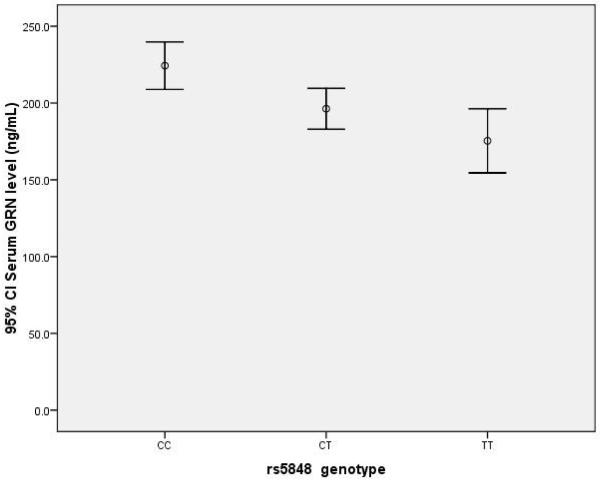
The demographic distribution of the subjects is shown in Table 1. Since the numbers in non-AD, non-FTD dementias were small, they were grouped as “other dementia” for further statistical analyses. Females were slightly overrepresented in all groups with the exception of the FTD-ALS (equal) and MCI subjects (male predominance). As expected, the proportion of APOE ε4 carriers was higher in the groups of AD and MCI subjects. The effect of rs5848 polymorphism on serum PGRN in our total cohort (excluding the GRN-mutation carriers) is shown in figure 1. TT genotype carriers had significantly lower serum PGRN levels (164 ng/mL, 95% C.I. 138-189), than the CT (191 ng/mL, 95% C.I. 177-206) and CC genotype carriers (222 ng/mL, 95% C.I. 205-238) (overall ANOVA p<0.005, with SNK p<0.05 for TT vs. CT, and CT vs. CC).
Table 1.
Demographics and ApoE allele status of study subjects
| Diagnoses | N | Age (mean, range) |
% female | % APOE e4 carriers |
|---|---|---|---|---|
| Controls | 36 | 46.7, 16-84 | 66.7 | 25 |
| MCI | 13 | 69.0, 51-83 | 38.5 | 69 |
| AD | 57 | 69.7, 51-86 | 56.1 | 62 |
| FTD-ALS | 12 | 67.1, 59-83 | 50 | 14.3 |
| Other dementias | 18 | 64.9, 59-71 | 66.7 | 35.3 |
|
GRN mutation carriers |
6 | 44.2, 27-63 | 83.3 | 16.7 |
Figure 1.
Serum progranulin levels and rs5848 genotype in entire cohort. The progranlulin level in TT genotype carriers is significantly lower than the CC carriers (p<0.005).
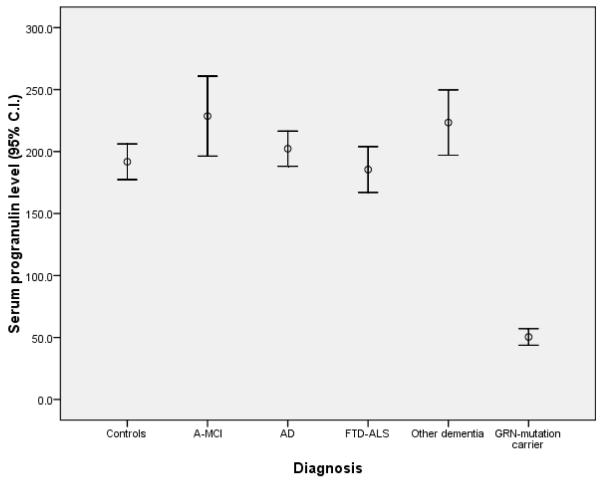
The comparison of serum PGRN levels across each clinical diagnostic category is shown in figure 2. The 6 GRN-mutation carriers had significantly lower serum PGRN levels (50.5 ng/mL, 95% C.I 43.8-57.2) than all other groups (p<10−7), with no overlap. However, there were no significant differences between any of the other groups; controls (195 ng/mL, 95% C.I. 180-211), amnestic mild cognitive impairment (228 ng/mL, 95% C.I. 196-261), probable AD (202 ng/mL, 95% C.I. 188-217), FTD-ALS (187 ng/mL, 95% C.I. 167-204), and other dementias (223 ng/mL, 95% C.I. 197-250). We have sequenced all the exons of the GRN gene in the FTD-ALS subjects but did not find any carrier of single base mutations.
Figure 2.
Serum progranulin levels across diagnostic subgroups
The role of other variables on serum PGRN level was also examined (excluding the 6 GRN mutation carriers). Males tended to have lower levels than females (191 ng/mL, 95% C.I. 180-204 vs. 215 ng/mL, 95% C.I. 202-229, respectively), however this was not significant (p=0.16). There were also no significant effects with age (p=0.19) or with APOE genotype (p=0.30). We further compared the measurements of PGRN in plasma and serum samples drawn from the same controls at the same time and found a strong correlation (R2 = 0.946, p=3×10−6), with the serum levels about 5% lower than plasma measurements (data not shown).
We also examined the influence of rs5848 genotype on serum PGRN levels within each diagnostic subgroup (Table 2). In every group there was a gradient with the TT genotype exhibiting the lowest PGRN level followed by CT genotype, with CC genotype the highest, although the differences only reached significance in the controls and other dementias groups.
Table 2.
Comparison of serum PGRN levels within each diagnostic subgroup and each rs5848 genotype
| N total |
Mean PGRN level within whole group |
N within each genotype |
Mean PGRN level in CC |
Mean PGRN level in CT |
Mean PGRN level in TT |
ANOVA P-valuve |
|
|---|---|---|---|---|---|---|---|
| Control | 36 | 198 | 13,16,7 | 242 | 182 | 155 | 0.002 |
| A-MCI | 13 | 229 | 5,6,2 | 265 | 203 | 183 | 0.051 |
| AD | 57 | 202 | 30,22,5 | 207 | 200 | 195 | 0.87 |
| FTD-ALS | 12 | 186 | 4,5,3 | 202 | 179 | 176 | 0.38 |
| Other Dementia |
18 | 223 | 6,9,3 | 271 | 202 | 191 | 0.01 |
Discussion
In this study, we have shown that the T allele of rs5848 is significantly associated with a lower PGRN levels in a gene-dose dependent fashion in populations beyond FTLD-TDP. In all diagnostic groups, those with the TT genotype were found to hae the lowest PGRN levels while CC carriers had the highest levels. The fact that these differences did not reach significance in several of the individual groups is likely a reflection of the small group sizes. Nonetheless, this supports the hypothesis that the rs5848 SNP affects PGRN expression through miR-659 dependent translational inhibition and that this mechanism likely has a significant effect on PGRN levels in normals as well as various disease populations.
Whether PGRN expression affects the risk of developing other neurodegenerative diseases remains to be determined. A study of patients with ALS found that some missense mutations and other genetic variations in GRN may be associated with earlier age of onset and shorter survival. GRN mRNA levels were found to be elevated in AD in one study, while in another, the rs5848 T allele was associated with reduction in GRN mRNA expression in AD brain tissue. In our study, we did not find any significant difference in serum PGRN levels in any of our disease groups compared with controls or with any of the other disease groups. Interestingly, the lowest mean PGRN serum level was found in patients with clinical FTD and ALS, a group with a high likelihood of the underlying pathology being FTLD-TDP. Although the small size of many of the groups might have affected our ability to detect statistical differences, there were no clear trends detected.
We also confirmed the findings of previous studies, that measuring PGRN in biological fluids is a sensitive and specific test for detecting GRN null mutations, even in unaffected carriers. Since the initial discovery of mutations in GRN as the cause of familial FTLD-TDP, nearly 150 genetic variabts in GRN have been described, of which many have unknown pathological significance. The identification of GRN mutations through sequencing of the GRN gene is a necessary but laborious step. Based on our findings, serum PGRN measurement may be used as an initial screen to identify subjects who are at-risk before expensive and labour-intensive sequencing is performed. No carrier of single base mutations in the GRN gene were found in any of our FTD subjects, whereas, the level of PGRN is significantly lower in the 6 subjects who carry a nonsense mutation. Others have reported that carriers of GRN missense mutations may have an intermediate level of PGRN between those of null GRN mutation carriers and non-carriers, although we have not identified any such carriers in our cohort. Since the clinical presentation of FTLD caused by GRN mutations often overlaps with other dementia phenotypes such as FTLD-tau, AD, corticobasal syndrome, and progressive supranuclear palsy, serum PGRN measurement, thus far, have demonstrated to be a valuable biomarker in the investigation of these neurodegenerative conditions.
Finally, although PGRN’s role as a neuronal survival factor is focused mainly on neurodegeneration, it is involved in a number of other important physiological processes that could also affect brain health. GRN peptides are known to be potent inflammatory mediators. Microglial activation has been implemented in a wide variety of neurodegenerative conditions and upregulation of PGRN expression is an important part of this process. In addition, vascular risk factors affect the risk of developing AD and a recent study found that serum PGRN concentrations are associated with visceral obesity, elevated plasma glucose, and dyslipidemia, and may be a biomarker for chronic inflammation in obesity and type 2 diabetes.
In summary, we found that the T allele of the rs5848 polymorphism is associated with a lower serum PGRN levels in populations beyond FTLD-TDP and may thereby increase the risk of other neurodegenerative conditions in which PGRN function plays a role. In addition, we have shown that serum PGRN measurement is a sensitive and specific screening test that can be used to identify patients with GRN null mutations.
Acknowledgements
This study is supported by grants from the Canadian Institute of Health Research (CIHR) #74580 & #179009, the Pacific Alzheimer Research Foundation #C06-01 (to IRAM & HHF), and the NIH R01 NS 065782 (RR), as well as donations to the UBC Clinic for Alzheimer and Related Disorders from the Townsend Family. Dr. Hsiung is supported by a Clinical Genetics Investigatorship award from the CIHR. We would like to thank Pheth Sengdy for coordinating and arranging clinical assessment for all at-risk FTD subjects enrolled in this project. We are also indebted to all the patients and families who donated their time and effort in their participation of this research.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Yin F, Banerjee R, Thomas B, Zhou P, Qian L, Jia T, et al. Exaggerated inflammation, impaired host defense, and neuropathology in progranulin-deficient mice. J Exp Med. 2010;207(1):117–28. S1–4. doi: 10.1084/jem.20091568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.He Z, Bateman A. Progranulin (granulin-epithelin precursor, PC-cell-derived growth factor, acrogranin) mediates tissue repair and tumorigenesis. J Mol Med. 2003;81(10):600–12. doi: 10.1007/s00109-003-0474-3. [DOI] [PubMed] [Google Scholar]
- 3.Tolkatchev D, Malik S, Vinogradova A, Wang P, Chen Z, Xu P, et al. Structure dissection of human progranulin identifies well-folded granulin/epithelin modules with unique functional activities. Protein Sci. 2008;17(4):711–24. doi: 10.1110/ps.073295308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Daniel R, He Z, Carmichael KP, Halper J, Bateman A. Cellular localization of gene expression for progranulin. J Histochem Cytochem. 2000;48(7):999–1009. doi: 10.1177/002215540004800713. [DOI] [PubMed] [Google Scholar]
- 5.Flanders Interuniversity Institute for Biotechnology B . Flanders Interuniversity Institute for Biotechnology; http://www.molgen.ua.ac.be/FTDMutations/ [Google Scholar]
- 6.Cruts M, Gijselinck I, van der Zee J, Engelborghs S, Wils H, Pirici D, et al. Null mutations in progranulin cause ubiquitin-positive frontotemporal dementia linked to chromosome 17q21. Nature. 2006;442(7105):920–4. doi: 10.1038/nature05017. [DOI] [PubMed] [Google Scholar]
- 7.Baker M, Mackenzie IR, Pickering-Brown SM, Gass J, Rademakers R, Lindholm C, et al. Mutations in progranulin cause tau-negative frontotemporal dementia linked to chromosome 17. Nature. 2006;442(7105):916–9. doi: 10.1038/nature05016. [DOI] [PubMed] [Google Scholar]
- 8.Sleegers K, Brouwers N, Van Damme P, Engelborghs S, Gijselinck I, van der Zee J, et al. Serum biomarker for progranulin-associated frontotemporal lobar degeneration. Ann Neurol. 2009 doi: 10.1002/ana.21621. [DOI] [PubMed] [Google Scholar]
- 9.Ghidoni R, Benussi L, Glionna M, Franzoni M, Binetti G. Low plasma progranulin levels predict progranulin mutations in frontotemporal lobar degeneration. Neurology. 2008;71(16):1235–9. doi: 10.1212/01.wnl.0000325058.10218.fc. [DOI] [PubMed] [Google Scholar]
- 10.Finch N, Baker M, Crook R, Swanson K, Kuntz K, Surtees R, et al. Plasma progranulin levels predict progranulin mutation status in frontotemporal dementia patients and asymptomatic family members. Brain. 2009;132(Pt 3):583–91. doi: 10.1093/brain/awn352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rademakers R, Eriksen JL, Baker M, Robinson T, Ahmed Z, Lincoln SJ, et al. Common variation in the miR-659 binding-site of GRN is a major risk factor for TDP43-positive frontotemporal dementia. Hum Mol Genet. 2008;17(23):3631–42. doi: 10.1093/hmg/ddn257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Simon-Sanchez J, Seelaar H, Bochdanovits Z, Deeg DJ, van Swieten JC, Heutink P. Variation at GRN 3′-UTR rs5848 is not associated with a risk of frontotemporal lobar degeneration in Dutch population. PLoS One. 2009;4(10):e7494. doi: 10.1371/journal.pone.0007494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rollinson S, Rohrer JD, van der Zee J, Sleegers K, Mead S, Engelborghs S, et al. No association of PGRN 3′UTR rs5848 in frontotemporal lobar degeneration. Neurobiol Aging. 2009 doi: 10.1016/j.neurobiolaging.2009.04.009. [DOI] [PubMed] [Google Scholar]
- 14.Brouwers N, Sleegers K, Engelborghs S, Maurer-Stroh S, Gijselinck I, van der Zee J, et al. Genetic variability in progranulin contributes to risk for clinically diagnosed Alzheimer disease. Neurology. 2008;71(9):656–64. doi: 10.1212/01.wnl.0000319688.89790.7a. [DOI] [PubMed] [Google Scholar]
- 15.Fenoglio C, Galimberti D, Cortini F, Kauwe JS, Cruchaga C, Venturelli E, et al. Rs5848 variant influences GRN mRNA levels in brain and peripheral mononuclear cells in patients with Alzheimer’s disease. J Alzheimers Dis. 2009;18(3):603–12. doi: 10.3233/JAD-2009-1170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.McKhann G, Drachman D, Folstein M, Katzman R, Price D, Stadlan EM. Clinical diagnosis of Alzheimer’s disease: report of the NINCDS-ADRDA Work Group under the auspices of Department of Health and Human Services Task Force on Alzheimer’s Disease. Neurology. 1984;34(7):939–44. doi: 10.1212/wnl.34.7.939. [DOI] [PubMed] [Google Scholar]
- 17.Neary D, Snowden JS, Gustafson L, Passant U, Stuss D, Black S, et al. Frontotemporal lobar degeneration: a consensus on clinical diagnostic criteria. Neurology. 1998;51(6):1546–54. doi: 10.1212/wnl.51.6.1546. [DOI] [PubMed] [Google Scholar]
- 18.Petersen RC, Smith GE, Waring SC, Ivnik RJ, Tangalos EG, Kokmen E. Mild cognitive impairment: clinical characterization and outcome. Arch Neurol. 1999;56(3):303–8. doi: 10.1001/archneur.56.3.303. [DOI] [PubMed] [Google Scholar]
- 19.Petersen RC, Doody R, Kurz A, Mohs RC, Morris JC, Rabins PV, et al. Current concepts in mild cognitive impairment. Arch Neurol. 2001;58(12):1985–92. doi: 10.1001/archneur.58.12.1985. [DOI] [PubMed] [Google Scholar]
- 20.Nasreddine ZS, Phillips NA, Bedirian V, Charbonneau S, Whitehead V, Collin I, et al. The Montreal Cognitive Assessment, MoCA: a brief screening tool for mild cognitive impairment. J Am Geriatr Soc. 2005;53(4):695–9. doi: 10.1111/j.1532-5415.2005.53221.x. [DOI] [PubMed] [Google Scholar]
- 21.Sleegers K, Brouwers N, Maurer-Stroh S, van Es MA, Van Damme P, van Vught PW, et al. Progranulin genetic variability contributes to amyotrophic lateral sclerosis. Neurology. 2008;71(4):253–9. doi: 10.1212/01.wnl.0000289191.54852.75. [DOI] [PubMed] [Google Scholar]
- 22.Coppola G, Karydas A, Rademakers R, Wang Q, Baker M, Hutton M, et al. Gene expression study on peripheral blood identifies progranulin mutations. Ann Neurol. 2008;64(1):92–6. doi: 10.1002/ana.21397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mackenzie IR, Rademakers R. The role of transactive response DNA-binding protein-43 in amyotrophic lateral sclerosis and frontotemporal dementia. Curr Opin Neurol. 2008;21(6):693–700. doi: 10.1097/WCO.0b013e3283168d1d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kelley BJ, Haidar W, Boeve BF, Baker M, Graff-Radford NR, Krefft T, et al. Prominent phenotypic variability associated with mutations in Progranulin. Neurobiol Aging. 2009;30(5):739–51. doi: 10.1016/j.neurobiolaging.2007.08.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kertesz A. Frontotemporal dementia: a topical review. Cogn Behav Neurol. 2008;21(3):127–33. doi: 10.1097/WNN.0b013e31818a8c66. [DOI] [PubMed] [Google Scholar]
- 26.Hsiung GY, Feldman H. GRN-Related Frontotemporal Dementia: in GeneReivews. NIH / University of Washington; Seattle, WA, USA: 2006. available at http://www.ncbi.nlm.nih.gov/sites/GeneTests/ [Google Scholar]
- 27.Carecchio M, Fenoglio C, De Riz M, Guidi I, Comi C, Cortini F, et al. Progranulin plasma levels as potential biomarker for the identification of GRN deletion carriers. A case with atypical onset as clinical amnestic Mild Cognitive Impairment converted to Alzheimer’s disease. J Neurol Sci. 2009;287(1-2):291–3. doi: 10.1016/j.jns.2009.07.011. [DOI] [PubMed] [Google Scholar]
- 28.Ahmed Z, Mackenzie IR, Hutton ML, Dickson DW. Progranulin in frontotemporal lobar degeneration and neuroinflammation. J Neuroinflammation. 2007;4:7. doi: 10.1186/1742-2094-4-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Youn BS, Bang SI, Kloting N, Park JW, Lee N, Oh JE, et al. Serum progranulin concentrations may be associated with macrophage infiltration into omental adipose tissue. Diabetes. 2009;58(3):627–36. doi: 10.2337/db08-1147. [DOI] [PMC free article] [PubMed] [Google Scholar]