Abstract
Polycystic ovaries and impaired fertility are the result of abnormal folliculogenesis. Our objective was to determine the role of four candidate folliculogenesis genes in the development of polycystic ovary syndrome (PCOS). Women with and without PCOS (335 cases, 198 controls) were genotyped for single nucleotide polymorphisms in GDF9, BMP15, AMH, and AMHR2. Variants in these genes were not associated with PCOS. Certain GDF9 variants were associated with hirsutism scores and parity in PCOS patients. GDF9 may thus serve as a modifier gene. These results suggest that inherited defects in folliculogenesis are not major factors in the genetic susceptibility to PCOS.
Keywords: folliculogenesis, polycystic ovary syndrome
Introduction
Polycystic ovary syndrome (PCOS) affects 6–8% of reproductive age women.1 PCOS is characterised by ovulatory dysfunction resulting in menstrual abnormalities and polycystic ovarian morphology that contribute to an increased risk of infertility.
Numerous factors affect the normal development of a follicle and subsequent ovulation. The transforming growth factor-beta (TGF-β) superfamily is a family of structurally related proteins that regulate basic biological processes including those having to do with growth and development. Several TGF-β superfamily members and their receptors are involved in folliculogenesis, including growth differentiation factor 9 (GDF9), bone morphogenetic protein 15 (BMP15), anti-müllerian hormone (AMH), and anti-müllerian hormone receptor type II (AMHR2).
During normal folliculogenesis, expression of GDF9, an early follicle growth promoter, begins in human oocytes at the primordial-primary follicle transition and increases with preantral follicle growth.2 BMP15 is also expressed in the oocytes and is thought to be involved in oocyte maturation and follicular development.2 Expression of AMH, an early follicle growth inhibitor, is normally low in primordial and primary follicles, increases to maximum levels in large preantral follicles and small antralstages, and then declines during final follicular maturation.3 AMH appears to be the only natural ligand of AMHR2; AMH and AMHR2 null mice both have the same phenotype.4
The current genetic study was inspired by studies that described dysregulation of several of these TGF-β proteins in the follicles of patients with PCOS. The first such study observed a decreased level of GDF9 mRNA in developing PCOS and PCO oocytes compared with normal, with no differences in BMP15 expression.5 Another study found reduced AMH expression during the initial stages of folliculogenesis in PCOS.3 Others found that AMH levels in follicular fluid and granulosa cell-conditioned media were higher for PCO compared to normal controls.6 Lastly, AMHR2 mRNA levels in granulosa cells of small follicles were elevated in PCOS.7 While the clinical implications of these abnormalities are not completely understood, this paracrine dysregulation of growth factors in PCOS can potentially alter the intrafollicular environment, disturb granulosa cell interactions, and impair cytoplasmic and/or nuclear maturation of the oocyte.3,6
Given the critical role of disordered folliculogenesis in PCOS, our goal in this study was to examine the genes for GDF9, BMP15, AMH and AMHR2 as candidate genes for PCOS and its component traits.
Methods
Subjects and Phenotyping
We studied 335 unrelated White PCOS patients and 198 White control women recruited at two centers, the University of Alabama at Birmingham (UAB; 287 PCOS and 187 controls) and Cedars-Sinai Medical Center (CSMC; 48 PCOS and 11 controls). Participation in research studies was offered to patients meeting inclusion criteria (premenopausal, non-pregnant, on no hormonal therapy for at least three months, and meeting diagnostic criteria for PCOS). Presence of PCOS was defined by the 1990 NIH consensus criteria.8 The specific parameters for defining hirsutism, hyperandrogenemia, ovulatory dysfunction and exclusion of related disorders were previously reported.1
Controls were healthy women, with regular menstrual cycles, and without a family history of hirsutism. These women had no evidence of hirsutism, acne, or alopecia, or endocrine dysfunction and had not taken hormonal therapy (including oral contraceptives) for at least 3 months prior to testing. Controls were recruited by word of mouth and advertisements, through a call for “healthy women” without detailing further the nature of the studies.
Subjects underwent a physical examination and hormone measurements, per a previously described protocol.1 In addition, we evaluated whether PCOS patients had severe oligomenorrhea (defined as > 3 months between menses), infertility (yes/no), and parity (yes/no). Clinical characteristics of the study cohort are given in Supporting Information S1.
All subjects gave written informed consent, and the study was performed according to the guidelines of the Institutional Review Boards of UAB and CSMC.
Genetic Analysis
We selected five GDF9 single nucleotide polymorphisms (SNPs) (rs10491279, rs254285, rs11739194, rs39830, rs30178), two BMP15 SNPs (rs3810682, rs3897937), three AMH SNPs (rs4807216, rs10407022, rs3746158), and three AMHR2 SNPs (rs11170547, rs2002555, rs11170553) using genotype data of the Caucasian population of the HapMap database (release 24, phase II, www.hapmap.org). These SNPs were selected because they are predicted to tag SNPs across the entirety of each gene, plus 5kb upstream and downstream. These SNPs capture 34 of 41 (83%) of the Caucasian HapMap SNP alleles at r2>0.8 for all four genes. The thirteen SNPs were genotyped using Applied Biosystems Taqman Assays on Demand according to manufacturer’s instructions.
The program Haploview v 4.1 (Broad Institute, Cambridge MA, USA) was used to calculate linkage disequilibrium (LD, the D′ statistic) between each pairwise combination of SNPs and determine haplotypes and their frequencies.9 The solid spine of LD algorithm in Haploview was used to determine haplotype blocks. Only subjects whose haplotype assignment was >95% certain were used in analyses.
Statistical Analysis
Unpaired T-tests and chi-square tests were used to compare clinical characteristics between women with and without PCOS; quantitative trait values were log- or square root-transformed as appropriate to reduce non-normality.
Data are expressed as the median (interquartile range). Association of SNPs or haplotypes with presence/absence of PCOS and qualitative traits was evaluated using logistic regression, adjusting for recruitment site, age and body mass index (BMI). Within the PCOS women, association with quantitative phenotypic traits was evaluated using analysis of covariance (ANCOVA), again adjusting for recruitment site, age and BMI in all analyses except those in which BMI was the dependent variable, wherein analyses were adjusted for recruitment site and age only. To handle multiple testing, significance of each analysis was taken as P<0.003 (=0.05/16), considering that we analyzed four linkage disequilibrium groups of SNPs against four families of traits (PCOS diagnosis, androgens, metabolic traits, reproductive endpoints), yielding a Bonferroni correction factor of 16 (i.e. 16 independent comparisons).
The sample size of 335 cases and 198 controls has excellent power (≥90%) to detect association of risk alleles of frequency ≥0.2 with PCOS at odds ratio ≥1.75, and has fair-to-good power (40–80%) to detect association at odds ratio 1.5. Detailed power calculations given in Supporting Information S2 reveal lower power to detect association of rare risk alleles (frequency ≤0.1) with PCOS at odds ratios less than 1.75.
Results
GDF9
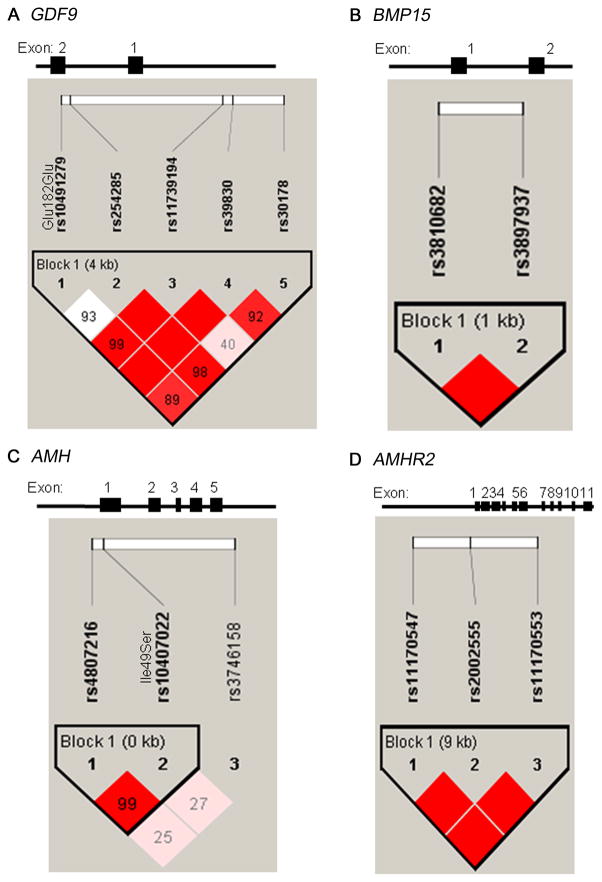
We genotyped five SNPs spanning GDF9 (Fig. 1A). SNP frequencies are shown in Supporting Information S3. All markers were in Hardy-Weinberg equilibrium. Linkage disequilibrium among these markers (D′) ranged from 0.40 to 1.0 (average pairwise D′ of 0.90). The common haplotypes are displayed in Supporting Information S4.
Figure 1.
Gene structure and linkage disequilibrium plot for A. GDF9, B. BMP15, C. AMH, and D. AMHR2. Gene structure is shown at top; GDF9 has 2 exons, is located on chromosome 5q31.1, and is 5.46 kb in size. BMP15 has 2 exons, is located on chromosome Xp11.2, and is 5.91 kb in size. AMH has 5 exons, is located on chromosome 19p13.3, and is 2.95 kb in size. AMHR2 has 11 exons, is located on chromosome 12q13.13, and is 7.67 kb in size. The locations of the genotyped SNPs relative to the exons are indicated. The linkage disequilibrium plot beneath each gene displays D′ values (%) for each pair of SNPs in the box at the intersection of the diagonals from each SNP. The solid blocks indicate D′ = 1 (100%) for the corresponding pair of variants with a logarithm of the odds (LOD) score of ≥2. For each gene, the SNPs were considered together in haplotype blocks as indicated.
GDF9 SNPs and haplotypes were not associated with PCOS susceptibility. Among women with PCOS, minor allele carriers of SNPs rs11739194 or rs30178 had significantly higher modified Ferriman-Gallwey (mFG) hirsutism scores than non-carriers with values of 8.0 (6.5) for carriers versus 7.0 (5.0) for non-carriers of rs11739194 and values of 8.0 (5.0) in carriers versus 7.0 (6.0) for non-carriers of rs30178 (P=0.001 and 0.001, respectively). In PCOS women, haplotype 3 carriers had significantly increased mFG scores compared to non-carriers with a score of 8.0 (6.0) versus 7.0 (6.0) (P=0.0002). This haplotype result was consistent with the SNP results in that this haplotype carries the minor alleles of both SNPs that displayed similar trait associations. Additionally, in PCOS patients, minor allele carries of SNP rs254285 had reduced parity (OR=0.19, 95% CI=0.06–0.57, P=0.003).
BMP15, AMH and AMHR2
The genetic structure and linkage disequilibrium for BMP15, AMH, and AMHR2 are displayed in Fig. 1B, 1C, and 1D. SNP frequencies (all in Hardy-Weinberg equilibrium) are shown in Supporting Information S3. The common haplotypes are displayed in Supporting Information S4. We did not observe any genotype-phenotype associations between BMP15, AMH or AMHR2 variants and PCOS or any of the qualitative or quantitative traits.
Discussion and Conclusion
We hypothesized that polymorphisms in genes involved in folliculogenesis would influence susceptibility to PCOS, but found no association of four key genes, GDF9, BMP15, AMH, and AMHR2, with PCOS susceptibility.
While we are not the first to conduct genetic association studies of BMP15 or AMH in PCOS, we are the first, to our knowledge, to evaluate GDF9 and AMHR2. In addition to the molecular studies evaluating the TGF-β members in PCOS, there have been several genetic studies that yielded conflicting results. In one study, two polymorphisms in BMP15 were associated with anovulation and infertility in PCOS.10 In contrast, we did not find that polymorphisms in BMP15 were associated with severe oligomenorrhea or infertility. Others found association of the AMH Ile49Ser variant (rs10407022) with reduced frequency of PCO and lower follicle numbers in PCOS women, concluding that genetic variation in AMH may contribute to abnormal ovarian morphology.11 We could not replicate their findings because ultrasounds were not routinely performed in our subjects. However, our data does suggest that the potential association of AMH genetic variation with ovarian morphology seen previously does not translate into a significant effect on reproductive function as evaluated by clinical reproductive endpoints. Lastly, gene sequencing in 38 women with PCOS found no missense mutation in any exons of GDF9 or BMP15.12
We found that variants in GDF9 were associated with hirsutism scores, potentially as a result of increased theca cell growth in the ovary leading to elevated androgens. Even though we did not see an association between variants in GDF9 and the serum androgen levels, androgens in women are difficult to measure, possibly obscuring this association. We also found that a variant in GDF9 was associated with reduced parity. Association of genetic variants with parity may be confounded by factors such as the relatively young age of our patients (median 27 yr) and unavailable information such as number of partners or who had attempted to conceive. Nevertheless, we speculate that variants in GDF9 may contribute to compromised oocyte development, leading to outcomes such as spontaneous abortion and consequently lower parity. With our current data, we cannot be sure of the functional role of these GDF9 variants or the broader significance of these findings.
The value of our study is that it reports on a group of genes, TGF-β superfamily members involved in folliculogenesis, which have not been extensively evaluated in PCOS. Our data demonstrate that polymorphisms in major folliculogenesis genes, GDF9, BMP15, AMH, and AMHR2, are not associated with PCOS susceptibility. While these data suggest that inherited defects in folliculogenesis are not major contributors to the development of PCOS, examination of other genes in this superfamily would be needed to fully support this conclusion.
Supplementary Material
Acknowledgments
Funding:
This study was supported in part by NIH grants R01-HD29364 and K24-HD01346 (to RA), R01-DK79888 (to M.O.G.), and M01-RR00425 (General Clinical Research Center Grant from the NCRR), the Cedars-Sinai Winnick Clinical Scholars Award (to M.O.G) and an endowment from the Helping Hand of Los Angeles, Inc.
Footnotes
Disclosure of Interests:
There are no conflicts of interest related to the content of this work.
Contribution to Authorship:
K. Sproul contributed to acquisition, analysis and interpretation of data, and drafted the article. M.R. Jones contributed to acquisition and interpretation of data, critical revision of the article. R. Mathur contributed to acquisition of data, critical revision of the article. R. Azziz contributed to acquisition and interpretation of data, critical revision of the article. M.O. Goodarzi contributed to conception and design, analysis and interpretation of data, critical revision of the article.
Details of Ethics Approval:
This study was performed according to the guidelines of the Institutional Review Boards of UAB and CSMC, under protocol numbers 6013 (approved 9/14/04) and 4190 (approved 1/22/04).
References
- 1.Azziz R, Woods KS, Reyna R, Key TJ, Knochenhauer ES, Yildiz BO. The prevalence and features of the polycystic ovary syndrome in an unselected population. J Clin Endocrinol Metab. 2004;89:2745–9. doi: 10.1210/jc.2003-032046. [DOI] [PubMed] [Google Scholar]
- 2.Yan C, Wang P, DeMayo J, DeMayo FJ, Elvin JA, Carino C, et al. Synergistic roles of bone morphogenetic protein 15 and growth differentiation factor 9 in ovarian function. Mol Endocrinol. 2001;15:854–66. doi: 10.1210/mend.15.6.0662. [DOI] [PubMed] [Google Scholar]
- 3.Stubbs SA, Hardy K, Da Silva-Buttkus P, Stark J, Webber LJ, Flanagan AM, et al. Anti-mullerian hormone protein expression is reduced during the initial stages of follicle development in human polycystic ovaries. J Clin Endocrinol Metab. 2005;90:5536–43. doi: 10.1210/jc.2005-0907. [DOI] [PubMed] [Google Scholar]
- 4.Mishina Y, Rey R, Finegold MJ, Matzuk MM, Josso N, Cate RL, et al. Genetic analysis of the Mullerian-inhibiting substance signal transduction pathway in mammalian sexual differentiation. Genes Dev. 1996;10:2577–87. doi: 10.1101/gad.10.20.2577. [DOI] [PubMed] [Google Scholar]
- 5.Teixeira Filho FL, Baracat EC, Lee TH, Suh CS, Matsui M, Chang RJ, et al. Aberrant expression of growth differentiation factor-9 in oocytes of women with polycystic ovary syndrome. J Clin Endocrinol Metab. 2002;87:1337–44. doi: 10.1210/jcem.87.3.8316. [DOI] [PubMed] [Google Scholar]
- 6.Pellatt L, Hanna L, Brincat M, Galea R, Brain H, Whitehead S, et al. Granulosa cell production of anti-Mullerian hormone is increased in polycystic ovaries. J Clin Endocrinol Metab. 2007;92:240–5. doi: 10.1210/jc.2006-1582. [DOI] [PubMed] [Google Scholar]
- 7.Catteau-Jonard S, Jamin SP, Leclerc A, Gonzales J, Dewailly D, di Clemente N. Anti-Mullerian hormone, its receptor, FSH receptor, and androgen receptor genes are overexpressed by granulosa cells from stimulated follicles in women with polycystic ovary syndrome. J Clin Endocrinol Metab. 2008;93:4456–61. doi: 10.1210/jc.2008-1231. [DOI] [PubMed] [Google Scholar]
- 8.Zawadzki JK, Dunaif A. Diagnostic criteria for polycystic ovary syndrome: towards a rational approach. In: Dunaif AJG, Haseltine F, Merriam GR, editors. Polycystic Ovary Syndrome. Boston: Blackwell Scientific; 1992. pp. 377–84. [Google Scholar]
- 9.Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21:263–5. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- 10.Gonzalez A, Ramirez-Lorca R, Calatayud C, Mendoza N, Ruiz A, Saez ME, et al. Association of genetic markers within the BMP15 gene with anovulation and infertility in women with polycystic ovary syndrome. Fertil Steril. 2008;90:447–9. doi: 10.1016/j.fertnstert.2007.06.083. [DOI] [PubMed] [Google Scholar]
- 11.Kevenaar ME, Laven JS, Fong SL, Uitterlinden AG, de Jong FH, Themmen AP, et al. A functional anti-mullerian hormone gene polymorphism is associated with follicle number and androgen levels in polycystic ovary syndrome patients. J Clin Endocrinol Metab. 2008;93:1310–6. doi: 10.1210/jc.2007-2205. [DOI] [PubMed] [Google Scholar]
- 12.Takebayashi K, Takakura K, Wang H, Kimura F, Kasahara K, Noda Y. Mutation analysis of the growth differentiation factor-9 and -9B genes in patients with premature ovarian failure and polycystic ovary syndrome. Fertil Steril. 2000;74:976–9. doi: 10.1016/s0015-0282(00)01539-9. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.