Abstract
NMR spectroscopists are hindered by the lack of standardization for spectral data among the file formats for various NMR data processing tools. This lack of standardization is cumbersome as researchers must perform their own file conversion in order to switch between processing tools and also restricts the combination of tools employed if no conversion option is available. The CONNJUR Spectrum Translator introduces a new, extensible architecture for spectrum translation and introduces two key algorithmic improvements. This first is translation of NMR spectral data (time and frequency domain) to a single in-memory data model to allow addition of new file formats with two converter modules, a reader and a writer, instead of writing a separate converter to each existing format. Secondly, the use of layout descriptors allows a single fid data translation engine to be used for all formats. For the end user, sophisticated metadata readers allow conversion of the majority of files with minimum user configuration. The open source code is freely available at http://connjur.sourceforge.net for inspection and extension.
Electronic supplementary material
The online version of this article (doi:10.1007/s10858-011-9497-1) contains supplementary material, which is available to authorized users.
Keywords: FID, Spectrum, Software, Conversion, CONNJUR, Reconstruction
Introduction
Over the past several decades NMR has proven itself to be a powerful and versatile tool for measuring many biophysical aspects of bio-molecules. It is an accepted technique for determining the three-dimensional structure of macromolecules and macromolecular complexes in solution (Williamson et al. 1985; Clore and Gronenborn 1998; Wuthrich 2003). It is also often relied on for measuring the internal dynamics of molecules, from picosecond to nanosecond motions by monitoring 15N relaxation (Barbato et al. 1992; Palmer 2004), microsecond to millisecond motions by measuring relaxation dispersion (Korzhnev et al. 2004; Neudecker et al. 2009), and millisecond-timescale and longer motions through observations of hydrogen exchange (Englander and Mayne 1992; Gryk et al. 1995) or other direct measurements made in real time. In the absence of a high-resolution structure, chemical shifts and chemical shift changes have been used for determining secondary (Wishart et al. 1992; Cornilescu et al. 1999) and tertiary structural elements (Cavalli et al. 2007; Shen et al. 2008) as well as the binding interfaces for ligand binding and macromolecular interactions (Shuker et al. 1996; Gryk et al. 2002). In addition, measurement of chemical shift changes upon titration of ligands or pH have proven effective for measuring binding constants and local pKa’s in solution (Malthouse 1999).
In contrast to these diverse applications of NMR, the primary measurement recorded on the spectrometer is essentially the same for all experiments—that of the precessing nuclear spin magnetization within the probe coil, termed the free induction decay (FID) (Bloch 1946). The diversity of information obtainable from such a uniform measurement type is gained through the large number of ways in which the physical sample can be perturbed, as well as those of the nuclear spin populations which exist within an otherwise static sample (Cavanagh et al. 1996). This diversity of information content comes at a cost; however, in that even though the primary data recording is essentially the same regardless of the design of the experiment, both the acquisition and processing paradigms for the data differ depending on the goal of the individual or set of experiments. Thus, the diversity in information content is achieved by adding complexity in the experimental setup and design, as well as a concomitant complexity in subsequent data analysis.
It is perhaps not surprising that with such a diversity in NMR applications, there has also grown a diversity in NMR data processing tools, spanning the arenas of spectral reconstruction (converting time domain data to the frequency domain), spectral analysis (peak identification, assignment and characterization), and biophysical characterization (converting NMR-specific measurements to biophysical insights) (Ellis et al. 2006b). These tools differ widely in their architecture, their underlying algorithms, the operating systems under which they perform and their internal and external data formats. It is not the case that one software tool is optimal for all given tasks and the others inferior. Rather, each of the various tools has pros and cons depending on the processing task to be performed as well as other competing concerns (speed, accuracy, cost of use, etc.) Thus, it is desirable to the NMR community that the diversity of tools be maintained and not eliminated. Unfortunately, the diversity in design and use of the tools, and principally of their respective file formats, creates an obstacle to tool interoperability.
The goal of the CONNJUR project is to alleviate the strain on the NMR user by increasing the interoperability of existing NMR processing tools (http://www.connjur.org). The overall strategy is (a) to provide a common data repository for all types of NMR data required and provided by the various third-party software tools; (b) provide software wrappers for the existing tools so they can make use of the common data repository; and (c) provide a user interface from which to drive all NMR processing tasks using any subset of existing NMR tools. Portions of the common data model under development have been published previously (Fox-Erlich et al. 2004; Ellis et al. 2006a; Gryk et al. 2010), and prototypes of the CONNJUR software integration environment have been demonstrated at recent Experimental NMR Conferences (http://www.enc-conference.org) and the 2008 NANUC NMR Bootcamp (http://www.nanuc.ca/resources/workshop2008.php). A similar but distinct effort has been undertaken by the Collaborative Computing Project for NMR (CCPN) (Fogh et al. 2002; Vranken et al. 2005). While sharing a similar goal of data integration, recent efforts of CCPN have been to redesign a powerful spectral analysis tool called CcpNmr Analysis. To date, CCPN does not support spectrum translation; however, CCPN does allow spectral data to be imported into the CCPN data model.
While detailing the functional requirements of CONNJUR, it was recognized that the CONNJUR integration platform would require the functionality to interconvert data between the different file formats utilized by the third party tools we intend to wrap. The requirements for such a universal spectrum translator are fourfold: (1) the spectrum translator should be able to translate between any two arbitrary file formats, (as within the context of CONNJUR, no directionality of the processing pipeline should be assumed), (2) the translator should use a common data model such that the data and metadata provide a portion of the fundamental data model used by CONNJUR, (3) the spectrum translator must be easily extensible to support the addition of new file formats as new tools emerge and additional tools are wrapped by CONNJUR, and (4), as CONNJUR is intended to be an open source project, the spectrum translator should also be open source.
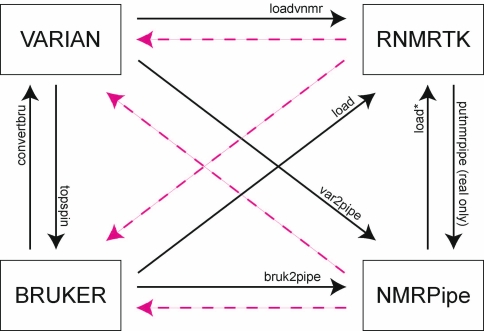
It became clear that such a spectrum translator would be of general use to the NMR community as a stand-alone tool, usable apart from the CONNJUR project. As shown in Fig. 1, while various third-party tools already exist for performing single-step format conversions, there is no single tool which supports the interconversion between all file formats. Also, some translation paths are not supported at all (magenta arrows in Fig. 1) while in other cases the existing conversion tools require multiple steps and significant configuration effort. For example, converting certain four dimension experiments from Varian to NMRPipe requires first running a script to reorder phases (Delaglio 2007), then determining the necessary metadata in order to execute the var2pipe utility, and finally executing the utility. Releasing the spectrum translator as a stand-alone tool added two more requirements to the design: (1) the spectrum translator should have a direct user-interface, and (2) the spectrum translator should provide direct interaction with files.
Fig. 1.
Schematic of conversion capabilities of existing and potential spectrum translator tools. The overall NMR processing workflow goes from left to right. Data is typically collected on either Varian or Bruker spectrometers. Spectral reconstruction can be done using the spectrometer software, or it can be processed using NMRPipe or the Rowland NMR Toolkit (RNMRTK). Translations for which existing tools exist are shown as solid black arrows. Bruker to Varian can be accomplished with the “convertbru” command in VNMR. Varian to Bruker can be accomplished within the Bruker Topspin software. Conversions to RNMRTK can be done using either the “loadvnmr” command for Varian data, or the generic “load” command for other data types. Configuring the load function for NMRPipe data is non-trivial. Conversions from Varian and Bruker to NMRPipe can be done with the ancillary programs “var2pipe” and “bruk2pipe”, respectively. RNMRTK supports export in NMRPipe format, but only for data which is REAL-only. The dashed magenta arrows represent translations for which no existing, third-party tool existed prior to the CONNJUR Spectrum Translator
The CONNJUR Spectrum Translator (CONNJUR-ST) represents our first official software release. CONNJUR-ST tackles a small but critical component of the overall NMR software integration problem—interconversion of NMR spectral data between the various data formats used by the various software processing tools. The current release supports six, vastly different data formats, four used by the spectrometer vendors, Varian (http://www.varianinc.com/) and Bruker (http://www.bruker-biospin.com/), as well as two NIH-supported spectral reconstruction suites, NMRPipe (Delaglio et al. 1995) and the Rowland NMR Toolkit (Hoch and Stern 2010). It is hoped that the establishment of CONNJUR-ST as an open source project will provide both the incentive and opportunity for the addition of conversion functionality for other useful time-domain data formats. The modular design of CONNJUR-ST should assist in such support of additional file formats.
Materials and methods
CONNJUR-ST (version 1.1) is written in Java and requires that the user has preinstalled the Java Virtual Machine (JVM v. 1.5) (http://www.Java.com) in order to execute. The JVM is multiplatform and typically comes preinstalled with most desktop computers. CONNJUR-ST has been developed using Eclipse (http://www.eclipse.org) as an integration development environment. A source code control system (Concurrent Versioning System, or CVS) (http://www.nongnu.org/cvs/) is used to track changes to the software and allow changes to be applied in a controlled manner. In the event testing finds a change has introduced errors, previous versions may be restored. A fully automated full extraction and build occurs daily. Regression test conversions against both existing tools and previous CONNJUR-ST versions are repeated to ensure data fidelity.
Validation of CONNJUR-ST conversions
CONNJUR-ST conversion was compared against existing conversion tools. Output was compared using one of three methods. First, visual examination of the converted spectra in display tools such as NmrDraw (Delaglio et al. 1995) was performed. Second, portions of output files containing NMR data were compared on a binary basis. (Some embedded non-NMR data, such as internal timestamps, vary on a binary basis. Therefore, a complete binary comparison as done with the Unix command ‘diff’ is not sufficient for testing.) Third, CONNJUR-ST has been alpha-tested by members of the CONNJUR team as well as collaborators at the UCONN Health Center.
File format documentation
Varian data file format information was primarily obtained from “VNMR User Programming” (Varian 1998). Kirk Marat provided supplementary information regarding use of binary bits (personal communication). Nmrpipe format was obtained from the “fdatap.html” and “fdatap.h” files distribution with NMRPipe (Delaglio et al. 1995). Rowland Toolkit information was found in the online manual (Hoch and Stern 2010). Bruker file information was obtained from “XWIN-NMR Software Manual” (Bruker 1995).
Test datasets
Initial datasets used included spectrum previously collected by the principal investigator, Dr. Michael Gryk, for Varian datasets and data collected at the University of Connecticut chemistry department for Bruker data. Additionally, a set of spectra of small file size was collected with various combinations of phasing and real/complex data. While the small datasets do not provide spectra usable for analysis they provide excellent test cases for possible variations in file formats. Synthetic datasets generated by the code allow tracking of specific values from data file format to another.
Results
The CONNJUR Spectrum Translator is available for download at http://www.connjur.org. The software and documentation, including assumptions, configuration parameter descriptions and quickstart instructions, as well as all test data can be found in the same location. The source code and manifesto for open source contributions will be released shortly following this publication. The CONNJUR spectral object is documented using standard Javadoc technology (http://www.Java.com). All Javadocs on the CONNJUR spectral object will be accessible through the CONNJUR website.
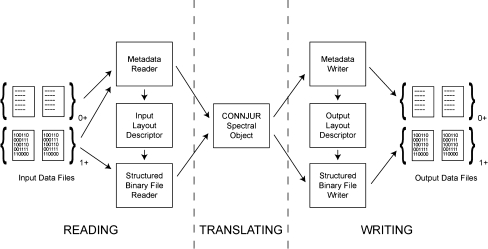
The overall design for the spectrum translator is to split the process of translation into three smaller tasks (Fig. 2). One, a common data model for NMR spectra was developed—in order to both facilitate the spectrum translator as well as for use within the greater CONNJUR initiative. Two, input modules are responsible for reading third party formats and translating them into the CONNJUR model. Three, output modules are responsible for translating data from the CONNJUR model and writing the data to the third party formats.
Fig. 2.
Process diagram for the CONNJUR Spectrum Translator. The overall process can be considered as occurring through three general phases: (1) Reading the input files, (2) Translating the spectrum into CONNJUR format, and (3) Writing the output files. As shown in the diagram, the input stage is further decomposed into three tasks: (i) Reading the input metadata, (ii) Creating a layout descriptor from the available metadata and knowledge of the input format, (iii) Reading the binary data with use of the input layout descriptor*. The output stage is broken down into three similar tasks: (i) Creating a layout descriptor* from the available metadata and knowledge of the output format, (ii) Writing the binary data with use of the output layout descriptor, and (iii) Writing the output meta data. The central portion retains the spectrum data in a CONNJUR format, within which semantic operations can be performed to the data such as those required when collecting spectra using Rance-Kay sensitivity enhancement. It is presupposed in the design of CONNJUR-ST that the input/output files consist of one or more binary file along with any number of optional ASCII files containing metadata. (See “Results” section for a discussion of the input/output layout descriptors.)
A major benefit of this modular approach is in the support of multiple, third-party file formats. If one simply supported one-to-one file converters, the number of converters necessary would be n(n−1), where n is the number of supported file formats. With our approach, however, the total number of translators required is only 2n as all translation is mediated through a common model, decoupling the input format from the output format. In addition, as existing file formats are altered, software bugs detected, or new features added, the maintenance cost amounts to only two converters needing to be updated rather than 2(n−1). For this release of CONNJUR-ST, this amounts to a twofold savings in implementation and a fourfold savings in support. This benefit will increase drastically as additional file formats are supported by CONNJUR-ST.
It is worth noting that this same computational savings would be achieved if the NMR community were to settle on any one file format as a standard, with a suite of importers and exporters built around that standard format. (See for example the solution offered by Olivia which offers a suite of importers/exporters (http://fermi.pharm.hokudai.ac.jp/olivia/api/index.php/Xyza2pipe_src) for the nmrPipe file format.) It is our estimation that each of the file formats supported by CONNJUR-ST has sufficient pros and cons associated with it that none would be ideal for such a standard. It is also true, that as a matter of computational efficiency, all translation should be done in memory, not through writing to disk, further discouraging such an approach.
An additional benefit arose in the CONNJUR-ST design, inspired by the manner in which the Rowland NMR Toolkit reads various third-party file formats. As there are a limited number of simple ways in which arrays of numbers may be ordered, the reading and writing portions of the translator are further broken down into three subtasks: reading (writing) metadata, reading (writing) data, and interpreting the binary data in terms of a layout descriptor. The layout descriptor—unique for each file format—dictates the ordering of the numbers in the binary file and can be thought of as a correspondence between the metadata in the file and the layout of the file as specified for each third party tool. Therefore, the maintenance tasks are simplified to the two main tasks of handling metadata and handling the layout descriptor—a vast savings in effort from supporting a stand-alone format converter for a given third-party format.
Model for time-domain NMR spectra
The NMR data model consists of the following three key components: (a) a set of attributes (metadata) related to the spectrum as a whole including the N dimensions for the experiment, where N is a positive integer, (b) a set of metadata related to each dimension of the experiment, and (c) the N-dimensional matrix of complex data points, representing the actual data collected in the experiment (that is, the magnetic induction measured within the spectrometer probe coil).
The core metadata for each dimension consist of the sweep width (which is inversely proportional to the spacing between collected time increments), the total number of points (either real or complex1 depending on data collection), and a Boolean descriptor as to whether the data are real or complex. A Boolean flag defining time domain versus frequency domain is also added for future support of frequency domain spectral data.
The model for the physical arrangement of NMR files includes indications of the data encoding: float versus integer, 32-bit versus16-bit, and little endian versus big endian, and the ordering of the complex data points (stored as real-imaginary pairs versus all reals followed by all imaginaries). The numerical, spectral data are assumed to be contained in one or more binary file, each binary file having optional headers, footers, and block headers. The metadata is assumed to be contained either within the binary data file, or within optional ASCII parameter files. The ordering of the data points within the file, and within multiple files, as well as the formatting of metadata in the parameter files are all included in our layout model.
The current implementation of CONNJUR-ST expects the numerical spectral data to be stored in binary, as is the case for each of the five file formats currently supported. If a third-party tool represents the data in ASCII rather than binary, the nature of the character encoding and numerical delimiters could be included as metadata to support such file formats. CONNJUR-ST does currently support output to ASCII, but has henceforth only been used for debugging purposes. It should also be noted that CONNJUR-ST expects the spectral data to be stored as files on the file system. However, as used within the CONNJUR project, CONNJUR-ST is also designed to accept input/output streams as well as files. This release of CONNJUR-ST (v 1.1) supports input/output data streams for the NMRPipe format—the only supported format consisting of a single, binary file.
Features
CONNJUR-ST converts time and frequency domain, spectral data from one file format to another while requiring minimal user input. Secondarily it provides semantic transformation operations on the data, including negating imaginary values on some or all dimensions, performing the Rance-Kay (Echo/Anti-Echo) sensitivity enhancement (Kay et al. 1992) correction, and removing leading data introduced by the Bruker digital filtering. Additionally it allows some metadata manipulation, e.g. data set comments and dimension labels.
CONNJUR-ST currently supports translation between the following five data formats for NMR spectral data with any arbitrary combination of quadrature settings:
Bruker acquisition format (FID for 1D, SER for higher dimensions);
Bruker processing format (ex., rr file);
NMRPipe: both the plane and cube representation;
Rowland NMR Toolkit standard layout only. RNMRTK is capable of reading and writing a limitless number of representations. By necessity, our translator is limited to reading/writing only the standard, default representation used by RNMRTK;
Varian acquisition format. The Varian format requires complex data in the first incremented dimension. Otherwise, arbitrary quadrature settings are supported.
Varian processed format (limited support). Varian software is not designed with the intention that frequency-domain should be stored or loaded from disk (George Gray, personal communication). Rather, the software expects the user to store/load only the time-domain data, and reprocess using VNMRJ if necessary. That said, many NMR users have learned to extract and use frequency-domain data, stored as the “phasefile” within the experiment directory. CONNJUR provides limited support for this type of data, allowing translation from the phasefile, but not to the phasefile (i.e. allowing import but not export). It should be noted that the data stored within the phasefile is not guaranteed to be complete, and so this type of translation should be done with caution.
In order to ensure that CONNJUR-ST is capable of handling all operations with minimal user input, it has been designed to self-configure based on some assumptions about the input and output layouts and metadata. These assumptions vary in confidence from the likely conjecture that the input metadata regarding sweep widths is correct to less certain assumptions such as that the input metadata regarding PPM referencing is correct. There are some configurations for which the translator makes no assumptions such as the output file format desired or the semantic conversions which the user may wish to implement.
The business logic that underlies the ease-of-use assumptions can be overridden by the user in cases where the user knows the metadata values and wishes to set them directly. For these cases, the user may either set metadata and configuration settings through command line flags, or may configure an XML file to be read by the spectrum translator. The latter is desirable for cases in which many different pieces of metadata need to be under user control, or for automated direction of the CONNJUR-ST either through external programs or through batch-processing scripts.
Some flags control multiple metadata settings. In the case of the Varian file format, there are groups of metadata associated with acquisition dimensions of the experiment (e.g., number of points, sweep width), while other groups are associated with amplifier channels on the spectrometer (e.g., amplifier frequency, observation nucleus). While the association between channels and dimensions may be ambiguous, all channel-specific parameters are expected to be associated with the same dimension. Thus, the user can specify a channel-dimension link, in which case all parameters associated with a specific channel will be linked to the desired dimension. The CONNJUR website provides a dynamic webpage for generating the appropriate XML file given desired channel-dimension associations. A more sophisticated graphical interface to serve this purpose is currently under development.
Limitations
There are two obvious use cases that CONNJUR-ST does not currently support. One is pseudo-multidimensional domain data, the other is non-uniformly spaced data. Both are of importance to the NMR community and will be prioritized developments for future releases.
Discussion and conclusions
CONNJUR-ST represents an important step forward in filling in for the lack of standardization in NMR file formats. This lack of standardization creates a constant hindrance to NMR spectroscopists, who must continually rely on multiple, third-party tools for reformatting data for input into NMR data processing and analysis tools. Some processing choices, such as reprocessing NMRPipe data within VNMR, have not been available because no tool existed for performing the required translation.
CONNJUR-ST currently supports the translation of NMR spectral data. CONNJUR-ST allows conversion to and from six, third-party formats: the two Varian file formats used in VNMR, the two Bruker formats used by TopSpin, the NMRPipe file format, and the standard file format for the Rowland NMR Toolkit (RNMRTK). We believe CONNJUR-ST will be primarily useful for back-converting data (for instance, converting NMRPipe datasets back to Bruker or Varian format for subsequent processing with the spectrometer software), cross-converting data between spectral reconstruction tools such as between NMRPipe and RNMRTK, and for cases in which the traditional translators (var2pipe, RNMRTK’s loader) are too tedious or cumbersome to configure.
CONNJUR-ST is implemented in Java, and therefore is multi-platform irrespective of the platform specificity of the underlying tools whose format it supports. This provides for the use case of translating spectra on computer systems which are incapable of supporting the underlying software tools and their built-in translators. CONNJUR-ST is provided free and open source, and contributions from the NMR and software development communities are encouraged. It is with the hope of such help that we envision CONNJUR-ST eventually supporting translation of all available data formats and additional semantic conversions.
Future enhancements to the CONNJUR-ST will be the addition of other tool file formats such as XEASY, Sparky and nmrViewJ. Other improvements would be the support of non-uniform datasets (datasets in which the time delay between collected datapoints is not uniform) and pseudo-multidimensional datasets—referring to datasets in which some of the dimensions do not represent frequency axes, but rather the variation of other parameters, such as relaxation delays.
Additional use cases will also be explored. Currently, CONNJUR-ST is a command-line driven application for converting individual spectra. CONNJUR-ST can translate multiple spectra if driven by a script, but CONNJUR-ST does not accept multiple spectra as direct input. Providing for this capability will be explored. Also, a GUI is planned to ease the configuration of the advanced XML options used to customize the translation of data. Of course, we will continue to explore how best to utilize CONNJUR-ST within the main CONNJUR framework.
To foster the open source development of CONNJUR-ST, all code will be stored on SourceForge under the project name CONNJUR. The CONNJUR spectral object is documented using standard Javadoc technology (http://www.Java.com) with Javadocs being available through the CONNJUR website. The code is free and open for the NMR community to download, share and use. We intend to provide a manifesto detailing our expectations of how developer contributions will be invited, screened, tested and eventually accepted into the CONNJUR-ST project. While adoption into the CONNJUR-ST project is encouraged, additions/improvements may be distributed independently, as in accordance with the GNU Public License structure which we have adopted for CONNJUR-ST. We encourage community engagement and feedback on all CONNJUR software as well as the project goals and direction.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgments
This research was funded by United States National Institutes of Health grants EB-001496 and GM-083072. The authors wish to thank Drs. Frank Delaglio, Jeffrey Hoch, Mark W. Maciejewski and Alan Stern for useful conversations and assistance with NMRPipe and the Rowland NMR Toolkit file formats. The authors also wish to thank Agilent Technologies and Bruker Biospin for assistance with understanding the Varian and Bruker file formats, with particular thanks to Drs. George Gray and Clemens Anklin.
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
Footnotes
In this context, complex refers to two orthogonal data collection channels—termed quadrature in signal processing. The two orthogonal channels may both be real data collected on independent probe coils, or can be virtual representing alteration of pulse phases during the experiment.
Ronald J. Nowling and Jay Vyas contributed equally to this work.
References
- Bruker (1995) XWIN-NMR software manual. Bruker Analytische Messtechnik GmbH
- Barbato G, Ikura M, Kay LE, Pastor RW, Bax A. Backbone dynamics of calmodulin studied by 15 N relaxation using inverse detected two-dimensional NMR spectroscopy: the central helix is flexible. Biochemistry. 1992;31:5269–5278. doi: 10.1021/bi00138a005. [DOI] [PubMed] [Google Scholar]
- Bloch F. Nuclear induction. Phys Review. 1946;70:460–474. doi: 10.1103/PhysRev.70.460. [DOI] [Google Scholar]
- Cavalli A, Salvatella X, Dobson CM, Vendruscolo M. Protein structure determination from NMR chemical shifts. Proc Natl Acad Sci U S A. 2007;104:9615–9620. doi: 10.1073/pnas.0610313104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavanagh J, Fairbrother WJ, Palmer AG, III, Skelton NJ. Protein NMR spectroscopy: principles and practice. San Diego: Academic Press; 1996. [Google Scholar]
- Clore GM, Gronenborn AM. NMR structure determination of proteins and protein complexes larger than 20 kDa. Curr Opin Chem Biol. 1998;2:564–570. doi: 10.1016/S1367-5931(98)80084-7. [DOI] [PubMed] [Google Scholar]
- Cornilescu G, Delaglio F, Bax A. Protein backbone angle restraints from searching a database for chemical shift and sequence homology. J Biomol NMR. 1999;13:289–302. doi: 10.1023/A:1008392405740. [DOI] [PubMed] [Google Scholar]
- Delaglio F (2007) Help needed processing 4D 13C-HMQC-NOESY-15 N-HSQC. http://tech.groups.yahoo.com/group/nmrpipe/message/1492
- Delaglio F, Grzesiek S, Vuister GW, Zhu G, Pfeifer J, Bax A. NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J Biomol NMR. 1995;6:277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- Ellis HJ, Fox-Erlich S, Martyn TO, Gryk MR (2006a). Development of an integrated framework for protein structure determinations: a logical data model for NMR data analysis. In: Information technology: new generations, 2006. ITNG 2006, IEEE Computer Society, Conference Publishing Services, Las Vegas, NV, pp 613–618. http://ieeexplore.ieee.org/stamp/stamp.jsp?tp=&arnumber=1611558
- Ellis HJC, Sinnot BD, Fox-Erlich S, Gryk MR (2006b) Undergraduate research in bioinformatics—the CONNJUR project. In: Frontiers in education conference, 36th annual, pp. 13–18
- Englander SW, Mayne L. Protein folding studied using hydrogen-exchange labeling and two-dimensional NMR. Annu Rev Biophys Biomol Struct. 1992;21:243–265. doi: 10.1146/annurev.bb.21.060192.001331. [DOI] [PubMed] [Google Scholar]
- Fogh R, Ionides J, Ulrich E, Boucher W, Vranken W, Linge JP, Habeck M, Rieping W, Bhat TN, Westbrook J, et al. The CCPN project: an interim report on a data model for the NMR community. Nat Struct Biol. 2002;9:416–418. doi: 10.1038/nsb0602-416. [DOI] [PubMed] [Google Scholar]
- Fox-Erlich S, Martyn TO, Ellis HJ, Gryk MR. Delineation and analysis of the conceptual data model implied by the “IUPAC Recommendations for Biochemical Nomenclature”. Protein Sci. 2004;13:2559–2563. doi: 10.1110/ps.04810204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gryk MR, Finucane MD, Zheng Z, Jardetzky O. Solution dynamics of the trp repressor: a study of amide proton exchange by T1 relaxation. J Mol Biol. 1995;246:618–627. doi: 10.1016/s0022-2836(05)80111-1. [DOI] [PubMed] [Google Scholar]
- Gryk MR, Marintchev A, Maciejewski MW, Robertson A, Wilson SH, Mullen GP. Mapping of the interaction interface of DNA polymerase beta with XRCC1. Structure. 2002;10:1709–1720. doi: 10.1016/S0969-2126(02)00908-5. [DOI] [PubMed] [Google Scholar]
- Gryk MR, Vyas J, Maciejewski MW. Biomolecular NMR data analysis. Prog Nucl Magn Reson Spectrosc. 2010;56:329–345. doi: 10.1016/j.pnmrs.2010.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoch J, Stern A (2010) The Rowland NMR toolkit. http://rnmrtk.uchc.edu/rnmrtk/RNMRTK.html
- Kay LE, Keifer P, Saarinen T. Pure absorption gradient enhanced heteronuclear single quantum correlation spectroscopy with improved sensitivity. J Am Chem Soc. 1992;114:10663–10665. doi: 10.1021/ja00052a088. [DOI] [Google Scholar]
- Korzhnev DM, Salvatella X, Vendruscolo M, Di Nardo AA, Davidson AR, Dobson CM, Kay LE. Low-populated folding intermediates of Fyn SH3 characterized by relaxation dispersion NMR. Nature. 2004;430:586–590. doi: 10.1038/nature02655. [DOI] [PubMed] [Google Scholar]
- Malthouse JP. Using NMR as a probe of protein structure and function. Biochem Soc Trans. 1999;27:701–713. doi: 10.1042/bst0270701. [DOI] [PubMed] [Google Scholar]
- Neudecker P, Lundstrom P, Kay LE. Relaxation dispersion NMR spectroscopy as a tool for detailed studies of protein folding. Biophys J. 2009;96:2045–2054. doi: 10.1016/j.bpj.2008.12.3907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmer AG., III NMR characterization of the dynamics of biomacromolecules. Chem Rev. 2004;104:3623–3640. doi: 10.1021/cr030413t. [DOI] [PubMed] [Google Scholar]
- Shen Y, Lange O, Delaglio F, Rossi P, Aramini JM, Liu G, Eletsky A, Wu Y, Singarapu KK, Lemak A, et al. Consistent blind protein structure generation from NMR chemical shift data. Proc Natl Acad Sci U S A. 2008;105:4685–4690. doi: 10.1073/pnas.0800256105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shuker SB, Hajduk PJ, Meadows RP, Fesik SW. Discovering high-affinity ligands for proteins: SAR by NMR. Science. 1996;274:1531–1534. doi: 10.1126/science.274.5292.1531. [DOI] [PubMed] [Google Scholar]
- Varian (1998) VNMR user programming VNMR 6.1B software. Varian Associates, Inc.
- Vranken WF, Boucher W, Stevens TJ, Fogh RH, Pajon A, Llinas M, Ulrich EL, Markley JL, Ionides J, Laue ED. The CCPN data model for NMR spectroscopy: development of a software pipeline. Proteins. 2005;59:687–696. doi: 10.1002/prot.20449. [DOI] [PubMed] [Google Scholar]
- Williamson MP, Havel TF, Wuthrich K. Solution conformation of proteinase inhibitor IIA from bull seminal plasma by 1H nuclear magnetic resonance and distance geometry. J Mol Biol. 1985;182:295–315. doi: 10.1016/0022-2836(85)90347-X. [DOI] [PubMed] [Google Scholar]
- Wishart DS, Sykes BD, Richards FM. The chemical shift index: a fast and simple method for the assignment of protein secondary structure through NMR spectroscopy. Biochemistry. 1992;31:1647–1651. doi: 10.1021/bi00121a010. [DOI] [PubMed] [Google Scholar]
- Wuthrich K. NMR studies of structure and function of biological macromolecules (Nobel lecture) J Biomol NMR. 2003;27:13–39. doi: 10.1023/A:1024733922459. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.