Metastases are the most common cause of cancer-related death in patients with solid tumors.[1–4] A considerable body of evidence indicates that tumor cells are shed from a primary tumor mass at the earliest stages of malignant progression[5–7]. These ‘break-away’ circulating tumor cells (CTCs)[8–11] enter the blood stream and travel to different tissues of the body, as a critical route for cancer metastasis. The current gold standard for determining tumor status requires invasive biopsy and subsequent genetic and proteomic analysis of biopsy samples. Alternatively, CTC measurement and analysis can be regarded as a “liquid biopsy” of the tumor, providing insight into tumor biology in the critical window where intervention could actually make a difference. However, detection and characterization of CTCs has been technically challenging due to their extremely low number in the bloodstream. CTCs are often found in the blood of patients with metastatic cancer (only up to hundreds of cells/mL) whereas common blood cells exist in high numbers (>109 cells/mL). Over the past decade, a diverse suite of technologies[8, 12–17] have been evolving to meet the challenge of counting and isolating CTCs from patient blood samples. Many employ different enrichment mechanisms such as immunomagnetic separation based on capture agent-labeled magnetic beads,[8, 16] microfluidics-based technologies[12, 14, 17] that enhance cell-surface contacts, and microfilter devices[13] that isolate CTCs based on size difference. The sensitivity of these emerging technologies, which is critical to their clinical utility for detecting early cancer progression (e.g., tumor invasion of vascular systems), relies on the degree of enrichment of CTCs.
Recently, we discovered that a 3D-nanostructured substrate[18] coated with cancer cell capture agents[19, 20] (i.e., epithelial cell adhesion molecule antibody, anti-EpCAM) exhibits significantly improved cell-capture efficiency due to its enhanced local topographic interactions[21] between the SiNP substrates and nanoscale cellular surface components (e.g., microvilli and filopodia). Such a high affinity cell assay can be employed to recover cancer cells from spiked whole blood samples, in a stationary device setting,[18] with cell-capture efficiency ranging from 40 to 70%. On the basis of this stationary cell-capture assay, we anticipated that further improvement of cell-capture performance can be achieved by increasing cell/substrates contact frequency. By integrating a simple but powerful fluidic handling system, namely a chaotic mixing channel,[22] with a patterned nanostructured substrate, highly efficient CTC capture can be realized by the synergistic effects of enhanced cell/substrate contact frequency as well as affinity. Although there are several microfluidic platforms[12, 14, 17] capable of achieving improved CTC-capture efficiency, the micropillar-based CTC-capture technologies[12, 17] suffer from depth of field issues thus requiring multiple cross-sectional imaging scans to avoid out-of-focus or superimposed images of device-immobilized CTCs due to the vertical depth of the device features. The microfluidic device[14] with an integrated conductivity sensor provides the significant advantage of label-free CTC detection. However, whether the lack of cellular morphology impacts pathologic characterization remains to be determined.
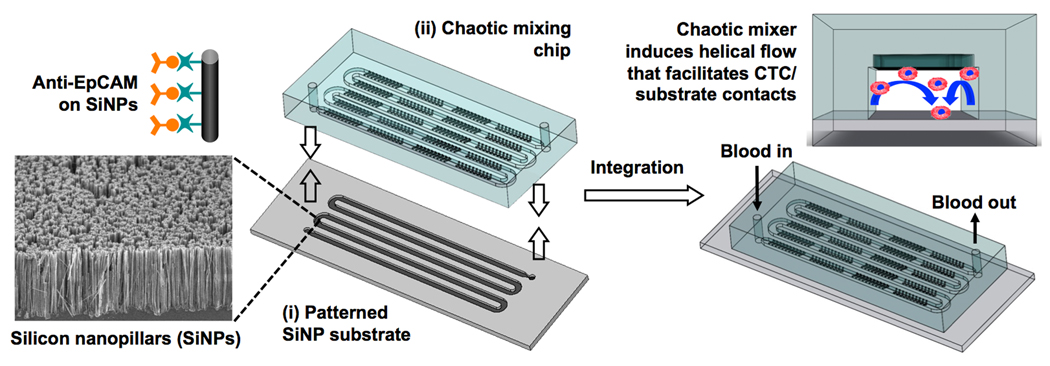
Here we introduce a new CTC-capture platform that integrates two functional components (Figure 1): (i) a patterned SiNP substrate with anti-EpCAM coating for recognizing/capturing EpCAM-expressing cells, and (ii) an overlaid polydimethylsiloxane (PDMS) chip with a serpentine chaotic mixing channel[22–27] that encourages cell/substrate contact frequency. When a blood sample containing CTCs flows through the device, the embedded chevron-shaped micropatterns on the channel roof induce vertical flow (Figure 1) in the microchannel. Consequently, the contact frequency between CTCs and the SiNP substrate increase, resulting in enhanced CTC capture. The performance of this integrated device was first characterized with a cell suspension (100 cells mL−1) of an EpCAM-positive breast cancer cell line (MCF7)[12, 14, 28] in cell culture medium (DMEM) or PBS at flow rates of 0.5–7 mL h−1. An optimal flow rate (1.0 mL h−1) was determined according to the resulting cell-capture efficiency. Finally, the optimal conditions were employed to capture and count CTCs from blood samples collected from prostate cancer patients with different degrees of tumor spread and with different sensitivity to treatments. The results observed by our integrated devices were compared with those observed by CellSearch® assay using immunomagnetic enrichment (the only commercially available approach).[8]
Figure 1.
Schematic representation of the configuration and operational mechanism of an integrated device for capturing circulating tumor cells (CTCs). The device is composed two functional components, (i) a patterned silicon nanopillar (SiNP) substrate with anti-EpCAM-coating exhibiting vastly enhanced CTC-capture affinity, and (ii) an overlaid microfluidic chaotic mixing chip capable of promoting cell/substrate contact frequency. The embedded chevron-shaped micropatterns on the roof of the chaotic mixing chip channel induce vertical flow that facilitates CTC/substrate contact. Thus, highly efficient CTC capture can be achieved by the synergistic effects associated with enhanced CTC/substrate affinity and contact frequency.
The patterned SiNP substrate (Figure 1) was fabricated by combining a lithographic method and a chemical etching process (see Supporting Information). SiNPs of 12 to 15 µm long were chemically etched onto a serpentine pattern defined by photolithography. According to the previously established protocol,[18] streptavidin coating was introduced onto the patterned SiNP substrate using N-hydroxysuccinimide (NHS)/maleimide chemistry[12, 29]. The PDMS chip with an 88-cm long chaotic mixing channel (w × h = 1000 × 100 µm) was produced via soft-lithography using a replicate on a silicon wafer. According to the theoretical model,[22] such a chaotic mixer can induce vertical flow of the blood, leading to significant enhancement in cell/substrate contact frequency compared to a static setting (see Supporting Information). A chip holder made of polyacrylate was designed and fabricated to sandwich the two functional components together. Prior to cell-capture experiments, 100 µL of biotinylated anti-EpCAM (10 µg mL−1, R&D Systems) was introduced onto the integrated device for antibody coating. A digital pressure regulator was utilized to control the flow rates of (i) cell suspensions or blood samples, (ii) fixation and permeabilization agents, and (iii) immuno- and nuclear staining agents introduced sequentially into the integrated devices during the studies.
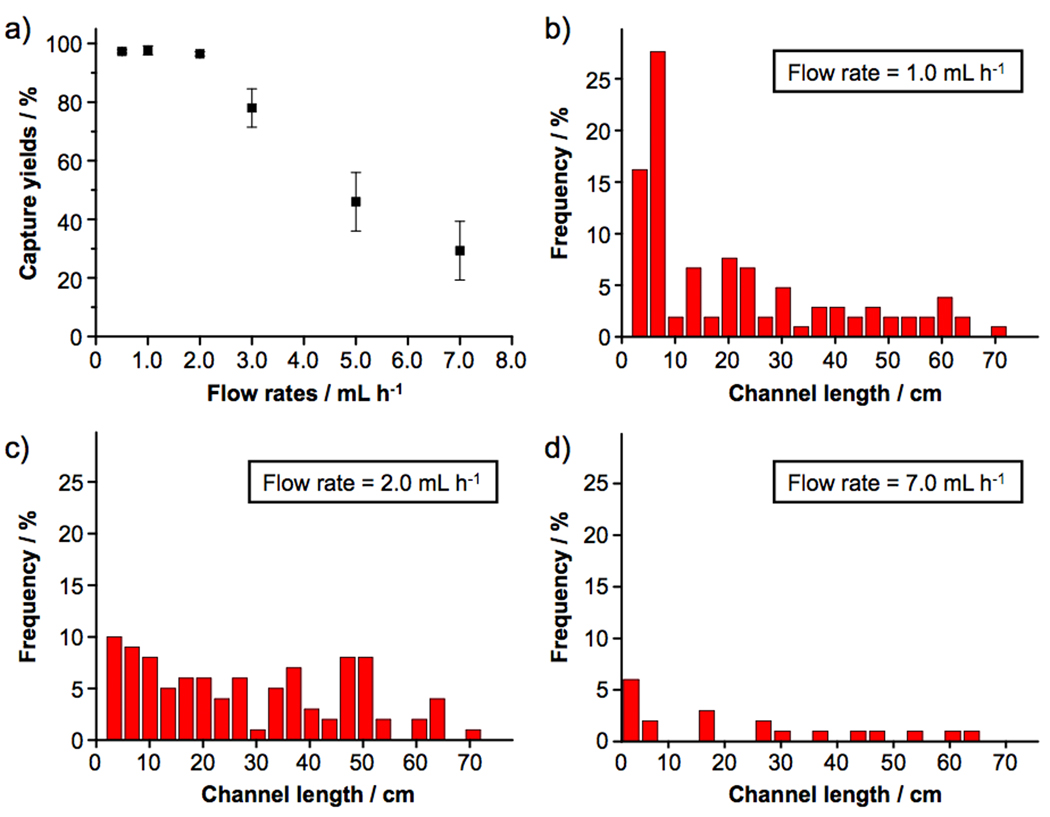
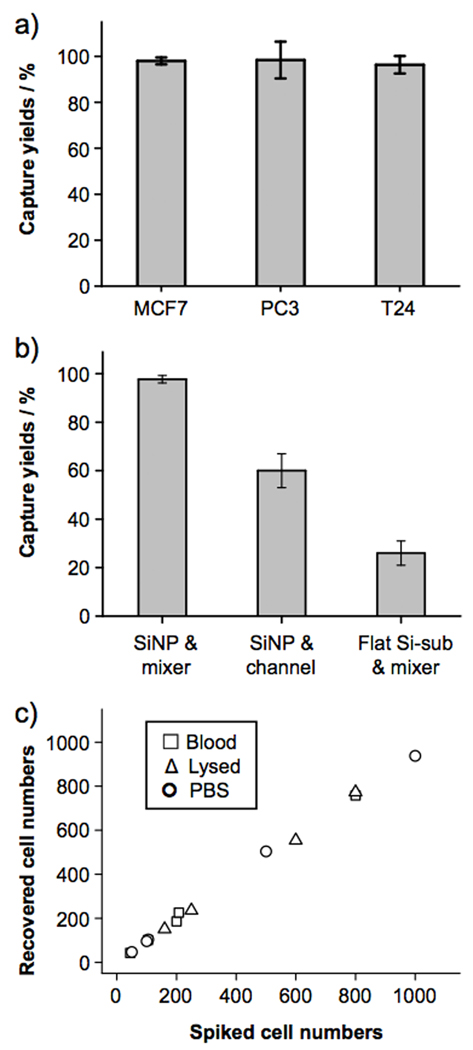
To test how the sample flow rates affect the capture efficiency of the device, cell suspensions (100 cells mL−1) containing EpCAM-positive MCF7 breast cancer cells in DMEM medium were introduced into the devices at flow rates of 0.5, 1, 2, 3, 5 and 7 mL h−1. After rinsing, fixing and DAPI nuclear staining, the PDMS component was detached from the SiNP substrate. Subsequently, substrate-immobilized cells were imaged and counted using a fluorescence microscope. As shown in Figure 2, superb cell capture efficiency (>95%) was accomplished at flow rates of 0.5, 1 and 2 mL h−1. Moreover, we were able to characterize the distribution of substrate-immobilized cells at different locations of the 88-cm serpentine pattern. At a flow rate of 1 mL h−1 (Figure 2b), 70% of the cells are captured in the first 25% of the SiNP-covered microchannels. With increasing flow rates (Figure 2c and d), immobilized cells are more spread out as a result of proportionally enhanced shear forces on the immobilized cells. The cell-capture efficiency decreased at higher flow rates due to (i) reduced duration that a cell interacts with the anti-EpCAM-coated SiNP substrate (making it less likely to develop “stable” cell adhesion) and (ii) increased flow-induced drag (sufficient to overcome “transient” cell adhesion). An optimal cell-capture flow rate of 1.0 mL h−1 was determined. To test the general applicability of these optimal cell-capture conditions, two additional EpCAM-positive cancer cell lines (PC3 prostate cancer and T24 bladder cancer cell lines) were tested in the devices with comparable cell-capture efficiency (see Figure 3a). Two control experiments based on identical design features (i) without SiNPs on the patterned substrate and (ii) without the chevron-shaped micropatterns in the microfluidic channels were also conducted separately. As shown in Figure 3b, device performance was dramatically compromised, suggesting that both SiNP-based high cell-capture efficiency and micropattern-generated chaotic mixing are crucial for enhanced device performance. Finally, the cell-capture efficiency of the optimal capture conditions was validated using artificial CTC samples containing MCF7 cells. A series of MCF7 cell-spiked blood samples was prepared by spiking Red-Dye-stained MCF7 cells into blood with cell densities of approximately 50, 100, 200, and 500–1000 cells mL−1. The results are summarized in Figure 3c. Regardless of whether the red blood cells were intact or lysed, optimal cell-capture conditions enabled more than 95% recovery of cancer cells from the artificial samples. For comparison, control studies in PBS were also examined with similar cell densities.
Figure 2.
a) Cell-capture efficiency of the integrated CTC-capture device at flow rates of 0.5, 1, 2, 3, 5 and 7 mL h−1. Error bars show standard deviations (n = 3). 1.0-mL Cell suspensions containing EpCAM-positive MCF7 breast cancer cells (100 cells mL−1) were employed as a model system. The error bars of the first three data points are very small. Spatial distribution of substrate-immobilized MCF7 cells along the serpentine microchannels at different flow rates of b) 1, c) 2, and d) 7 mL h−1.
Figure 3.
a) Cell-capture efficiency of the integrated CTC-capture device using suspensions of breast (MCF7), prostate (PC3) and bladder (T-24) cell lines. Error bars show standard deviations (n = 3). b) Comparison of cell-capture between the integrated CTC-capture device and two control devices (i) without SiNPs on the patterned substrate and (ii) without the chevron-shaped micropatterns in the microfluidic channels. c) Comparison of cell-capture efficiency at different cell numbers ranging from 50–1000 cells mL−1. Three different types of samples, including whole blood (□), lysed blood (△) and PBS buffer (○) were examined.
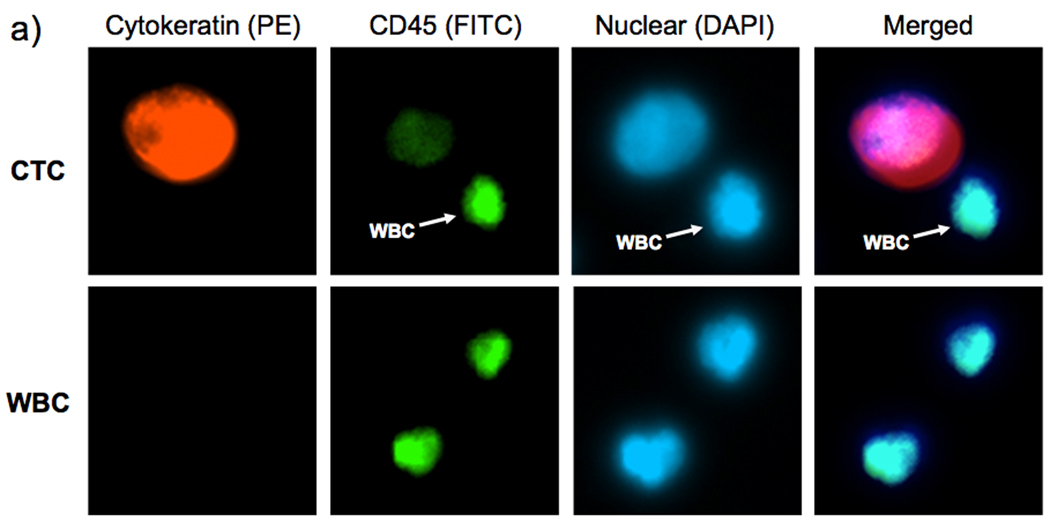
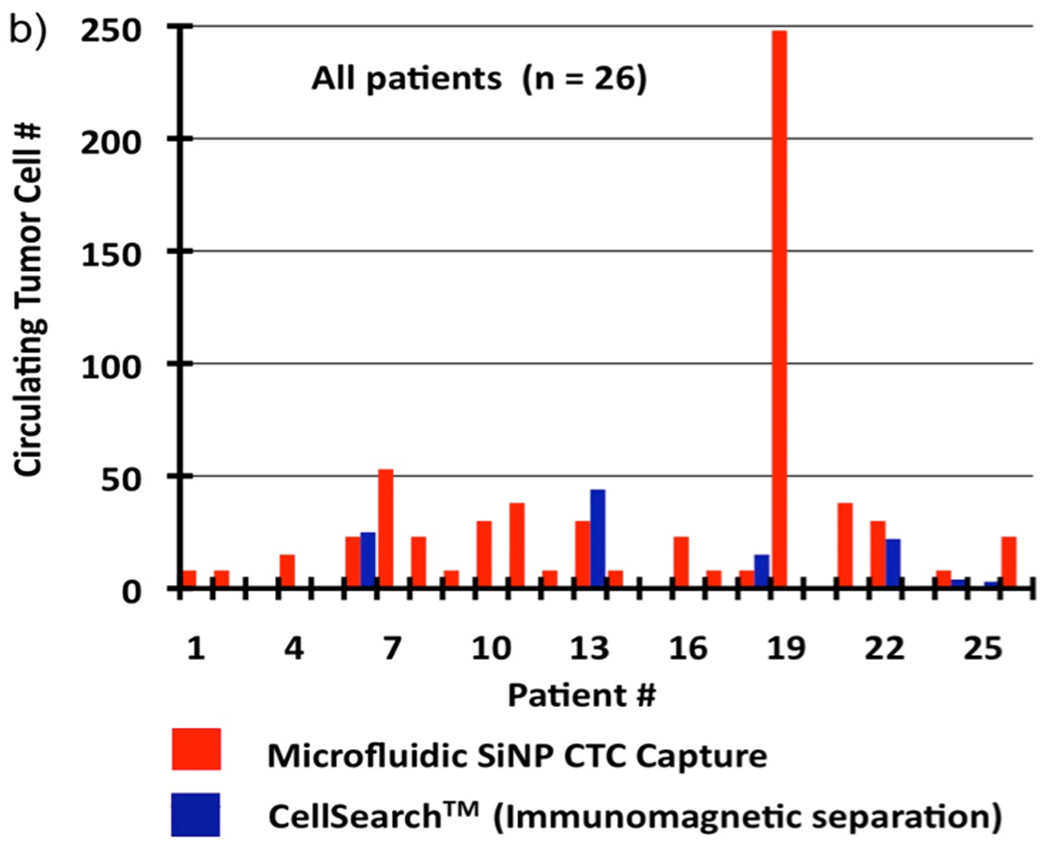
We applied the optimized cell-capture conditions to study prostate cancer patient samples (Figure 4). Specifically, we attempted to validate the performance of our integrated CTC-capture platform by carrying out side-by-side comparisons with the CellSearch® assay[30]. The peripheral blood samples were obtained from prostate cancer patients with different stages of the disease and under different treatments and preserved in CellSave® Tubes (containing fixation agents). In each study, 1.0-mL blood was introduced into integrated devices at a back pressure ranging from 1.5 to 3.0 psi depending on the sample viscosity. After rinsing with PBS, fixation and permeabilization agents were introduced into the devices followed by 30-min incubation. Subsequently, a commonly used three-color immunocytochemistry protocol was applied to identify and enumerate CTCs from non-specifically trapped white blood cells (WBCs), including FITC-labeled anti-CD45 (a marker for WBCs) and PE-labeled anti-CK (Cytokeratin, a protein marker for epithelial cells) as well as DAPI nuclear staining. Fluorescence microscopy was employed to quantify[31, 32] DAPI intensities and expression levels of CK and CD45 in individual cells. As shown in Figure 4a, CTCs exhibit strong CK expression and negligible CD45 signals. In contrast, WBCs present low CK and high CD45 expression levels. DAPI staining validates that the captured cells retain intact nuclei. Furthermore, the morphology and footprint sizes of the cells offer another layer of confinement for cross checking the observation from the perspectives of pathology and cytology. The combined information was utilized to delineate CTCs (DAPI+/CK+/CD45−, footprint sizes: 40 µm > sizes > 10 µm) from WBCs (DAPI+/CK−/CD45+, sizes < 15 µm) and cellular debris. The side-by-side comparison data of the integrated microfluidic SiNP platform vs. CellSearch® is summarized in Figure 4b. Since our CTC counts were obtained from measurements using 1.0-mL blood samples, the results were normalized to CTC counts per 7.5 mL blood (the quantity routinely tested and units reported by CellSearch® assay) to facilitate comparison. In 17 out of 26 patient blood samples, our platform captured significantly greater CTC numbers compared to the CellSearch® assay. Please see Supporting Information for more details on the method comparison and clinical correlation.
Figure 4.
a) Fluorescent micrographs of CTCs captured from blood samples from a prostate cancer (PC) patients. Three-color immunocytochemistry protocol based on PE-labeled anti-Cytokeratin (a protein marker for epithelium cells), FITC-labeled anti-CD45 (a marker for WBCs) and DAPI nuclear staining was applied to identify and enumerate CTCs from non-specifically trapped WBCs on the SiNP substrates. Side-by-side comparison of CTC enumeration results obtained from b) our integrated CTC-capture technology (red columns, normalized to 7.5 mL of blood) and a CellSearch® assay (blue columns) on matched samples from 26 PC patients. **The raw data without normalization from 1.0 to 7.5 mL using our integrated devices and by CellSearch® assay are summarized in Supporting Information (Table S2).
In conclusion, we have demonstrated a new CTC-capture platform that combines a high-affinity cell enrichment assay based on cell capture agent-coated nanostructured substrates and a chaotic mixing chip capable of improving CTC/substrate contact frequency. The resulting synergistic effects led to the superb CTC-capture performance observed for both spiked and clinical blood samples. We envision that the significantly improved sensitivity of our new CTC capture technology will open up opportunities for (i) early detection of cancer metastasis and (ii) for isolation of rare populations of cells that cannot feasibly be done using existing technologies.
Footnotes
This research was supported by NIH IMAT Program (R21CA151159-01) and Prostate Cancer Foundation Creativity Award. We thank Professor Allan Pantuck from the UCLA Urology Department for providing CTC blood samples.
Contributor Information
Shutao Wang, Beijing National Laboratory for Molecular Sciences, Key Laboratory of Organic Solids, Institute of Chemistry, Chinese Academy of Sciences, Beijing (P. R. China), stwang@iccas.ac.cn; Department of Molecular and Medical Pharmacology, Crump Institute for Molecular Imaging (CIMI), California NanoSystems Institute (CNSI), Institute for Molecular Medicine (IMED), University of California, Los Angeles, 570 Westwood Plaza, Building 114, Los Angeles, CA 90095-1770 (USA).
Kan Liu, College of Electronics and Information Engineering, Wuhan Textile University, Wuhan (P. R. China); Department of Molecular and Medical Pharmacology, Crump Institute for Molecular Imaging (CIMI), California NanoSystems Institute (CNSI), Institute for Molecular Medicine (IMED), University of California, Los Angeles, 570 Westwood Plaza, Building 114, Los Angeles, CA 90095-1770 (USA).
Jian Liu, Uro-oncology Research Program, Samuel Oschin Comprehensive Cancer Institute, Cedars Sinai Medical Center, Los Angeles, CA (USA); Department of Molecular and Medical Pharmacology, Crump Institute for Molecular Imaging (CIMI), California NanoSystems Institute (CNSI), Institute for Molecular Medicine (IMED), University of California, Los Angeles, 570 Westwood Plaza, Building 114, Los Angeles, CA 90095-1770 (USA).
Zeta T.-F. Yu, Department of Molecular and Medical Pharmacology, Crump Institute for Molecular Imaging (CIMI), California NanoSystems Institute (CNSI), Institute for Molecular Medicine (IMED), University of California, Los Angeles, 570 Westwood Plaza, Building 114, Los Angeles, CA 90095-1770 (USA)
Xiaowen Xu, Department of Molecular and Medical Pharmacology, Crump Institute for Molecular Imaging (CIMI), California NanoSystems Institute (CNSI), Institute for Molecular Medicine (IMED), University of California, Los Angeles, 570 Westwood Plaza, Building 114, Los Angeles, CA 90095-1770 (USA).
Libo Zhao, Department of Molecular and Medical Pharmacology, Crump Institute for Molecular Imaging (CIMI), California NanoSystems Institute (CNSI), Institute for Molecular Medicine (IMED), University of California, Los Angeles, 570 Westwood Plaza, Building 114, Los Angeles, CA 90095-1770 (USA).
Tom Lee, Department of Molecular and Medical Pharmacology, Crump Institute for Molecular Imaging (CIMI), California NanoSystems Institute (CNSI), Institute for Molecular Medicine (IMED), University of California, Los Angeles, 570 Westwood Plaza, Building 114, Los Angeles, CA 90095-1770 (USA).
Eun Kyung Lee, Department of Molecular and Medical Pharmacology, Crump Institute for Molecular Imaging (CIMI), California NanoSystems Institute (CNSI), Institute for Molecular Medicine (IMED), University of California, Los Angeles, 570 Westwood Plaza, Building 114, Los Angeles, CA 90095-1770 (USA).
Jean Reiss, Department of Pathology and Laboratory Medicine, University of California, Los Angeles (USA).
Yi-Kuen Lee, Department of Mechanical Engineering, The Hong Kong University of Science and Technology (Hong Kong).
Leland W. K. Chung, Uro-oncology Research Program, Samuel Oschin Comprehensive Cancer Institute, Cedars Sinai Medical Center, Los Angeles, CA (USA)
Jiaoti Huang, Department of Pathology and Laboratory Medicine, University of California, Los Angeles (USA).
Matthew Rettig, Department of Urology, University of California, Los Angeles (USA).
David Seligson, Department of Pathology and Laboratory Medicine, University of California, Los Angeles (USA).
Kumaran N. Duraiswamy, Department of Molecular and Medical Pharmacology, Crump Institute for Molecular Imaging (CIMI), California NanoSystems Institute (CNSI), Institute for Molecular Medicine (IMED), University of California, Los Angeles, 570 Westwood Plaza, Building 114, Los Angeles, CA 90095-1770 (USA).
Clifton K.-F. Shen, Department of Molecular and Medical Pharmacology, Crump Institute for Molecular Imaging (CIMI), California NanoSystems Institute (CNSI), Institute for Molecular Medicine (IMED), University of California, Los Angeles, 570 Westwood Plaza, Building 114, Los Angeles, CA 90095-1770 (USA).
Hsian-Rong Tseng, Department of Molecular and Medical Pharmacology, Crump Institute for Molecular Imaging (CIMI), California NanoSystems Institute (CNSI), Institute for Molecular Medicine (IMED), University of California, Los Angeles, 570 Westwood Plaza, Building 114, Los Angeles, CA 90095-1770 (USA).
References
- 1.Pantel K, Brakenhoff RH. Nat. Rev. Cancer. 2004;4:448. doi: 10.1038/nrc1370. [DOI] [PubMed] [Google Scholar]
- 2.Steeg PS. Nat. Med. 2006;12:895. doi: 10.1038/nm1469. [DOI] [PubMed] [Google Scholar]
- 3.Klein CA. Science. 2008;321:1785. doi: 10.1126/science.1164853. [DOI] [PubMed] [Google Scholar]
- 4.Fidler IJ. Nat. Rev. Cancer. 2003;3:453. doi: 10.1038/nrc1098. [DOI] [PubMed] [Google Scholar]
- 5.Bernards R, Weinberg RA. Nature. 2002;418:823. doi: 10.1038/418823a. [DOI] [PubMed] [Google Scholar]
- 6.Chiang AC, Massague J. New Eng. J. Med. 2008;359:2814. doi: 10.1056/NEJMra0805239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bos PD, Zhang XHF, Nadal C, Shu WP, Gomis RR, Nguyen DX, Minn AJ, van de Vijver MJ, Gerald WL, Foekens JA, Massague J. Nature. 2009;459:1005. doi: 10.1038/nature08021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cristofanilli M, Budd GT, Ellis MJ, Stopeck A, Matera J, Miller MC, Reuben JM, Doyle GV, Allard WJ, Terstappen LWMM, Hayes DF. New Eng. J. Med. 2004;351:781. doi: 10.1056/NEJMoa040766. [DOI] [PubMed] [Google Scholar]
- 9.Pantel K, Brakenhoff RH, Brandt B. Nat. Rev. Cancer. 2008;8:329. doi: 10.1038/nrc2375. [DOI] [PubMed] [Google Scholar]
- 10.Nguyen DX, Bos PD, Massague J. Nat. Rev. Cancer. 2009;9:274. doi: 10.1038/nrc2622. [DOI] [PubMed] [Google Scholar]
- 11.Marrinucci D, Bethel K, Luttgen M, Bruce RH, Nieva J, Kuhn P. Arch. Pathol. Lab. Med. 2009;133:1468. doi: 10.1043/1543-2165-133.9.1468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nagrath S, Sequist LV, Maheswaran S, Bell DW, Irimia D, Ulkus L, Smith MR, Kwak EL, Digumarthy S, Muzikansky A, Ryan P, Balis UJ, Tompkins RG, Haber DA, Toner M. Nature. 2007;450:1235. doi: 10.1038/nature06385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zheng S, Lin H, Liu JQ, Balic M, Datar R, Cote RJ, Tai YC. J. Chromatogr. A. 2007;1162:154. doi: 10.1016/j.chroma.2007.05.064. [DOI] [PubMed] [Google Scholar]
- 14.Adams A, Okagbare PI, Feng J, Hupert ML, Patterson D, Göttert J, McCarley RL, Nikitopoulos D, Murphy MC, Soper SA. J. Am. Chem. Soc. 2008;130:8633. doi: 10.1021/ja8015022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gascoyne PR, Noshari J, Anderson TJ, Becker FF. Electrophoresis. 2009;30:1388. doi: 10.1002/elps.200800373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Talasaz AH, Powell AA, Huber DE, Berbee JG, Roh K-H, Yu W, Xiao W, Davis MM, Pease RF, Mindrinos MN, Jeffrey SS, Davis RW. Proc. Natl. Acad. Sci. USA. 2009;106:3970. doi: 10.1073/pnas.0813188106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gleghorn JP, Pratt ED, Denning D, Liu H, Bander NH, Tagawa ST, Nanus DM, Giannakakou PA, Kirby BJ. Lab on a Chip. 2010;10:27. doi: 10.1039/b917959c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang S, Wang H, Jiao J, Chen KJ, Owens GE, Kamei K, Sun J, Sherman DJ, Behrenbruch CP, Wu H, Tseng HR. Angew. Chem. 2009;121:9132. doi: 10.1002/anie.200901668. ; Angew. Chem. Int. Ed. Engl.2009, 48, 8970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Went PTH, Lugli A, Meier S, Bundi M, Mirlacher M, Sauter G, Dirnhofer S. Human Pathology. 2004;35:122. doi: 10.1016/j.humpath.2003.08.026. [DOI] [PubMed] [Google Scholar]
- 20.Munz M, Baeuerle PA, Gires O. Cancer Research. 2009;69:5627. doi: 10.1158/0008-5472.CAN-09-0654. [DOI] [PubMed] [Google Scholar]
- 21.Fischer KE, Aleman BJ, Tao SL, Daniels RH, Li EM, Bunger MD, Nagaraj G, Singh P, Zettl A, Desai TA. Nano Letters. 2009;9:716. doi: 10.1021/nl803219f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Stroock AD, Dertinger SK, Ajdari A, Mezic I, Stone HA, Whitesides GM. Science. 2002;295:647. doi: 10.1126/science.1066238. [DOI] [PubMed] [Google Scholar]
- 23.Liu RH, Stremler MA, Sharp KV, Olsen MG, Santiago JG, Adrian RJ, Aref H, Beebe DJ. J. Microelectromechanical Systems. 2000;9:190. [Google Scholar]
- 24.Niu XZ, Lee YK. J. Micromechanics and Microengineering. 2003;13:454. [Google Scholar]
- 25.Liu J, Williams BA, Gwirtz RM, Wold BJ, Quake S. Angew. Chem. 2006;118:3700. doi: 10.1002/anie.200503830. ; Angew. Chem. Int. Ed. Engl.2006, 45, 3618. [DOI] [PubMed] [Google Scholar]
- 26.Wang J, Sui G, Mocharla VP, Lin RJ, Phelps ME, Kolb HC, Tseng HR. Angew. Chem. 2006;118:5402. doi: 10.1002/anie.200601677. ; Angew. Chem. Int. Ed. Engl.2006, 45, 5276. [DOI] [PubMed] [Google Scholar]
- 27.Foley JO, Mashadi-Hossein A, Fu E, Finlayson BA, Yager P. Lab on a Chip. 2008;8:557. doi: 10.1039/b713644g. [DOI] [PubMed] [Google Scholar]
- 28.Rao CG, Chianese D, Doyle GV, Miller MC, Russell T, Sanders RA, Jr, Terstappen LW. Int. J. Oncol. 2005;27:49. [PubMed] [Google Scholar]
- 29.Murthy SK, Sin A, Tompkins RG, Toner M. Langmuir. 2004;20:11649. doi: 10.1021/la048047b. [DOI] [PubMed] [Google Scholar]
- 30.Shaffer DR, Leversha MA, Danila DC, Lin O, Gonzalez-Espinoza R, Gu B, Anand A, Smith K, Maslak P, Doyle GV, Terstappen LW, Lilja H, Heller G, Fleisher M, Scher HI. Clin Cancer Res. 2007;13:2023. doi: 10.1158/1078-0432.CCR-06-2701. [DOI] [PubMed] [Google Scholar]
- 31.Sun J, Masterman-Smith MD, Graham NA, Jiao J, Mottahedeh J, Laks DR, Ohashi M, DeJesus J, Kamei K, Lee KB, Wang H, Yu ZTF, Lu YT, Hou SA, Li KY, Liu M, Zhang NG, Wang ST, Angenieux B, Panosyan E, Samuels ER, Park J, Williams D, Konkankit V, Nathanson D, van Dam RM, Phelps ME, Wu H, Liau LM, Mischel PS, Lazareff JA, Kornblum HI, Yong WH, Graeber TG, Tseng HR. Cancer Research. 2010;70:6128. doi: 10.1158/0008-5472.CAN-10-0076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kamei KI, Ohashi M, Gschweng E, Ho Q, Suh J, Tang JH, Yu ZTF, Clark AT, Pyle AD, Teitell MA, Lee KB, Witte ON, Tseng HR. Lab on a Chip. 2010;10:1113. doi: 10.1039/b922884e. [DOI] [PMC free article] [PubMed] [Google Scholar]