Abstract
Background
The low incidence of sarcomas in the general population makes heritable contribution to disease risk difficult to discern beyond highly penetrant Mendelian syndromes.
Methods
The Utah Cancer Registry (UCR) and Utah Population Database (UPBD) were interrogated for sarcoma diagnostic codes grouped by genetic type, either complex genotype/karyotype sarcoma or balanced translocation-associated sarcoma. The Genealogical Index for Familialty (GIF) was calculated and relative risks (RR) of disease estimated for 1st, 2nd, and 3rd degree relatives of sarcoma probands. Cancer patterns in pedigrees of sarcoma probands were examined to rule out known hereditary cancer syndromes.
Results
229 balanced translocation type and 1,161 complex genotype type sarcomas with at least 3 generations of ancestral genealogy data were identified in the UCR. There was no evidence for excess relatedness for the balanced translocation group using the GIF test (p=0.657) and no significantly elevated RRs. In the complex genotype group, we observed significantly elevated GIF (p=0.03). Modest RRs corroborated the GIF analysis, in which excess relatedness existed in distant relationships. No recognized cancer syndromes were identified among high risk pedigrees.
Discussion
We identified strong familiality among complex genotype sarcomas, independent from known cancer predisposition syndromes. In the absence of significantly elevated RRs for close relatives, the high GIF argues for a strong genetic--rather than environmental--component to complex genotype sarcoma risk. We observed no significant familial risk of developing balanced translocation sarcomas, but the sample was small.
Impact
There exists yet to be deciphered heritable risk for development of complex genotype sarcomas.
Keywords: sarcoma, familial risk, heritable risk, balanced-translocation, complex karyotype
INTRODUCTION
Sarcomas, cancers of mesenchymal tissues, remain challenging diseases to treat. Sarcomas are rare cancers, and directly affect a small portion of the general population(1). However, their impact is heightened by their deadly incidence among adolescents and young adults.
Mesenchymal tissues line neither the body surface nor ingesting/inhaling organ cavities exposed directly to environmental toxins. The development of cancer in mesenchyme therefore may depend more on biologically intrinsic factors than environmental exposures. This thought is supported by the relative rarity of sarcomas, despite the fact that mesenchymal tissues comprise a strong majority of tissue volumes and body mass percentages in the human body(2). The major determinant of tissue-intrinsic characteristics beyond the chance accrual of replication errors, mis-recombinations, and erroneous chromosomal segregations, is the inherited genome from which each cell begins. This raises the possibility for heritable risks for sarcomagenesis.
The major challenge to studying the familiality of sarcoma is its scarcity in the general population. If the brother or sister of a sarcoma patient had even a 5-fold relative risk for sarcoma, that risk would not be readily detected unless the patient had tens or hundreds of thousands of siblings. The Utah population database (UPDB) has proven valuable to the study of heritability, especially for rare diseases, given the depth of genealogies recorded and its careful linking to the Utah Cancer Registry (UCR), which is part of the Surveillance, Epidemiology and End Results (SEER) Program and has been maintained for the last 50 years(3). Nonetheless, an investigation of familiality for any individual subtype of sarcoma is likely to be underpowered even over this 50-year population accrual due to insufficient case numbers. A potential route forward from this challenge is the meaningful grouping of sarcoma subtypes. While individual sarcoma subtypes tend to derive their identities from known or presumed tissues of origin, there is a variety of ways to lump subtypes together. For example, sarcomas may be grouped according to the population affected, adolescents and young adults versus the elderly. Alternatively, they can be grouped according to bone versus soft-tissue locations; many current treatment paradigms roughly follow this crude grouping, with chemotherapeutic adjuvants for bone sarcomas and adjuvant radiation for soft-tissue sarcomas, generally.
Sarcomas can also be classified according to tumor cell genetics. Many are associated with balanced chromosomal translocations, which generate subtype specific fusion oncogenes, such as EWS-FLI1 in Ewing’s sarcoma and SYT-SSX1 in synovial sarcoma(4). Other sarcomas can be termed complex karyotype sarcomas. This latter group exhibits genomic and chromosomal instability, with mutations and copy number alterations common throughout the genome and wild non-diploid karyotypes frequent(4). Familiality has been suspected, but not proven, for both types of sarcoma. It has been most carefully explored in the bone sarcomas, osteosarcoma and Ewing’s sarcoma.
Osteosarcoma, the prototype complex genotype sarcoma, arises more frequently in three heritable Mendelian cancer predisposition syndromes, Li Fraumeni(5), hereditary retinoblastoma(6), and Rothmund Thompson syndrome(7). However, these syndromes contribute only a scant number of cases to the overall population incidence of osteosarcoma. Beyond these sydnromes, there may be other complex heritable predispositions not yet recognized that engender the genomic instability resulting in complex genotype sarcomas like osteosarcoma.
Ewing’s sarcoma, the most common balanced translocation-associated sarcoma does not arise commonly in any heritable cancer predisposition syndrome. Individual cases have been reported following diagnosis and treatment for retinoblastoma(8-9). The general association between Ewing’s sarcoma and other cancers in families has been suggested by a few small series only (10). Four sibling pairs with Ewing’s sarcoma have been described(11-13). Ewing’s sarcoma also has been associated in families with both umbilical and inguinal hernias(14-16). Finally, Ewing’s sarcoma has a much lower incidence among American individuals of African descent than among Americans of European or Asian ancestry(17). These epidemiologic findings all suggest a modest but discernable genetic contribution to disease risk, despite the lack of Ewing’s sarcoma with any known hereditary cancer syndrome(18).
There are two hypothesized heritable risks for the group of balanced translocation associated sarcomas like Ewing’s sarcoma. First, there may be a heritable predisposition to generate the translocations themselves. Such heritable predispositions to generate translocations could be either generalized or locus (sarcoma sub-type) specific. The latter, are obviously difficult to detect without large numbers of cases and deep genealogies. Second, there may be heritable tendencies for a cell that has undergone such a translocation to complete transformation, rather than apoptose. Silencing of the p53 pathway, for example, is common even among balanced translocation sarcomas(19).
METHODS
The Utah Population Database (UPDB) is a computerized data resource consisting of genealogical and demographic data representing the Utah population(3). The genealogical data in the UPDB has been record linked to the Utah Cancer Registry. The Utah genealogy database was created in the 1970s to investigate the familial aggregation of cancer and now spans up to 12 generations in some Utah pedigrees(20). Several studies using the genealogy data linked to the Utah Cancer Registry (UCR) have defined familial cancer predispositions and syndromes(21-27). The UCR was established in 1966, and became part of the NCI Surveillance, Epidemiology, and End Results (SEER) program in 1973. All cancers occurring in the state are reportable by law to the UCR; follow-up rates exceed 95%. The UCR data include primary site, histology, stage, grade, survival months, and age at diagnosis data for each cancer. The Utah population is genetically representative of northern Europe and has low inbreeding levels, similar to the rest of the United States(28-29). Our analysis was restricted to the 2.3 million subjects in the UPDB with at least three generations of genealogy, ranging in time from approximately 1850 to the present. Use of the data for this study was approved by the University of Utah Institutional Review Board.
All individuals in the UCR with a sarcoma diagnosis who also had at least 3 generations of genealogy data in the UPDB were identified by searching for ICD-O codes, grouped into translocation-associated sarcomas and complex karyotype sarcomas as shown in Table 1. Malignant peripheral nerve sheath tumors (MPNSTs), although fitting into the complex karyotype sarcomas, were excluded from the analysis due to fact that roughly half of these malignancies arise in the setting of the Mendelian syndrome, neurofibromatosis type I, which might bias the data set toward familiality. Dermatofibrosarcoma protuberans and gastrointestinal stromal tumor cases were also excluded. These patients typically bear point mutations in specific genes, rather than balanced translocations, but are otherwise simple genetic sarcomas.
Table 1.
Diagnoses included in each group
| Diagnosis | Histology ICD-O codes | number |
|---|---|---|
| Complex Genotype/Karyotype Sarcoma | ||
| Pleiomorphic Liposarcoma | 8854/3; 8850/3 | 124 |
| Low-Grade Myofibroblastic Sarcoma | 8825/3 | 1 |
| Myxofibrosarcoma | 8811/3 | 6 |
| Pleiomorphic Sarcoma Nos/Malignant Fibrous Histiocytoma | 8830/3 | 223 |
| Leiomyosarcoma | 8890/3 | 300 |
| Embryonal Rhabdomyosarcoma | 8910/3; 8912/3 | 43 |
| Pleiomorphic Rhabdomyosarcoma | 8901/3 | 4 |
| Angiosarcoma | 9120/3 | 72 |
| Epithelioid Sarcoma | 8804/3 | 15 |
| Malignant Mesenchymoma | 8990/3 | 13 |
| Chondrosarcoma | 9220/3; 9221/3; 9240/3; 9242/3; 9243/3 | 102 |
| Osteosarcoma | 9180/3; 9181/3; 9182/3; 9183/3; 9184/3; 9185/3; 9187/3; 9192/3; 9193/3; 9194/3; 9195/3 |
120 |
| Fibrosarcoma | 8810/3 | 116 |
| Chordoma | 9370/3; 9371/3; 9372/3 | 23 |
| Adamantinoma | 9261/3 | 0 |
| Balanced translocation-associated sarcoma | ||
| Synovial Sarcoma | 9040/3; 9041/3; 9043/3 | 51 |
| Alveolar rhabomyosarcoma | 8920/3 | 23 |
| Myxoid Liposarcoma | 8852/3 | 36 |
| Round Cell Liposarcoma | 8853/3 | 4 |
| Ewing’s Sarcoma Family Tumors | 9260/3; 9364/3; 9365/3 | 102 |
| Infantile Fibrosarcoma | 8814/3 | 1 |
| Extraskeletal Myxoid Chondrosarcoma | 9231/3 | 3 |
| Alveolar Soft Parts Sarcoma | 9581/3 | 6 |
| Clear Cell Sarcoma | 9044/3 | 2 |
| Desmoplastic Small Round Cell Tumor | 8806/3 | 1 |
Genealogical Index of Familiality Method
The Genealogical Index of Familiality (GIF) statistic was designed to identify familial aggregation of specific traits within the Utah genealogy(30). The GIF method of analysis has been used in previous studies of familiality for cancers(21-22, 26-27). A similar method of analysis has been used in extended Icelandic genealogies(31).
The GIF statistic measures the average relatedness between all pairs of individuals with a specific phenotype (cases). The Malecot coefficient of kinship is used to measure the degree of relatedness between all pairs of cases. The coefficient of kinship is defined as the probability that randomly selected homologous genes from two individuals are identical by descent from a common ancestor(24). The coefficient is ½ for parent/offspring, ¼ for siblings, 1/8 for grandparent/grandchild, and so forth. The case GIF is the mean of all coefficients of kinship between all possible pairs of cases. The coefficient of kinship for any two individuals in a population is expected to be close to zero. The case GIF is multiplied by 105 for ease of presentation. The GIF statistic takes into account all genetic relationships between all cases.
To test the null hypothesis of no excess relatedness among cases, we created an empirical control distribution. For each sarcoma case, we selected a control at random from the UPDB genealogy resource (also limited to individuals with at least 3 generations recorded), matched on sex, five-year birth cohort, and place of birth (in or out of Utah), resulting in a control set of the same size as the case set. The matching strategy is employed to account for potential differences in kinship based on differences in birth year, sex and place of birth. One thousand independent 1:1 matched control sets were selected and the GIF measured for each set to create an empirical distribution of average relatedness under the null hypothesis of no excess relatedness among cases. We then tested this hypothesis by comparing the case GIF to the empirical distribution of the 1000 control groups’ GIFs.
The degree of shared genetic composition between pairs of cases representing different genetic distances is quantifiable through the GIF analysis. We assume that the degree of shared environment among individuals diminishes to a population level of sharing beyond second or third degree relatives for the Utah population. Among close relationships, it is difficult to determine whether excess familiality is due to shared environment or to shared genetics, or to a combination of both. Among more distant relationships, however, significant excess relatedness most likely indicates a genetic contribution. The empirical significance of the GIF test tells us whether overall excess familiality is observed. When this same test is performed excluding all close relationships (first and second degree relatives) it allows us to determine whether significant excess familiality exists in distant relationships, further supporting the hypothesis of a genetic contribution (this test is termed the distant or dGIF).
Relative Risks (RR)
Estimation of RRs in relatives is an alternative approach to test the hypothesis of a familial contribution to disease. While the GIF analysis utilizes all relationships between all cases, regardless of genetic distance, the RR analysis typically relies on comparisons in close relatives only. The RR, or the risk of the disease in the relatives compared to the risk in the general population (also termed standardized morbidity ratio) also was calculated. This ratio is directly related to the power to identify disease predisposition genes and is typically a primary test for a genetic component to a disease.The RR approach compares observed rates of sarcoma in relatives of probands with the expected rates of sarcoma as estimated in the UPDB. All individuals in the UPDB with genealogy were used to estimate cohort-specific cancer rates.
We estimated RR as follows: All 2.3 million individuals in the UPDB with at least 3 generations of genealogy were assigned to one of 132 cohorts based on birthplace (in/out Utah), sex and five-year birth-year cohorts. For each cohort, internal cohort-specific cancer rates are calculated by summing the number of individuals in each cohort with sarcoma and dividing by the total number of individuals in the cohort. These internal cohort-specific sarcoma rates estimate the expected rate of sarcoma for each cohort. The expected number of sarcoma cases among first degree relatives of sarcoma cases is calculated by multiplying the number of first degree relatives of sarcoma cases in each cohort by the cohort-specific internal rate of sarcoma, and then summing over all cohorts. The number of observed sarcoma cases among relatives is a count (without duplication) of all of the relatives of sarcoma cases who also were diagnosed with sarcoma. The first degree RR statistic is the ratio of the number of observed sarcoma cases among first degree relatives of sarcoma cases to the number of expected sarcoma cases among first degree relatives of sarcoma cases. The RR was similarly estimated for second and third degree relatives.
The RR is assumed to follow a Poisson distribution with the mean value equal to the number of expected sarcoma cases among relatives of sarcoma cases. The Poisson distribution is an approximation to a sum of multiple binomial distributions, representing the number of expected deaths in each cohort. This Poisson approximation is appropriate for both rare and common phenotypes, being more conservative for common diseases. Probability values for one-sided tests of significance and 95% CIs for the RR statistic can be calculated by Poisson distribution under the null hypothesis that the RR is equal to unity. While significantly elevated risks in first-degree relatives suggest a genetic contribution to disease, they may result from shared environment, or from a combination of both genes and environment. However, significantly elevated risks for second or third degree relatives would strongly suggest a heritable component.
High-Risk Pedigrees
We also identified as high-risk pedigrees those families with an excess of cases identified among the progeny by RR and a total of more than 5 sarcoma cases. Among these high-risk pedigrees we examined co-aggregation of each sarcoma group with other cancers. This analysis was used to rule out the inclusion of recognizable cancer predisposition syndromes in the final dataset.
These analytic methods to identify familial contribution to disease (average relatedness, relative risk, and high risk pedigree identification) previously have been shown to be unbiased, but conservative(22-23),(27),(32-36).
RESULTS
Genealogical Index of Familiality
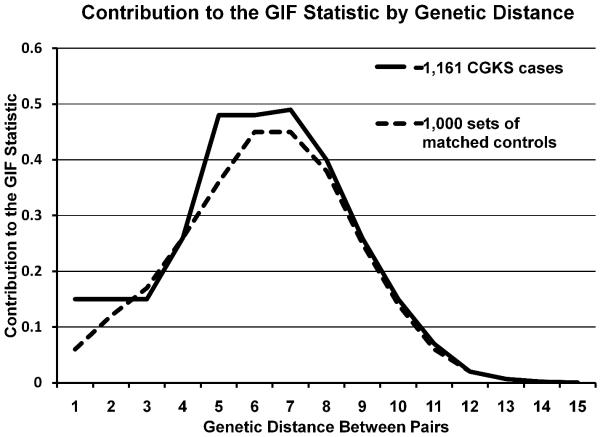
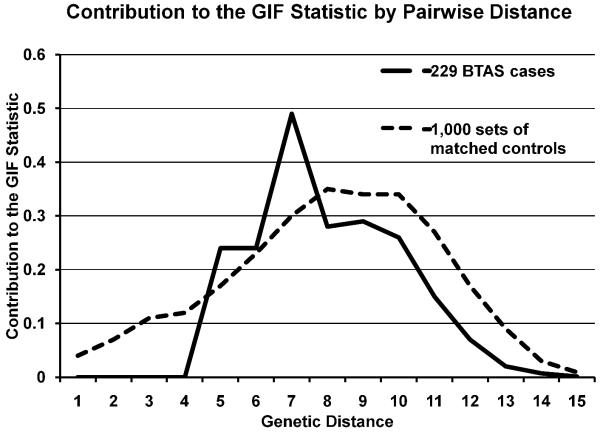
There were 229 individuals with balanced translocation or simple genetic sarcomas identified in the UCR with at least 3 generations of ancestral genealogy recorded in the UPDB. The breakdown into specific diagnoses represented is offered in Table 1. There was no overall excess relatedness for the balanced translocation group using the GIF test, with case GIF of 2.05 and mean control GIF of 2.32 (GIF empirical p = 0.657), and no excess distant relatedness (dGIF empirical p = 0.530) as shown in Figure 1. The complex genotype karyotype sarcoma group of 1,161 patients, however, did show excess overall relatedness, with case GIF of 3.05 and mean control GIF of 2.73 (GIF empirical p = 0.034), and borderline significance when close relationships were ignored (dGIF empirical p = 0.054), as shown in Figure 2.
Figure 1.
Chart showing the contribution to the GIF statistic for the 1,161 complex genotype/karyotype sarcoma cases compared to the mean distribution for 1,000 sets of matched controls. The genetic distance (x axis) represents relationships: 1 represents parent/offspring, 2 represents siblings, 3 represents avunculars, 4 first cousins, and so forth. The excess related is most apparent in the 5th to 7th degree relatives.
Figure 2.
Chart displaying the contribution to the GIF statistic for 229 balanced translocation sarcomas, compared to the mean distribution for 1,000 sets of matched controls. The genetic distance (x-axis) represents relationships: 1 represents parent/offspring, 2 represents siblings, 3 represents avunculars, 4 first cousins, and so forth. There is no significant excess relatedness among balanced translocation sarcomas compared to control data, but statistical power is low given the small number of cases.
Relative Risks
For translocation sarcomas, the RR estimates corrobated the GIF data; no translocation sarcoma cases were observed among the first-, second-, or third-degree relatives of balanced translocation probands.
For complex genotype karyotype sarcomas, RR estimates were greater than 1.0 for both first and second-degree relatives, but were not significantly elevated. The RR for third degree relatives was estimated to be less than 1.0, but again, not significantly. These RRs (Table 2) corroborate the GIF analysis, in which excess relatedness exists primarily at more distant relationships.
Table 2.
Estimates of RR in first-, second-, and third-degree relatives for the same type of sarcoma
| Group | Relationship | Number of Relatives |
Cases Observed |
Cases Expected |
Relative Risk |
1 T 95% CI |
|---|---|---|---|---|---|---|
| Balanced translocation | ||||||
| first-degree | 1454 | 0 | 0.22 | N/A | N/A | |
| second-degree | 4235 | 0 | 0.45 | N/A | N/A | |
| third-degree | 10982 | 0 | 0.94 | N/A | N/A | |
| Complex genotype/karyotype | ||||||
| first-degree | 10,201 | 12 | 8.95 | 1.34 | 0.77, 2.17 | |
| second-degree | 33,483 | 22 | 20.22 | 1.09 | 0.74, 1.55 | |
| third-degree | 88,900 | 51 | 53.14 | 0.96 | 0.75, 1.21 | |
High-Risk Pedigrees
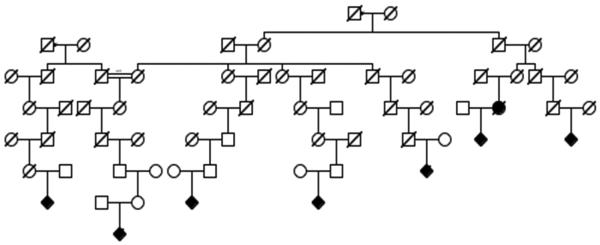
We identified “high-risk” pedigrees for complex genotype/karyotype sarcomas defined as having a significant excess of complex genetic sarcoma cases among all descendants of the founding couples compared to calculated general rates across the UPDB population. 20 pedigrees in this category had more than 5 complex genotype sarcomas in related individuals (Example shown in Figure 3). Interestingly, in these 20 high-risk complex genetic sarcoma pedigrees, other cancer types were also observed in excess. These associated cancers and the number of high risk pedigrees in which they were found in excess are listed in Table 3. Notably, none of the pedigrees had inheritance patterns and cancer types that would fit with any of the well-documented cancer predisposition syndromes such as: Li Fraumeni, hereditary retinoblastoma, or Werner, Bloom or Rothmund Thompson syndromes.
Figure 3.
Example of a high-risk complex genotype/karyotype sarcoma pedigree, using standard pedigree symbols. Diamonds are males and circles females. Filled shapes are individual diagnosed with a complex genotype sarcoma. Slashes indicate deceased individuals. The 8 cases identified in this pedigree were among 5,705 descendants from the couple noted at the top, in which population 2.30 cases were expected (p = 0.0026).
Table 3.
Other cancers common in high-risk complex genotype sarcoma pedigrees
| Cancer type | Number of Pedigrees (of 20) with an excess of cases |
|---|---|
| Lymphoma | 9 |
| Prostate Cancer | 6 |
| Esophageal Cancer | 5 |
| Lip Cancer | 4 |
| Gall Bladder Cancer | 4 |
| Testicular Cancer | 4 |
| Renal Cell Cancer | 3 |
| Ewing’s Sarcoma Family of Tumors | 3 |
| Ovarian Cancer | 2 |
| Pharyngeal Cancer | 2 |
| Brain/CNS Cancer | 2 |
| Liver Cancer | 2 |
| Lung Cancer | 2 |
DISCUSSION
Recognizing the heritable genetic contribution to the risk of rare diseases such as individual sarcoma subtypes is very difficult due to relatively low patient numbers and insufficient statistical power. Traditional means of suspecting familial cancer risk depend on clinically-oriented family histories, which rarely probe deeper than two generations beyond the proband. These methods have identified familial sarcomas as potential manfestations of otherwise penetrant and recognizable heritable disorders. Examples, include Li Fraumeni syndrome(5), arising from inherited disruption of p53, recognized for the strong predisposition to a variety of carcinomas, but also including osteosarcoma and pleiomorphic rhabdomyosarcoma; hereditary retinoblastoma(6), in which syndrome osteosarcoma is the second most common cancer but the first is almost universally penetrant; and neurofibromatosis type I(37), which is recognized from a broad array of non-oncologic clinical manifestations, but also predisposes patients to MPNST. Beyond these, familiality has been suspected, but never explored for sarcomas.
In this study, therefore, we used two methods to strengthen our general detection of familiality among relatively rare sarcomas: the probing of deep genealogies (at least 3 generations and often many more) and the pooling of specific sarcoma diagnoses into genetically-defined categories. This dual approach utilizing the unique resource of the UPDB allowed us to identify evidence for excess familiality among complex genotype sarcomas. This excess familiality was particularly strong in 5th through 7th degree relatives, suggesting a genetic component to risk for complex genotype sarcomas rather than an environmental influence from nuclear family surroundings and occupations. We presume that this genetic risk is inherited in the form of multiple minor susceptibility loci, but we have not strictly ruled out unrecognized, strong, single gene susceptibilities. That these complex genotype sarcomas co-localize in families with other cancers in patterns that do not fit any known syndromes suggests a broader relevance to our findings. Identifying the susceptibility loci in these high-risk families may impact cancers beyond sarcoma and increase our over-all understanding of oncogenesis more generally. We actively participate in a new intercontinental collaboration to collect germline genetic samples from sarcoma patients in order to genetically define genetic loci carrying risk for sarcomagenesis.
Of interest are the specific cancers that arise more commonly in the families with an excess prevalence of complex genotype sarcomas. An excess of Ewing’s sarcoma family of tumors cases was apparent in 3 of the 20 pedigrees with more than 5 complex genotype sarcomas. Certainly, the development of an osteosarcoma or pleiomorphic soft-tissue sarcoma as a radiation-induced secondary cancer following treatments for Ewing’s sarcoma has been reported(38), but these associations in the pedigrees were not in the same patients, and merit further exploration. The excess of lymphoma cases in half of the pedigrees with an excess of complex genotype sarcomas follows to the extent that lymphoma is one of very few solid tumors that is neither a carcinoma nor a sarcoma. Interestingly, species more prone to sarcomas than humans, such as rodents, canines, and felines, are also more prone to lymphomas(39). Lymphoma and sarcoma have previously been linked in terms of risk factors in the settings of acquired immunodeficiency(40) and herbicide exposure(41). Beyond these, a few families with both sarcomas and lymphomas have been reported in isolation in the literature(42-44). Again, further investigation is required.
Similar evidence for familiality was not identified in the smaller set of balanced translocation sarcomas. While we did not see evidence for familial clustering, the small sample size may have affected our power to see such an effect for a rare disease. Furthermore, our negative findings in these families for balanced translocation sarcomas as a pooled group gives no suggestion for or against the possibility of familiality for any given specific subtype of sarcoma even in the same population. Pooling of all of the balanced translocation sarcomas together in one analysis would also dilute the effect seen if any single balanced translocation sarcoma subtype was familial, independent from the group. For example, the suggestion of familiality in prior studies of Ewing’s sarcoma is not significantly challenged by these data, as it was not tested individually. Efforts are ongoing to gather resources that will permit the specific testing of familial risk for Ewing’s sarcoma family of tumors.
Although the accumulation of random genetic changes is certainly involved in sarcomagenesis, our study indicates that perhaps more than chance plays a role in complex genotype sarcomagenesis. We report evidence for the contribution of familiality to the risk of developing one from this group of sarcomas, which risk, if deciphered, could lead to important insights into sarcomagenesis and perhaps oncogenesis more generally.
ACKNOWLEDGEMENTS
We recognize support, although not project specific, from the Huntsman Cancer Foundation (K.B.J., J.S., R.L.R., and S.L.L), the Deparment of Orthopaedics (K.B.J. and R.L.R.), NIH/NCI K08138764 (K.B.J.), and the S.A.R.C. Foundation Career Development Award (J.S.).
No specific funding supported this research.
REFERENCES
- 1.Lahat G, Lazar A, Lev D. Sarcoma epidemiology and etiology: potential environmental and genetic factors. Surg Clin North Am. 2008 Jun;88(3):451–481. v. doi: 10.1016/j.suc.2008.03.006. [DOI] [PubMed] [Google Scholar]
- 2.Clarys JP, Martin AD, Marfell-Jones MJ, Janssens V, Caboor D, Drinkwater DT. Human body composition: A review of adult dissection data. Am J Hum Biol. 1999;11(2):167–174. doi: 10.1002/(SICI)1520-6300(1999)11:2<167::AID-AJHB4>3.0.CO;2-G. [DOI] [PubMed] [Google Scholar]
- 3.Skolnick MH. The Utah Genealogical Database: a Resource for Genetic Epidemiology. In: Cairns J, Lyon JL, Skolnick MH, editors. Banbury report No. 4: cancer incidence in defined populations. Cold Spring Harbor Laboratories; New York: 1980. pp. 285–297. [Google Scholar]
- 4.Fletcher CDM, Unni KK, Mertens F. Pathology and Genetics of Tumours of Soft Tissue and Bone. IARC Press; Lyon: 2002. [Google Scholar]
- 5.Li FP, Fraumeni JF, Jr., Mulvihill JJ, et al. A cancer family syndrome in twenty-four kindreds. Cancer Res. 1988 Sep 15;48(18):5358–5362. [PubMed] [Google Scholar]
- 6.Francois J. Retinoblastoma and osteogenic sarcoma. Ophthalmologica. 1977;175(4):185–191. doi: 10.1159/000308656. [DOI] [PubMed] [Google Scholar]
- 7.Larizza L, Roversi G, Volpi L. Rothmund-Thomson syndrome. Orphanet J Rare Dis. 2010;5:2. doi: 10.1186/1750-1172-5-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Helton KJ, Fletcher BD, Kun LE, Jenkins JJ, 3rd, Pratt CB. Bone tumors other than osteosarcoma after retinoblastoma. Cancer. 1993 May 1;71(9):2847–2853. doi: 10.1002/1097-0142(19930501)71:9<2847::aid-cncr2820710928>3.0.co;2-e. [DOI] [PubMed] [Google Scholar]
- 9.Mittal R, Al Awadi S, Sahar O, Behbehani AM. Ewing’s sarcoma as second malignant neoplasm after retinoblastoma: a case report. Med Princ Pract. 2008;17(1):84–85. doi: 10.1159/000109597. [DOI] [PubMed] [Google Scholar]
- 10.Ji J, Hemminki K. Familial risk for histology-specific bone cancers: an updated study in Sweden. Eur J Cancer. 2006 Sep;42(14):2343–2349. doi: 10.1016/j.ejca.2005.11.043. [DOI] [PubMed] [Google Scholar]
- 11.Hutter RV, Francis KC, Foote FW., Jr. Ewing’s Sarcoma in Siblings: Report of the Second Known Occurrence. Am J Surg. 1964 Apr;107:598–603. doi: 10.1016/0002-9610(64)90328-9. [DOI] [PubMed] [Google Scholar]
- 12.Joyce MJ, Harmon DC, Mankin HJ, Suit HD, Schiller AL, Truman JT. Ewing’s sarcoma in female siblings. A clinical report and review of the literature. Cancer. 1984 May 1;53(9):1959–1962. doi: 10.1002/1097-0142(19840501)53:9<1959::aid-cncr2820530926>3.0.co;2-9. [DOI] [PubMed] [Google Scholar]
- 13.Zamora P, Garcia de Paredes ML, Gonzalez Baron M, et al. Ewing’s tumor in brothers. An unusual observation. Am J Clin Oncol. 1986 Aug;9(4):358–360. doi: 10.1097/00000421-198608000-00017. [DOI] [PubMed] [Google Scholar]
- 14.Ferris ITJ, Berbel Tornero O, Ortega Garcia JA, et al. Risk factors for pediatric malignant bone tumors. An Pediatr (Barc) 2005 Dec;63(6):537–547. doi: 10.1016/s1695-4033(05)70254-x. [DOI] [PubMed] [Google Scholar]
- 15.Valery PC, Holly EA, Sleigh AC, Williams G, Kreiger N, Bain C. Hernias and Ewing’s sarcoma family of tumours: a pooled analysis and meta-analysis. Lancet Oncol. 2005 Jul;6(7):485–490. doi: 10.1016/S1470-2045(05)70242-4. [DOI] [PubMed] [Google Scholar]
- 16.Valery PC, McWhirter W, Sleigh A, Williams G, Bain C. A national case-control study of Ewing’s sarcoma family of tumours in Australia. Int J Cancer. 2003 Jul 20;105(6):825–830. doi: 10.1002/ijc.11129. [DOI] [PubMed] [Google Scholar]
- 17.Jawad MU, Cheung MC, Min ES, Schneiderbauer MM, Koniaris LG, Scully SP. Ewing sarcoma demonstrates racial disparities in incidence-related and sex-related differences in outcome: an analysis of 1631 cases from the SEER database, 1973-2005. Cancer. 2009 Aug 1;115(15):3526–3536. doi: 10.1002/cncr.24388. [DOI] [PubMed] [Google Scholar]
- 18.Randall RL, Lessnick SL, Jones KB, et al. Is There a Predisposition Gene for Ewing’s Sarcoma? J Oncol. 2010;2010:397632. doi: 10.1155/2010/397632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ghule P, Kadam PA, Jambhekar N, et al. p53 gene gets altered by various mechanisms: studies in childhood sarcomas and retinoblastoma. Med Sci Monit. 2006 Dec;12(12):BR385–396. [PubMed] [Google Scholar]
- 20.Skolnick MH. Prospects for population oncogenetics. In: Mulvihill JJ, Miller RW, Fraumeni JF, editors. Genetics of human cancer. Raven Press; New York: 1977. pp. 19–25. [Google Scholar]
- 21.Bishop DT, Skolnick MH. Genetic epidemiology of cancer in Utah genealogies: a prelude to the molecular genetics of common cancers. J Cell Physiol Suppl. 1984;3:63–77. doi: 10.1002/jcp.1041210409. [DOI] [PubMed] [Google Scholar]
- 22.Cannon-Albright LA, Thomas A, Goldgar DE, et al. Familiality of cancer in Utah. Cancer Res. 1994 May 1;54(9):2378–2385. [PubMed] [Google Scholar]
- 23.Goldgar DE, Easton DF, Cannon-Albright LA, Skolnick MH. Systematic population-based assessment of cancer risk in first-degree relatives of cancer probands. J Natl Cancer Inst. 1994 Nov 2;86(21):1600–1608. doi: 10.1093/jnci/86.21.1600. [DOI] [PubMed] [Google Scholar]
- 24.Malecot G. Les Mathematiques de l’Heredite. Masson et Cie; Paris: 1948. [Google Scholar]
- 25.Thomas A, Cannon-Albright L, Bansal A, Skolnick MH. Familial associations between cancer sites. Comput Biomed Res. 1999 Dec;32(6):517–529. doi: 10.1006/cbmr.1999.1525. [DOI] [PubMed] [Google Scholar]
- 26.Albright LA, Schwab A, Camp NJ, Farnham JS, Thomas A. Population-based risk assessment for other cancers in relatives of hereditary prostate cancer (HPC) cases. Prostate. 2005 Sep 1;64(4):347–355. doi: 10.1002/pros.20248. [DOI] [PubMed] [Google Scholar]
- 27.Allen-Brady K, Camp NJ, Ward JH, Cannon-Albright LA. Lobular breast cancer: excess familiality observed in the Utah Population Database. Int J Cancer. 2005 Nov 20;117(4):655–661. doi: 10.1002/ijc.21236. [DOI] [PubMed] [Google Scholar]
- 28.McLellan T, Jorde LB, Skolnick MH. Genetic distances between the Utah Mormons and related populations. Am J Hum Genet. 1984 Jul;36(4):836–857. [PMC free article] [PubMed] [Google Scholar]
- 29.Jorde LB. Inbreeding in the Utah Mormons: an evaluation of estimates based on pedigrees, isonymy, and migration matrices. Ann Hum Genet. 1989 Oct;53(Pt 4):339–355. doi: 10.1111/j.1469-1809.1989.tb01803.x. [DOI] [PubMed] [Google Scholar]
- 30.Hill JR. A survey of cancer sites by kinship in the Utah Mormon population. In: Cairns J, Lyon JL, Skolnick M, editors. Banbury repor No. 4: cancer incidence in defined populations. Cold Spring Harbor Laboratories; New York: 1980. pp. 299–318. [Google Scholar]
- 31.Stefansson H, Geirsson RT, Steinthorsdottir V, et al. Genetic factors contribute to the risk of developing endometriosis. Hum Reprod. 2002 Mar;17(3):555–559. doi: 10.1093/humrep/17.3.555. [DOI] [PubMed] [Google Scholar]
- 32.Albright FS, Orlando P, Pavia AT, Jackson GG, Cannon Albright LA. Evidence for a heritable predisposition to death due to influenza. J Infect Dis. 2008 Jan 1;197(1):18–24. doi: 10.1086/524064. [DOI] [PubMed] [Google Scholar]
- 33.Cannon Albright LA. Utah family-based analysis: past, present and future. Hum Hered. 2008;65(4):209–220. doi: 10.1159/000112368. [DOI] [PubMed] [Google Scholar]
- 34.Cannon Albright LA, Camp NJ, Farnham JM, MacDonald J, Abtin K, Rowe KG. A genealogical assessment of heritable predisposition to aneurysms. J Neurosurg. 2003 Oct;99(4):637–643. doi: 10.3171/jns.2003.99.4.0637. [DOI] [PubMed] [Google Scholar]
- 35.Horne BD, Camp NJ, Muhlestein JB, Cannon-Albright LA. Identification of excess clustering of coronary heart diseases among extended pedigrees in a genealogical population database. Am Heart J. 2006 Aug;152(2):305–311. doi: 10.1016/j.ahj.2005.12.028. [DOI] [PubMed] [Google Scholar]
- 36.Horne BD, Camp NJ, Muhlestein JB, Cannon-Albright LA. Evidence for a heritable component in death resulting from aortic and mitral valve diseases. Circulation. 2004 Nov 9;110(19):3143–3148. doi: 10.1161/01.CIR.0000147189.85636.C3. [DOI] [PubMed] [Google Scholar]
- 37.Williams VC, Lucas J, Babcock MA, Gutmann DH, Korf B, Maria BL. Neurofibromatosis type 1 revisited. Pediatrics. 2009 Jan;123(1):124–133. doi: 10.1542/peds.2007-3204. [DOI] [PubMed] [Google Scholar]
- 38.Ginsberg JP, Goodman P, Leisenring W, et al. Long-term survivors of childhood Ewing sarcoma: report from the childhood cancer survivor study. J Natl Cancer Inst. 2010 Aug 18;102(16):1272–1283. doi: 10.1093/jnci/djq278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.MacEwen EG. Spontaneous tumors in dogs and cats: models for the study of cancer biology and treatment. Cancer Metastasis Rev. 1990 Sep;9(2):125–136. doi: 10.1007/BF00046339. [DOI] [PubMed] [Google Scholar]
- 40.Biggar RJ, Horm J, Goedert JJ, Melbye M. Cancer in a group at risk of acquired immunodeficiency syndrome (AIDS) through 1984. Am J Epidemiol. 1987 Oct;126(4):578–586. doi: 10.1093/oxfordjournals.aje.a114697. [DOI] [PubMed] [Google Scholar]
- 41.Hoar SK, Blair A, Holmes FF, et al. Agricultural herbicide use and risk of lymphoma and soft-tissue sarcoma. JAMA. 1986 Sep 5;256(9):1141–1147. [PubMed] [Google Scholar]
- 42.Buehler SK, Firme F, Fodor G, Fraser GR, Marshall WH, Vaze P. Common variable immunodeficiency, Hodgkin’s disease, and other malignancies in a Newfoundland family. Lancet. 1975 Jan 25;1(7900):195–197. doi: 10.1016/s0140-6736(75)91361-6. [DOI] [PubMed] [Google Scholar]
- 43.Meisner LF, Gilbert E, Ris HW, Haverty G. Genetic mechanisms in cancer predisposition: report of a cancer family. Cancer. 1979 Feb;43(2):679–689. doi: 10.1002/1097-0142(197902)43:2<679::aid-cncr2820430240>3.0.co;2-5. [DOI] [PubMed] [Google Scholar]
- 44.Pastore G, Mosso ML, Carli M, et al. Cancer mortality among relatives of children with soft-tissue sarcoma: a national survey in Italy. Cancer Lett. 1987 Oct;37(1):17–24. doi: 10.1016/0304-3835(87)90141-8. [DOI] [PubMed] [Google Scholar]