Abstract
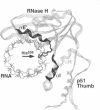
Amino acid sequences homologous to 259KLVGKL (X)16KLLR284 of human immunodeficiency virus type 1 reverse transcriptase (HIV-1 RT) are conserved in several nucleotide polymerizing enzymes. This amino acid motif has been identified in the crystal structure model as an element of the enzyme's nucleic acid binding apparatus. It is part of the helix-turn-helix structure, alpha H-turn-alpha I, within the 'thumb' region of HIV-1 RT. The motif grasps the complexed nucleic acid at one side. Molecular modeling studies on HIV-1 RT in complex with a nucleic acid fragment suggest that the motif has binding function in the p66 subunit as well as in the p51 subunit, acting as a kind of 'helix clamp'. Given its wide distribution within the nucleic acid polymerases, the helix clamp motif is assumed to be a structure of general significance for nucleic acid binding.
Full text
PDF








Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Argos P. A sequence motif in many polymerases. Nucleic Acids Res. 1988 Nov 11;16(21):9909–9916. doi: 10.1093/nar/16.21.9909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnold E., Jacobo-Molina A., Nanni R. G., Williams R. L., Lu X., Ding J., Clark A. D., Jr, Zhang A., Ferris A. L., Clark P. Structure of HIV-1 reverse transcriptase/DNA complex at 7 A resolution showing active site locations. Nature. 1992 May 7;357(6373):85–89. doi: 10.1038/357085a0. [DOI] [PubMed] [Google Scholar]
- Basu A., Ahluwalia K. K., Basu S., Modak M. J. Identification of the primer binding domain in human immunodeficiency virus reverse transcriptase. Biochemistry. 1992 Jan 21;31(2):616–623. doi: 10.1021/bi00117a045. [DOI] [PubMed] [Google Scholar]
- Blanco L., Bernad A., Blasco M. A., Salas M. A general structure for DNA-dependent DNA polymerases. Gene. 1991 Apr;100:27–38. doi: 10.1016/0378-1119(91)90346-d. [DOI] [PubMed] [Google Scholar]
- Claessens M., Van Cutsem E., Lasters I., Wodak S. Modelling the polypeptide backbone with 'spare parts' from known protein structures. Protein Eng. 1989 Jan;2(5):335–345. doi: 10.1093/protein/2.5.335. [DOI] [PubMed] [Google Scholar]
- Davies J. F., 2nd, Hostomska Z., Hostomsky Z., Jordan S. R., Matthews D. A. Crystal structure of the ribonuclease H domain of HIV-1 reverse transcriptase. Science. 1991 Apr 5;252(5002):88–95. doi: 10.1126/science.1707186. [DOI] [PubMed] [Google Scholar]
- Delarue M., Poch O., Tordo N., Moras D., Argos P. An attempt to unify the structure of polymerases. Protein Eng. 1990 May;3(6):461–467. doi: 10.1093/protein/3.6.461. [DOI] [PubMed] [Google Scholar]
- Gopalakrishnan V., Peliska J. A., Benkovic S. J. Human immunodeficiency virus type 1 reverse transcriptase: spatial and temporal relationship between the polymerase and RNase H activities. Proc Natl Acad Sci U S A. 1992 Nov 15;89(22):10763–10767. doi: 10.1073/pnas.89.22.10763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hostomsky Z., Hostomska Z., Fu T. B., Taylor J. Reverse transcriptase of human immunodeficiency virus type 1: functionality of subunits of the heterodimer in DNA synthesis. J Virol. 1992 May;66(5):3179–3182. doi: 10.1128/jvi.66.5.3179-3182.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobo-Molina A., Arnold E. HIV reverse transcriptase structure-function relationships. Biochemistry. 1991 Jul 2;30(26):6351–6356. doi: 10.1021/bi00240a001. [DOI] [PubMed] [Google Scholar]
- Jacobo-Molina A., Ding J., Nanni R. G., Clark A. D., Jr, Lu X., Tantillo C., Williams R. L., Kamer G., Ferris A. L., Clark P. Crystal structure of human immunodeficiency virus type 1 reverse transcriptase complexed with double-stranded DNA at 3.0 A resolution shows bent DNA. Proc Natl Acad Sci U S A. 1993 Jul 1;90(13):6320–6324. doi: 10.1073/pnas.90.13.6320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson M. S., McClure M. A., Feng D. F., Gray J., Doolittle R. F. Computer analysis of retroviral pol genes: assignment of enzymatic functions to specific sequences and homologies with nonviral enzymes. Proc Natl Acad Sci U S A. 1986 Oct;83(20):7648–7652. doi: 10.1073/pnas.83.20.7648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamer G., Argos P. Primary structural comparison of RNA-dependent polymerases from plant, animal and bacterial viruses. Nucleic Acids Res. 1984 Sep 25;12(18):7269–7282. doi: 10.1093/nar/12.18.7269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohlstaedt L. A., Wang J., Friedman J. M., Rice P. A., Steitz T. A. Crystal structure at 3.5 A resolution of HIV-1 reverse transcriptase complexed with an inhibitor. Science. 1992 Jun 26;256(5065):1783–1790. doi: 10.1126/science.1377403. [DOI] [PubMed] [Google Scholar]
- Kumar A., Kim H. R., Sobol R. W., Becerra S. P., Lee B. J., Hatfield D. L., Suhadolnik R. J., Wilson S. H. Mapping of nucleic acid binding in proteolytic domains of HIV-1 reverse transcriptase. Biochemistry. 1993 Jul 27;32(29):7466–7474. doi: 10.1021/bi00080a018. [DOI] [PubMed] [Google Scholar]
- Kyte J., Doolittle R. F. A simple method for displaying the hydropathic character of a protein. J Mol Biol. 1982 May 5;157(1):105–132. doi: 10.1016/0022-2836(82)90515-0. [DOI] [PubMed] [Google Scholar]
- Larder B. A., Purifoy D. J., Powell K. L., Darby G. Site-specific mutagenesis of AIDS virus reverse transcriptase. 1987 Jun 25-Jul 1Nature. 327(6124):716–717. doi: 10.1038/327716a0. [DOI] [PubMed] [Google Scholar]
- Maxfield F. R., Scheraga H. A. Status of empirical methods for the prediction of protein backbone topography. Biochemistry. 1976 Nov 16;15(23):5138–5153. doi: 10.1021/bi00668a030. [DOI] [PubMed] [Google Scholar]
- Metzger W., Hermann T., Schatz O., Le Grice S. F., Heumann H. Hydroxyl radical footprint analysis of human immunodeficiency virus reverse transcriptase-template.primer complexes. Proc Natl Acad Sci U S A. 1993 Jul 1;90(13):5909–5913. doi: 10.1073/pnas.90.13.5909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ollis D. L., Brick P., Hamlin R., Xuong N. G., Steitz T. A. Structure of large fragment of Escherichia coli DNA polymerase I complexed with dTMP. 1985 Feb 28-Mar 6Nature. 313(6005):762–766. doi: 10.1038/313762a0. [DOI] [PubMed] [Google Scholar]
- Pearson W. R., Lipman D. J. Improved tools for biological sequence comparison. Proc Natl Acad Sci U S A. 1988 Apr;85(8):2444–2448. doi: 10.1073/pnas.85.8.2444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peliska J. A., Benkovic S. J. Mechanism of DNA strand transfer reactions catalyzed by HIV-1 reverse transcriptase. Science. 1992 Nov 13;258(5085):1112–1118. doi: 10.1126/science.1279806. [DOI] [PubMed] [Google Scholar]
- Poch O., Sauvaget I., Delarue M., Tordo N. Identification of four conserved motifs among the RNA-dependent polymerase encoding elements. EMBO J. 1989 Dec 1;8(12):3867–3874. doi: 10.1002/j.1460-2075.1989.tb08565.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rost B., Sander C. Prediction of protein secondary structure at better than 70% accuracy. J Mol Biol. 1993 Jul 20;232(2):584–599. doi: 10.1006/jmbi.1993.1413. [DOI] [PubMed] [Google Scholar]
- Schatz O., Cromme F. V., Grüninger-Leitch F., Le Grice S. F. Point mutations in conserved amino acid residues within the C-terminal domain of HIV-1 reverse transcriptase specifically repress RNase H function. FEBS Lett. 1989 Nov 6;257(2):311–314. doi: 10.1016/0014-5793(89)81559-5. [DOI] [PubMed] [Google Scholar]
- Sobol R. W., Suhadolnik R. J., Kumar A., Lee B. J., Hatfield D. L., Wilson S. H. Localization of a polynucleotide binding region in the HIV-1 reverse transcriptase: implications for primer binding. Biochemistry. 1991 Nov 5;30(44):10623–10631. doi: 10.1021/bi00108a004. [DOI] [PubMed] [Google Scholar]
- Sousa R., Chung Y. J., Rose J. P., Wang B. C. Crystal structure of bacteriophage T7 RNA polymerase at 3.3 A resolution. Nature. 1993 Aug 12;364(6438):593–599. doi: 10.1038/364593a0. [DOI] [PubMed] [Google Scholar]
- Yadav P. N., Yadav J. S., Arnold E., Modak M. J. A computer-assisted analysis of conserved residues in the three-dimensional structures of the polymerase domains of Escherichia coli DNA polymerase I and HIV-1 reverse transcriptase. J Biol Chem. 1994 Jan 7;269(1):716–720. [PubMed] [Google Scholar]