Abstract
Toll/interleukin-1 receptor (TIR) domain-containing proteins play important roles in defense against pathogens in both animals and plants, connecting the immunity signaling pathways via a chain of specific protein–protein interactions. Among them is SARM, the only TIR domain-containing adaptor that can negatively regulate TLR signaling. By extensive phylogenetic analysis, we show here that SARM is closely related to bacterial proteins with TIR domains, suggesting that this family has a different evolutionary history from other animal TIR-containing adaptors, possibly emerging via a lateral gene transfer from bacteria to animals. We also show evidence of several similar, independent transfer events, none of which, however, survived in vertebrates. An evolutionary relationship between the animal SARM adaptor and bacterial proteins with TIR domains illustrates the possible role that bacterial TIR-containing proteins play in regulating eukaryotic immune responses and how this mechanism was possibly adapted by the eukaryotes themselves.
Keywords: Toll-like receptor, host–commensal interaction, host–pathogen interaction, lateral gene transfer, commensal microflora, innate immunity
1. Introduction
In humans, Toll-like receptors (TLRs) play a critical role in defense against microbial infection, initiating the host innate immune response upon binding microbe-derived molecules (Medzhitov, 2007; West et al., 2006). TLRs use a C-terminal TIR protein–protein interaction domain to connect to downstream adaptor molecules. The TIR domain is also present in the interleukin-1 receptor (IL-1R) family, where it plays a similar role. The signal is carried further by TIR-domain containing adaptors, with receptor TIR domains interacting directly with adaptor TIR domains. Five TIR-containing adaptors have been discovered in human, including four positive regulators—myeloid differentiation factor 88 (MyD88), TIR domain-containing adaptor protein (TIRAP, also known as MAL), TIR domain-containing adaptor inducing interferon-β (TRIF), TRIF-related adaptor molecule (TRAM)—and one negative regulator—sterile α and HEAT-Armadillo motifs containing protein (SARM) (O'Neill and Bowie, 2007). TIR domains are also present in plant pathogen resistance (R) proteins, which elicit hypersensitive responses against pathogen infection, but it is not clear what their downstream targets are (Burch-Smith and Dinesh-Kumar, 2007).
TIR domain-containing proteins have been also found in bacteria (Koonin and Aravind, 2002), and, recently, several of them, mostly from pathogenic bacteria, have been studied in detail. TlpA from Salmonella enterica serovar enteritidis (Newman et al., 2006), TcpC from uropathogenic Escherichia coli strain CFT073 (Cirl et al., 2008), TcpB from Brucella melitensis 16M (Cirl et al., 2008), and Btp1 from Brucella melitensis biovar Abortus 2308 (Salcedo et al., 2008) can all interfere with the host TLR signaling, thus forming a new class of virulence factors. A number of questions arise from these observations. How widely is this interfering mechanism used by bacteria? Is it strictly a virulence mechanism or could it represent a more general mechanism of interaction between the plant or animal hosts and their indigenous microbiota? What is the relationship between TIR-containing proteins from different kingdoms? To address these questions, we searched the NCBI non-redundant (nr) protein database and various metagenomic protein datasets for homologs of well-characterized animal TIR domains and performed a detailed phylogenetic analysis of the TIR family in all kingdoms of life, as well as a structural comparison of the recently solved representatives. Our results show that the TIR family had a complicated evolutionary history that included several independent bacteria-eukaryotes lateral gene transfer events, and moreover, our findings suggest that bacterial TIR domain-containing proteins may play important roles in interactions between bacteria and eukaryotes.
2. Materials and methods
2.1. Database search and sequence analysis
The non-redundant (nr) protein database was downloaded from the NCBI FTP site. TIR domain-containing proteins were identified using a cascade of PSI-BLAST searches (Altschul et al., 1997) against the nr protein database by using known human, Drosophila, or C. elegans TIR domain amino acid sequences as seeds. Up to five iterations of PSI-BLAST were run, and proteins with an e-value below 0.005 were taken for further consideration. The selected proteins were then checked by reciprocal BLAST analysis, Pfam protein searches (Bateman et al., 2004), and Conserved Domain Search (CD-Search) (Marchler-Bauer and Bryant, 2004).
Similar strategies were used to identify the TIR domain-containing proteins from human gut (Gill et al., 2006; Kurokawa et al., 2007), soil (Tringe et al., 2005), and global ocean (Venter et al., 2004; Rusch et al., 2007) metagenomic datasets. Taxonomic assignment of the retrieved TIR-containing metagenomic proteins was performed according to the best-hit pairs in the BLASTP analysis against the NCBI nr protein database. To tentatively assign the taxonomic origins of the metagenomic proteins, we adopted a 90% BLASTP identity threshold and the hit length coverage should be more than 60% of the query sequence.
The SEED database (Overbeek et al., 2005) was used to check the gene neighborhood information for bacterial proteins identified in this analysis.
2.2. Multiple sequence alignments, phylogeny reconstructions, and cluster analysis
The extend of the TIR domain for the purpose of multiple alignments was defined by the Pfam 21.0 model of the TIR domain (Bateman et al., 2004). Multiple sequence alignments were produced by MAFFT 6.240 (localpair, maxiterate 1000) (Katoh et al., 2005). Multiple sequence alignment columns with a gap in more than 50% of sequences were deleted and not used in further analysis. Phylogenetic trees were calculated using FastME 1.1 (Desper and Gascuel, 2002) and RAxML 7.0.4 (Stamatakis, 2006), with the best-fit model inferred by ProtTest (Abascal et al., 2005). TreeDyn 198.3 (Chevenet et al., 2006) was used for tree visualization. All sequence, alignment, and phylogeny files are available upon request.
Cluster analysis of all retrieved TIR domains was based on pairwise sequence similarities using the clustering program CLANS (Frickey and Lupas, 2004) with a P-value cut off of 1e-7.
2.3. Multiple structural alignment
Partial Order Structure Alignment (POSA) (Ye and Godzik, 2005) was used to build the multiple structural alignment of the available TIR domain structures, including bacteria TIR (from Paracoccus denitrificans, PDB 3H16 (Chan et al., 2009)), plant TIR (from Arabidopsis thaliana, PDB 3JRN (Chan et al., 2010)), and animal TIRs (from human: TLR1, PDB 1FYV (Xu et al., 2000); TLR2, PDB 1FYW (Xu et al., 2000); TLR10, PDB 2J67 (Nyman et al., 2008); IL-1RAPL, PDB 1T3G (Khan et al., 2004); MyD88, PDB 2JS7). The superimposed structures were displayed with Discovery Studio (http://accelrys.com).
3. Results
3.1. Animal SARM is closely related to bacterial TIR domain-containing proteins
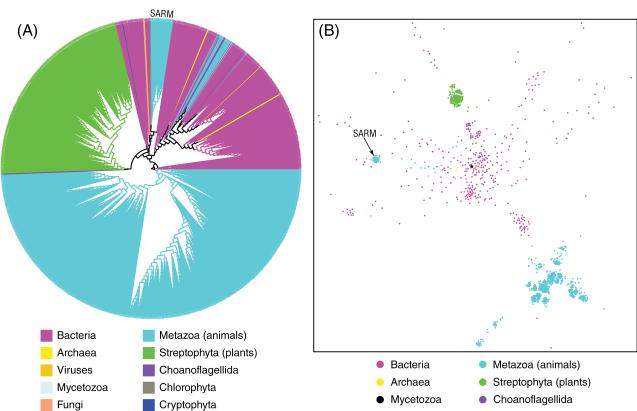
Following a protocol described in the Methods section, we identified a total of 1688 TIR domain-containing proteins, including 483 from bacteria, 11 from archaea, 1193 from eukaryotes, and 1 from viruses (a bacteriophage). The TIR domains from these proteins were aligned and used to calculate their evolutionary history, as presented in Fig. 1A. The evolutionary tree consists of three main branches, which mostly correspond to the phylogenetic division between the animal (cyan), the plant (green), and the bacteria (magenta) branches (see the discussion below for the exceptions to this rule). The topology of the tree suggests a very ancient origin of this family, followed by massive lineage-specific expansions in each branch. Internal structure of the plant and the animal branches of the tree shows early emergence of few major lineages of TIR domains, followed mostly by speciation events. In contrast, the bacterial branch of the tree has clearly evolved by multiple lateral transfer events (see Fig. 2 and the discussion below for more details).
Fig. 1.
Phylogeny and CLANS clustering of the TIR protein family. (A) Phylogeny of the TIR protein family. On the phylogenetic tree, branches are assigned different colors according to different taxonomic groups. (B) Two-dimensional CLANS clustering of the TIR domain sequences. TIR domains from the animal SARM adaptors are illustrated. The TIR domain of TirA from the social amoeba Dictyostelium discoideum, a member of Mycetozoa, is shown as a black dot in the middle of the picture. For clarity, only six taxonomic groups are shown.
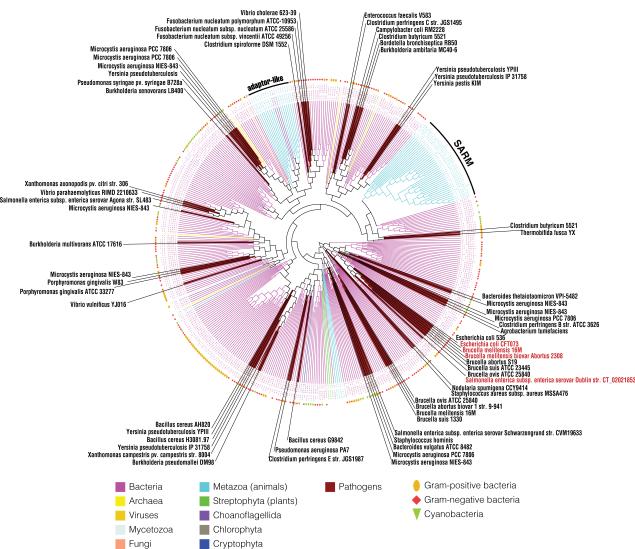
Fig. 2.
Phylogeny of the bacterial TIR branch. The branch of the bacterial TIR from Fig. 1A is selected for a closer view. Clades of pathogenic bacteria are colored in brown with species name noted. The four experimentally studied TIR-containing proteins from pathogens (TcpC from Escherichia coli strain CFT073, TcpB from Brucella melitensis 16M, Btp1 from Brucella melitensis biovar Abortus 2308, and TlpA from Salmonella enteric) are labeled in red. The classification of each bacterium is shown in the outer concentric circle with Gram-positive bacteria in the gold oval, Gram-negative bacteria in the brown diamond, and cyanobacteria in the olive triangle. TIR domains from the family of animal SARM adaptors, as well as several other adaptor-like sequences from invertebrates and choanoflagellates, are clustered with bacterial TIRs.
The tree presented in Fig. 1A is built on a multiple alignment of a very divergent protein family. For this reason, the statistical significance of some specific branches is not high, and trees with slightly different topologies can also be built. However, the main observations discussed here are supported by all alternative tree topologies we investigated. To further verify these observations, we used the protein clustering program CLANS (Frickey and Lupas, 2004), which exploits all-against-all similarity, so it does not depend on the approximations inherent in building the phylogenetic tree. CLANS presents the internal structure of a protein family as a two dimensional graph. While CLANS result cannot be interpreted in the context of phylogeny, it can support (as in this case) the overall features of the phylogenetic tree. The plant and animal TIR domains form very tight clusters (green and cyan clusters on the top and bottom of Fig. 1B, respectively).
On closer inspection it can be seen that several eukaryotic TIR domain subfamilies are found within the bacterial branch of the tree (Fig. 2). Most notable is the presence of the TIR domains from the family of animal SARM innate immunity adaptors deep in the bacterial branch of the tree. This is also seen in the clustering results (Fig. 1B), where the SARM family forms a small patch (center left in Fig. 1B), clearly separate from the main cluster of animal TIR domains (bottom right in Fig. 1B). This suggests that the evolutionary history of the animal SARM family is different from that of other adaptors, with this family likely representing a lateral gene transfer between bacteria and animals. A multiple sequence alignment of selected TIR domains from animal SARM adaptors and bacterial TIR-containing proteins that are close in the TIR phylogeny (Fig. 1A and Fig. 2) are presented in Supplementary Figure 1 to illustrate the similarity between them at amino acid level. In humans, SARM is the only TIR adaptor that negatively regulates TLR signaling, blocking TRIF-dependent NF-κB and interferon-regulatory factor 3 (IRF3) activation (Carty et al., 2006). It can also inhibit TRIF- and MyD88-mediated activator protein 1 (AP-1) activation and p38 phosphorylation (Peng et al., 2010). Amphioxus SARM—bbtSARM—can attenuate the TLR signaling via interaction with amphioxus MyD88 and tumor necrosis receptor associated factor 6 (TRAF6) (Yuan et al., 2010). CrSARM, the ortholog of human SARM from horseshoe crab, can inhibit the TLR signaling pathway via TRIF, suggesting the conservation of the negative regulatory function of SARM from arthropod to human (Belinda et al., 2008). It is intriguing to speculate that the negative regulatory mechanism of SARM may reflect evolutionary conserved function of bacterial TIR-containing proteins of inhibiting their hosts' immune response.
3.2. SARM is not alone–other animal TIR-containing proteins in bacterial branch
Besides the SARM family, several other adaptor-like proteins from invertebrate- and choanoflagellate-specific families are found on the bacterial TIR branch of the phylogenetic tree (Fig. 2), including: reversed SARM-like proteins from sea urchin, sea anemone, and Trichoplax adhaerens (Srivastava et al., 2008); MyD88-like proteins form sea urchin; and CARD–TIR domains containing protein from Trichoplax adhaerens. Again, this observation is supported by the clustering results, where these eukaryotic TIR domains (cyan dots) are found scattered among the bacterial ones (magenta) (Fig. 1B). This suggests that there might have been several independent lateral gene transfer events involving TIR domain containing proteins between bacteria and eukaryotes. However, these families have no orthologs in vertebrates, and none were characterized by experiment.
Another interesting finding is that the TIR-containing protein TirA from the social amoeba Dictyostelium discoideum is also clustered with bacterial TIR domains (the black dot in the middle of Fig. 1B). This soil-dwelling amoeba is a single-celled organism but aggregates to form a migrating slug when starved. A gene coding for the TirA protein is required for Dictyostelium to feed on live bacteria and to defend it against bacterial pathogens (Chen et al., 2007). While the specific function of the TirA protein is unknown, our results suggest it is likely of bacterial origin.
3.3. The ubiquity of TIR domain-containing proteins in bacteria
Almost 2000 TIR domains are currently identified, and it is likely that additional distant homologs could be retrieved with less-restrictive significance thresholds and that the real number of TIR domain proteins, especially in bacteria in which sequences are more divergent, is much larger that the ~500 proteins we report here.
In bacteria, TIR domain-containing proteins are usually found in a single copy in a genome. In completely sequenced bacterial genomes that have TIR-containing proteins, 68% of them have only one copy. There are only a few species in which multiple copies are found, such as Gram-positive Frankia alni ACN14a with 25 copies, Gram-negative Sorangium cellulosum `So ce 56' with 10 copies, or cyanobacteria Cyanothece sp. PCC 7424 with 17 copies. Interestingly, many microbes containing multiple copies of TIR domain proteins are plant symbionts. A detailed list of bacterial TIR-containing proteins can be found in Supplementary table 1.
It is interesting to note that the domain architectures of TIR domain-containing proteins in bacteria are very diverse. Some of them are similar to those found in proteins involved with immune response in animals and plants, such as the LRRs–TIR domain combination (similar to animal Toll-like receptors) and the TIR–NBARC–TPRs and TIR–NACHT–WD40s combinations (similar to plant R proteins). In general, in bacterial TIR-containing proteins, TIR domains are often associated with various repeat domains, including LRR (Leucine rich repeat), WD40 (Trp-Asp repeat), TPR (Tetratricopeptide repeat), Sel1 (Sel1 repeat), HEAT_PBS (PBS lyase HEAT-like repeat), and Ank (Ankyrin repeat).
3.4. TIR domain-containing proteins from pathogenic bacteria
Among the 483 bacterial TIR domain-containing proteins, 65 come from pathogenic bacteria using the NCBI Entrez Genome Project classification—including 54 from human or animal pathogens; 6 from opportunistic human or animal pathogens; 4 from plant pathogens; and 1 from a shared pathogen of human and plant (details in Table 1). On the TIR domain phylogenetic tree, proteins from pathogenic bacteria form several distinct small branches (shown with the brown background in Fig. 2). It is interesting to note that four of these clusters, containing, among others, proteins from several subspecies of Clostridia, Yersinia, and Thermobifida, are grouped together with the SARM group (Fig. 2). In the CLANS clustering result, the TIR-containing protein from bacterium Thermobifida fusca YX, whose spores are known to cause allergic respiratory diseases called mushroom worker disease and farmer's lung, is also one of the closest bacteria grouped together with the animal SARM group. Unfortunately, none of these proteins were studied by experiment, and the only ones that were, (TlpA, TcpC, TcpB, and Btp1, labeled in red in Fig. 2) belong to a branch that also contains several proteins from non-pathogenic bacteria and is phylogenetically more distant from the SARM group.
Table 1.
Distribution of the TIR domain-containing proteins in pathogenic bacteria.
| Bacteria species | Gi number of TIR domain-containing protein | |
|---|---|---|
| Pathogens in human, animal | Bordetella bronchiseptica RB50 | 33599474 |
| Bacillus cereus AH820 | 166997552 | |
| Bacillus cereus G9842 | 168142494 | |
| Bacillus cereus H3081.97 | 166980602 | |
| Brucella abortus biovar 1 str. 9-941 | 62289692 | |
| Brucella abortus S19 | 189023503 | |
| Brucella melitensis 16M | 17987499, 17987957 | |
| Brucella melitensis biovar Abortus 2308 | 82699179 | |
| Brucella ovis ATCC 25840 | 148559127, 148559442 | |
| Brucella suis 1330 | 23501622 | |
| Brucella suis ATCC 23445 | 163842531 | |
| Burkholderia ambifaria MC40-6 | 172063457 | |
| Burkholderia multivorans ATCC 17616 | 161524091 | |
| Campylobacter coli RM2228 | 57504994 | |
| Clostridium butyricum 5521 | 182418236, 182418248 | |
| Clostridium perfringens B str. ATCC 3626 | 168210190 | |
| Clostridium perfringens C str. JGS1495 | 169342429 | |
| Clostridium perfringens E str. JGS1987 | 168204813 | |
| Clostridium spiroforme DSM 1552 | 169350767 | |
| Escherichia coli 536 | 110642087 | |
| Escherichia coli CFT073 | 26248250 | |
| Fusobacterium nucleatum polymorphum ATCC-10953 | 167007634 | |
| Fusobacterium nucleatum subsp. nucleatum ATCC 25586 | 19704057 | |
| Fusobacterium nucleatum subsp. vincentii ATCC 49256 | 34762589 | |
| Microcystis aeruginosa PCC 7806 | 159026969, 159029511, 159029883, 159030587 | |
| Microcystis aeruginosa NIES-843 | 166363533, 166364872, 166364873, 166366472, 166368365, 166368885 | |
| Nodularia spumigena CCY9414 | 119512412 | |
| Porphyromonas gingivalis ATCC 33277 | 188995660 | |
| Porphyromonas gingivalis W83 | 34541475 | |
| Salmonella enterica subsp. enterica serovar Agona str. SL483 | 168219791 | |
| Salmonella enterica subsp. enterica serovar Dublin str. CT_02021853 | 167560111 | |
| Salmonella enterica subsp. enterica serovar Schwarzengrund str. CVM19633 | 167982220 | |
| Staphylococcus aureus subsp. aureus MSSA476 | 49484950 | |
| Staphylococcus hominis | 18148891 | |
| Thermobifida fusca YX | 72161132 | |
| Vibrio cholerae 623-39 | 153829970 | |
| Vibrio parahaemolyticus RIMD 2210633 | 28898586 | |
| Yersinia pestis KIM | 22126310 | |
| Yersinia pseudotuberculosis IP 31758 | 153947653, 153948345 | |
| Yersinia pseudotuberculosis YPIII | 170024518, 170025361 | |
| Yersinia pseudotuberculosis | 49658892 | |
|
| ||
| Opportunistic pathogens in human, animal | Bacteroides thetaiotaomicron VPI-5482 | 29348412 |
| Bacteroides vulgatus ATCC 8482 | 150006151 | |
| Burkholderia pseudomallei DM98 | 167717459 | |
| Enterococcus faecalis V583 | 29376482 | |
| Pseudomonas aeruginosa PA7 | 152988748 | |
| Vibrio vulnificus YJ016 | 37680110 | |
|
| ||
| Pathogens in plants | Agrobacterium tumefaciens | 10955119 |
| Pseudomonas syringae pv. syringae B728a | 66043375 | |
| Xanthomonas axonopodis pv. citri str. 306 | 21244022 | |
| Xanthomonas campestris pv. campestris str. 8004 | 66571812 | |
|
| ||
| Pathogen in both human and plant | Burkholderia xenovorans LB400 | 91781261 |
3.5. Lateral gene transfer of TIR domain-containing proteins
Phylogenetic analysis shows that the clustering pattern of the bacterial TIR domains does not follow the phylogenetic clustering, with Gram-positive bacteria, Gram-negative bacteria, and cyanobacteria often mixed together in the same cluster. This strongly suggests that bacterial TIR-containing genes are distributed by lateral gene transfer. This is also supported by the observation that many bacterial TIR-containing proteins are found in genomic regions with phage origins, close to genes of phage integrase, site-specific recombinases, and transposases. The positions of the animal SARM group, along with other eukaryotic TIR proteins within the bacterial branch (see Fig. 1 and Fig. 2), implies that at least two independent lateral gene transfer events between bacteria and eukaryotes took place: the lateral gene transfer of the SARM group between bacteria and the ancestor of ecdysozoa (molting animals, which include arthropods and nematodes) and deuterostomes, which is estimated to have occurred no later than 728 million years ago (Hedges et al., 2006); and the transfer of the other eukaryotic adaptor-like TIR proteins in Fig. 2 between bacteria and the ancestor of choanoflagellates and animals, which is estimated to have happened no later than 1020 million years ago (Hedges et al., 2006).
3.6. Comparative metagenomic analyses of TIR domain-containing proteins in human gut, global ocean, and soil metagenomes
Proteins containing TIR domains were also found in both human gut (Gill et al., 2006; Kurokawa et al., 2007) and environmental (Venter et al., 2004; Tringe et al., 2005; Rusch et al., 2007) metagenomics datasets. For instance, a total of 25 TIR domain-containing proteins were identified in the 15 human gut metagenomics datasets searched (see Supplementary table 2 for details). Three proteins could be assigned to particular genus with a 90% BLASTP identity threshold, including one with 99% sequence identity to TcpC (Cirl et al., 2008) from uropathogenic Escherichia coli strain CFT073, and another two similar to proteins from members of the normal human gut microbiota—Clostridium bolteae ATCC BAA-613 and Faecalibacterium prausnitzii M21/2, while all others represent novel proteins.
TIR-containing proteins are also found in ocean and soil microbiota. The total number of TIR domain-containing proteins retrieved from the global ocean and the soil metagenomes are 41 and 53 respectively (Supplementary table 2), but none of them can be assigned to a specific genus. The relative abundance of TIR-containing proteins in different metagenomes as compared to the sample size is shown in Table 2, suggesting that the TIR domain-containing proteins are most abundant in soil microbiota, while being rare in ocean microbiota. This observation is consistent with the relative abundance of TIR domain proteins in plant symbionts, and could be explained by a relatively high frequency of interactions between bacteria and eukaryotes in the soil environment.
Table 2.
Relative abundance of TIR domain-containing proteins in different metagenomes.
| Metagenome | TIR-containing protein count | Metagenome protein count | Relative abundance |
|---|---|---|---|
| Human Gut Metagenome | 25 | 709051 | 1 |
| Ocean Metagenome | 41 | 6115750 | 0.19 |
| Soil Metagenome | 53 | 184374 | 8.15 |
The richness of TIR domain-containing proteins in each metagenome is normalized by its database size. The relative abundance of TIR domain-containing proteins in human gut metagenome is set to one.
3.7. Structural analysis of TIR domains
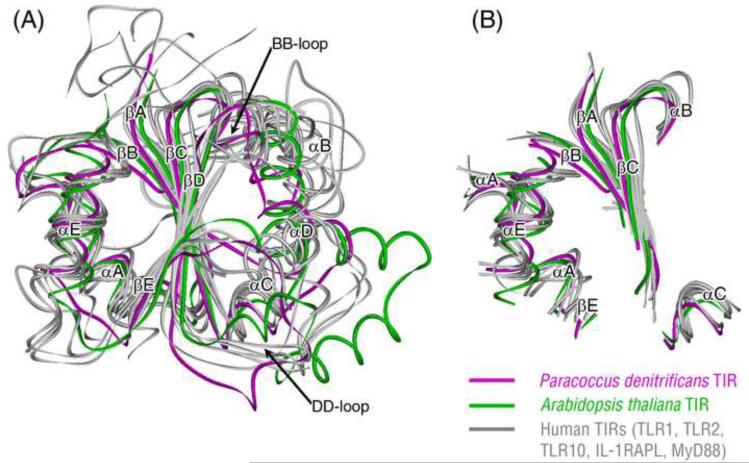
Structural information is available for representatives from all major branches of the TIR family tree. Here we compare the structure of bacteria TIR (from Paracoccus denitrificans (Chan et al., 2009)) to both plant (from Arabidopsis thaliana (Chan et al., 2010)) and animal (from human: TLR1 (Xu et al., 2000), TLR2 (Xu et al., 2000), TLR10 (Nyman et al., 2008), IL-1RAPL (Khan et al., 2004), and MyD88) TIRs. Partial Order Structure Alignment (POSA) (Ye and Godzik, 2005) was used to build the multiple structural alignment of the above TIRs, and the superimposed structures are shown in Fig. 3A. All TIR structures belong to the flavodoxin-like fold according to the SCOP (Structural Classification of Proteins) classification, with a central five-stranded parallel β-sheet (βA–βE) surrounded by a total of 4–5 α helices (αA–αE) (Fig. 3A). The conserved structural core of TIR domains include the following segments (βA, αA, βB, βC, αE, part of the αB, αC and βE) (Fig. 3B), with an average RMSD of 2.78 Å. With the representation of such a common core, the TIR structure can be divided into two parts—the two α helices conserved side (left portion of Fig. 3A) and the three α helices divergent side (right portion of Fig. 3A). Germ-line mutagenesis studies in mice implicated that the BB-loop is used for receptor:adaptor interaction, whereas the αE-helix supports adaptor:adaptor and receptor:receptor oligomerization (Jiang et al., 2006). In our superimposed TIR structures, the BB-loop belongs to the divergent side, while the αE-helix resides in the conserved side, so these two sides may play separate roles in hetero- versus homotypic TIR domain oligomerization. Structural comparison shows that the BB-loops are most divergent between different TIR groups, with Paracoccus's TIR, human MyD88 TIR, and human receptors TIRs adopting three different conformations in this region. Both BB-loop and DD-loop have been suggested as major contributors to the TIR dimer interface (Tao et al., 2002; Ronni et al., 2003; Gautam et al., 2006; Jiang et al., 2006; Nyman et al., 2008; Chan et al., 2009), so it is possible that these two loops can adopt different conformations in different TIR complexes. The overall structural similarity between bacterial and eukaryotic TIR domains along with the available experimental data of bacterial TIR proteins suggests that this domain could play important roles in interactions between bacteria and their eukaryotic host cells.
Fig. 3.
Structural comparison of various TIR domains. (A) Superposition of the structures of the bacterial TIR domain (magenta), the plant TIR domain (green), and the human TIR domains (gray). (B) The conserved structural core of the superimposed TIR domains shown in (A).
4. Discussion
The evolutionary history of the TIR protein family shows that TIR domains generally form separate plant, animal, and bacterial/archeal branches, suggesting an ancient origin of this domain. However, the group of animal SARM adaptors, as well as a few other small groups present only in invertebrates, are found on the bacterial branch of the phylogenetic tree, suggesting at least two later gene transfer events between bacteria and animals. Among the five TIR domain-containing adaptors, SARM is the only negative regulator of TLR signaling, which opens the interesting possibility that the bacterial TIR-containing proteins may share the negative regulatory mechanism of SARM to inhibit their eukaryotic hosts' immune response.
Experimentally characterized bacterial TIR domain proteins, including TlpA from Salmonella, TcpC from E. coli, TcpB from Brucella, and PdTIR from Paracoccus, were reported to interact with the host MyD88 (Newman et al., 2006; Cirl et al., 2008; Chan et al., 2009) and TIRAP (Radhakrishnan et al., 2009; Sengupta et al., 2010). However, they all belong to a very small cluster, which includes homologs from several other pathogenic and non-pathogenic bacteria (the small cluster with four red-labeled pathogenic bacteria in Fig. 2). All members in this cluster have conserved domain structures, consisting of the TIR domain and a coiled-coil N-terminal domain. Therefore, it is not clear how far their function is conserved in other bacterial TIR domains. The specific function of other bacterial TIR proteins, including other pathogenic bacterial TIR clusters, especially for those clustering together with the SARM group, still needs to be tested.
With the exception of four proteins from human pathogens mentioned earlier, none of the bacterial TIR-containing proteins have been experimentally characterized, and in particular we don't know if they interact with the eukaryotic TIR signaling proteins. However, an indirect argument can be made that these proteins may have no intrinsic role in bacteria and therefore could be specifically targeting eukaryotic signaling pathways. First, according to our genomic analysis results, most (68%) of the bacterial genomes that contain a TIR domain protein have only a single such protein. Since its role is to form complexes with other TIR domains, such proteins have no obvious partners to form complexes with. In addition, our gene neighborhood analysis results show that TIR-containing genes from bacteria are also often co-located with gene members of the type IV secretion system, suggesting that they may be secreted and thus play a role in interactions between bacteria and their environment.
TIR-containing proteins are present in various environments, including human gut, ocean, and soil microbiomes. From the 25 TIR-containing proteins identified in human gut metagenomes, only one originated from known pathogenic bacteria. Many TIR proteins are present in members of the normal human gut microbiota (see more details in Supplementary table1), which implies that the interactions between TIR-containing proteins could also happen between the host and its commensal microflora. It is intriguing to speculate about the possibly important role of TIR-containing proteins among human commensal bacteria, which are known to stimulate the development of the host immune system (Mazmanian et al., 2005) and the recognition of commensal bacteria by host TLRs is required for intestinal homeostasis (Rakoff-Nahoum et al., 2004). Therefore, a continuous low level of activity of the innate immune system is essential to host health, and the TIR-containing proteins in commensal bacteria may help maintain such simulation in the host at an appropriate level.
Some bacteria, including many plant symbionts, have large numbers of TIR-containing proteins, from 10 to 25 copies (Supplementary table1). It will be interesting to test if these proteins can interact with their host plants' TIR-containing R proteins and what will happen if all TIR-containing genes are removed from these plant commensal bacteria.
With superposition of multiple TIR structures and illustration of their common core, the TIR structure can be divided into a conserved and a divergent part (Fig. 3). This partition is in agreement with the model suggested by Jiang et al. in which a TIR domain may use two separate regions for homotypic (adaptor:adaptor or receptor:receptor) and heterotypic (receptor:adaptor) interactions (Jiang et al., 2006). Because the structure of a bacterium TIR in complex with a host TIR is not available, further experimentation is needed to clarify the exact binding mechanism between them.
Here we show that not only pathogenic bacteria, but a broad range of non-pathogenic, both free-living and commensal bacteria also contain TIR domain proteins. The sheer size and broad distribution of such proteins in bacteria challenge the view of their primary function as virulence factors. This result is in agreement with the recent opinion raised by Spear et al (Spear et al., 2009). Considering the available experimental data for pathogenic TIRs (Newman et al., 2006; Cirl et al., 2008; Salcedo et al., 2008; Radhakrishnan et al., 2009; Sengupta et al., 2010) and all other available evidence we hypothesize that bacterial TIR domain-containing proteins may play important roles in commensal or mutualistic interactions between bacteria and eukaryotes. However, with so little experimental data available, the puzzle of bacterial TIR-containing proteins is still mostly unresolved.
Supplementary Material
Acknowledgements
We thank Jaime Pascual for very helpful discussions regarding this work and Marcin Feder for initial sequence searches and alignments. This work was supported by grant GM076221 (JCMM) from the National Institutes of Health.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Abascal F, Zardoya R, Posada D. ProtTest: selection of best-fit models of protein evolution. Bioinformatics. 2005;21:2104–2105. doi: 10.1093/bioinformatics/bti263. [DOI] [PubMed] [Google Scholar]
- Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bateman A, Coin L, Durbin R, Finn RD, Hollich V, Griffiths-Jones S, et al. The Pfam protein families database. Nucleic Acids Res. 2004;32:D138–141. doi: 10.1093/nar/gkh121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belinda LW, Wei WX, Hanh BT, Lei LX, Bow H, Ling DJ. SARM: a novel Toll-like receptor adaptor, is functionally conserved from arthropod to human. Mol Immunol. 2008;45:1732–1742. doi: 10.1016/j.molimm.2007.09.030. [DOI] [PubMed] [Google Scholar]
- Burch-Smith TM, Dinesh-Kumar SP. The functions of plant TIR domains. Sci STKE. 2007;2007:pe46. doi: 10.1126/stke.4012007pe46. [DOI] [PubMed] [Google Scholar]
- Carty M, Goodbody R, Schroder M, Stack J, Moynagh PN, Bowie AG. The human adaptor SARM negatively regulates adaptor protein TRIF-dependent Toll-like receptor signaling. Nat Immunol. 2006;7:1074–1081. doi: 10.1038/ni1382. [DOI] [PubMed] [Google Scholar]
- Chan SL, Low LY, Hsu S, Li S, Liu T, Santelli E, et al. Molecular Mimicry in Innate Immunity: CRYSTAL STRUCTURE OF A BACTERIAL TIR DOMAIN. J Biol Chem. 2009;284:21386–21392. doi: 10.1074/jbc.C109.007591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan SL, Mukasa T, Santelli E, Low LY, Pascual J. The crystal structure of a TIR domain from Arabidopsis thaliana reveals a conserved helical region unique to plants. Protein Sci. 2010;19:155–161. doi: 10.1002/pro.275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen G, Zhuchenko O, Kuspa A. Immune-like phagocyte activity in the social amoeba. Science. 2007;317:678–681. doi: 10.1126/science.1143991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chevenet F, Brun C, Banuls AL, Jacq B, Christen R. TreeDyn: towards dynamic graphics and annotations for analyses of trees. BMC Bioinformatics. 2006;7:439. doi: 10.1186/1471-2105-7-439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cirl C, Wieser A, Yadav M, Duerr S, Schubert S, Fischer H, et al. Subversion of Toll-like receptor signaling by a unique family of bacterial Toll/interleukin-1 receptor domain-containing proteins. Nat Med. 2008;14:399–406. doi: 10.1038/nm1734. [DOI] [PubMed] [Google Scholar]
- Desper R, Gascuel O. Fast and accurate phylogeny reconstruction algorithms based on the minimum-evolution principle. J Comput Biol. 2002;9:687–705. doi: 10.1089/106652702761034136. [DOI] [PubMed] [Google Scholar]
- Frickey T, Lupas A. CLANS: a Java application for visualizing protein families based on pairwise similarity. Bioinformatics. 2004;20:3702–3704. doi: 10.1093/bioinformatics/bth444. [DOI] [PubMed] [Google Scholar]
- Gautam JK, Ashish, Comeau LD, Krueger JK, Smith MF., Jr. Structural and functional evidence for the role of the TLR2 DD loop in TLR1/TLR2 heterodimerization and signaling. J Biol Chem. 2006;281:30132–30142. doi: 10.1074/jbc.M602057200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gill SR, Pop M, Deboy RT, Eckburg PB, Turnbaugh PJ, Samuel BS, et al. Metagenomic analysis of the human distal gut microbiome. Science. 2006;312:1355–1359. doi: 10.1126/science.1124234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hedges SB, Dudley J, Kumar S. TimeTree: a public knowledge-base of divergence times among organisms. Bioinformatics. 2006;22:2971–2972. doi: 10.1093/bioinformatics/btl505. [DOI] [PubMed] [Google Scholar]
- Jiang Z, Georgel P, Li C, Choe J, Crozat K, Rutschmann S, et al. Details of Toll-like receptor:adapter interaction revealed by germ-line mutagenesis. Proc Natl Acad Sci U S A. 2006;103:10961–10966. doi: 10.1073/pnas.0603804103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh K, Kuma K, Toh H, Miyata T. MAFFT version 5: improvement in accuracy of multiple sequence alignment. Nucleic Acids Res. 2005;33:511–518. doi: 10.1093/nar/gki198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan JA, Brint EK, O'Neill LA, Tong L. Crystal structure of the Toll/interleukin-1 receptor domain of human IL-1RAPL. J Biol Chem. 2004;279:31664–31670. doi: 10.1074/jbc.M403434200. [DOI] [PubMed] [Google Scholar]
- Koonin EV, Aravind L. Origin and evolution of eukaryotic apoptosis: the bacterial connection. Cell Death Differ. 2002;9:394–404. doi: 10.1038/sj.cdd.4400991. [DOI] [PubMed] [Google Scholar]
- Kurokawa K, Itoh T, Kuwahara T, Oshima K, Toh H, Toyoda A, et al. Comparative metagenomics revealed commonly enriched gene sets in human gut microbiomes. DNA Res. 2007;14:169–181. doi: 10.1093/dnares/dsm018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchler-Bauer A, Bryant SH. CD-Search: protein domain annotations on the fly. Nucleic Acids Res. 2004;32:W327–331. doi: 10.1093/nar/gkh454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazmanian SK, Liu CH, Tzianabos AO, Kasper DL. An immunomodulatory molecule of symbiotic bacteria directs maturation of the host immune system. Cell. 2005;122:107–118. doi: 10.1016/j.cell.2005.05.007. [DOI] [PubMed] [Google Scholar]
- Medzhitov R. Recognition of microorganisms and activation of the immune response. Nature. 2007;449:819–826. doi: 10.1038/nature06246. [DOI] [PubMed] [Google Scholar]
- Newman RM, Salunkhe P, Godzik A, Reed JC. Identification and characterization of a novel bacterial virulence factor that shares homology with mammalian Toll/interleukin-1 receptor family proteins. Infect Immun. 2006;74:594–601. doi: 10.1128/IAI.74.1.594-601.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nyman T, Stenmark P, Flodin S, Johansson I, Hammarstrom M, Nordlund P. The crystal structure of the human toll-like receptor 10 cytoplasmic domain reveals a putative signaling dimer. J Biol Chem. 2008;283:11861–11865. doi: 10.1074/jbc.C800001200. [DOI] [PubMed] [Google Scholar]
- O'Neill LA, Bowie AG. The family of five: TIR-domain-containing adaptors in Toll-like receptor signalling. Nat Rev Immunol. 2007;7:353–364. doi: 10.1038/nri2079. [DOI] [PubMed] [Google Scholar]
- Overbeek R, Begley T, Butler RM, Choudhuri JV, Chuang HY, Cohoon M, et al. The subsystems approach to genome annotation and its use in the project to annotate 1000 genomes. Nucleic Acids Res. 2005;33:5691–5702. doi: 10.1093/nar/gki866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng J, Yuan Q, Lin B, Panneerselvam P, Wang X, Luan XL, et al. SARM inhibits both TRIF- and MyD88-mediated AP-1 activation. Eur J Immunol. 2010;40:1738–1747. doi: 10.1002/eji.200940034. [DOI] [PubMed] [Google Scholar]
- Radhakrishnan GK, Yu Q, Harms JS, Splitter GA. Brucella TIR Domain-containing Protein Mimics Properties of the Toll-like Receptor Adaptor Protein TIRAP. J Biol Chem. 2009;284:9892–9898. doi: 10.1074/jbc.M805458200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rakoff-Nahoum S, Paglino J, Eslami-Varzaneh F, Edberg S, Medzhitov R. Recognition of commensal microflora by toll-like receptors is required for intestinal homeostasis. Cell. 2004;118:229–241. doi: 10.1016/j.cell.2004.07.002. [DOI] [PubMed] [Google Scholar]
- Ronni T, Agarwal V, Haykinson M, Haberland ME, Cheng G, Smale ST. Common interaction surfaces of the toll-like receptor 4 cytoplasmic domain stimulate multiple nuclear targets. Mol Cell Biol. 2003;23:2543–2555. doi: 10.1128/MCB.23.7.2543-2555.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rusch DB, Halpern AL, Sutton G, Heidelberg KB, Williamson S, Yooseph S, et al. The Sorcerer II Global Ocean Sampling expedition: northwest Atlantic through eastern tropical Pacific. PLoS Biol. 2007;5:e77. doi: 10.1371/journal.pbio.0050077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salcedo SP, Marchesini MI, Lelouard H, Fugier E, Jolly G, Balor S, et al. Brucella control of dendritic cell maturation is dependent on the TIR-containing protein Btp1. PLoS Pathog. 2008;4:e21. doi: 10.1371/journal.ppat.0040021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sengupta D, Koblansky A, Gaines J, Brown T, West AP, Zhang D, et al. Subversion of innate immune responses by Brucella through the targeted degradation of the TLR signaling adapter, MAL. J Immunol. 2010;184:956–964. doi: 10.4049/jimmunol.0902008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spear AM, Loman NJ, Atkins HS, Pallen MJ. Microbial TIR domains: not necessarily agents of subversion? Trends Microbiol. 2009;17:393–398. doi: 10.1016/j.tim.2009.06.005. [DOI] [PubMed] [Google Scholar]
- Srivastava M, Begovic E, Chapman J, Putnam NH, Hellsten U, Kawashima T, et al. The Trichoplax genome and the nature of placozoans. Nature. 2008;454:955–960. doi: 10.1038/nature07191. [DOI] [PubMed] [Google Scholar]
- Stamatakis A. RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics. 2006;22:2688–2690. doi: 10.1093/bioinformatics/btl446. [DOI] [PubMed] [Google Scholar]
- Tao X, Xu Y, Zheng Y, Beg AA, Tong L. An extensively associated dimer in the structure of the C713S mutant of the TIR domain of human TLR2. Biochem Biophys Res Commun. 2002;299:216–221. doi: 10.1016/s0006-291x(02)02581-0. [DOI] [PubMed] [Google Scholar]
- Tringe SG, von Mering C, Kobayashi A, Salamov AA, Chen K, Chang HW, et al. Comparative metagenomics of microbial communities. Science. 2005;308:554–557. doi: 10.1126/science.1107851. [DOI] [PubMed] [Google Scholar]
- Venter JC, Remington K, Heidelberg JF, Halpern AL, Rusch D, Eisen JA, et al. Environmental genome shotgun sequencing of the Sargasso Sea. Science. 2004;304:66–74. doi: 10.1126/science.1093857. [DOI] [PubMed] [Google Scholar]
- West AP, Koblansky AA, Ghosh S. Recognition and signaling by toll-like receptors. Annu Rev Cell Dev Biol. 2006;22:409–437. doi: 10.1146/annurev.cellbio.21.122303.115827. [DOI] [PubMed] [Google Scholar]
- Xu Y, Tao X, Shen B, Horng T, Medzhitov R, Manley JL, Tong L. Structural basis for signal transduction by the Toll/interleukin-1 receptor domains. Nature. 2000;408:111–115. doi: 10.1038/35040600. [DOI] [PubMed] [Google Scholar]
- Ye Y, Godzik A. Multiple flexible structure alignment using partial order graphs. Bioinformatics. 2005;21:2362–2369. doi: 10.1093/bioinformatics/bti353. [DOI] [PubMed] [Google Scholar]
- Yuan S, Wu K, Yang M, Xu L, Huang L, Liu H, et al. Amphioxus SARM involved in neural development may function as a suppressor of TLR signaling. J Immunol. 2010;184:6874–6881. doi: 10.4049/jimmunol.0903675. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.