Abstract
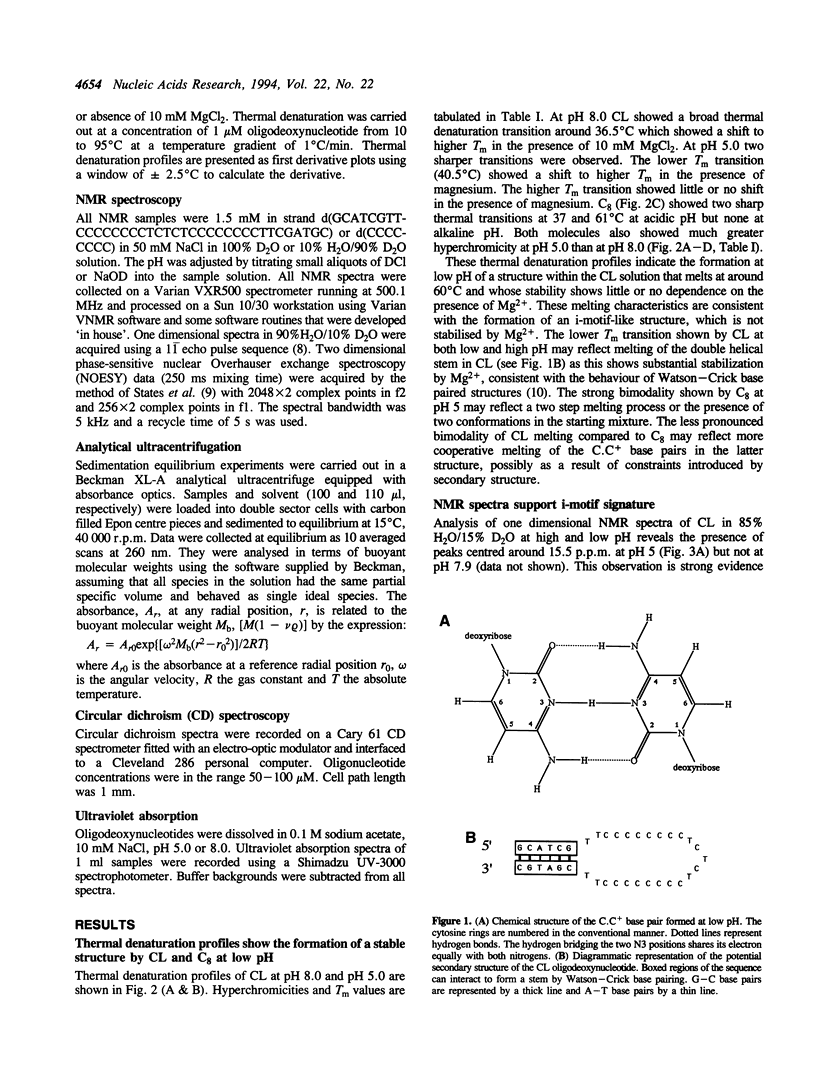
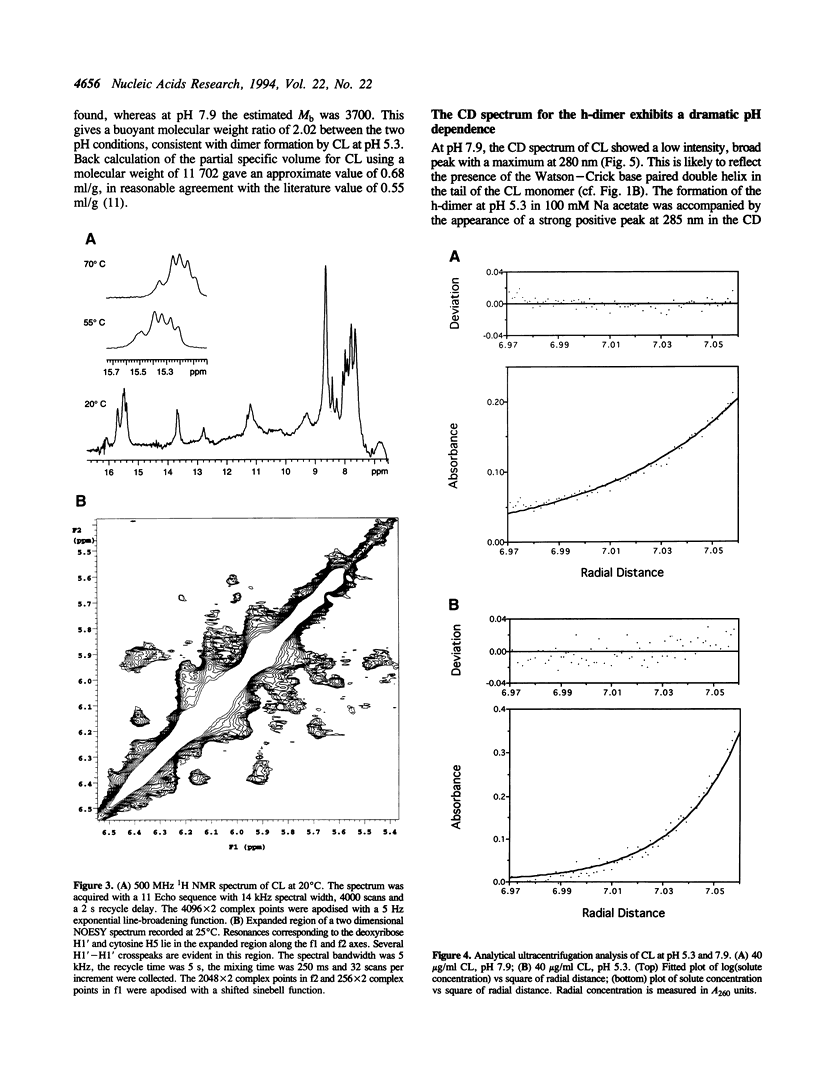
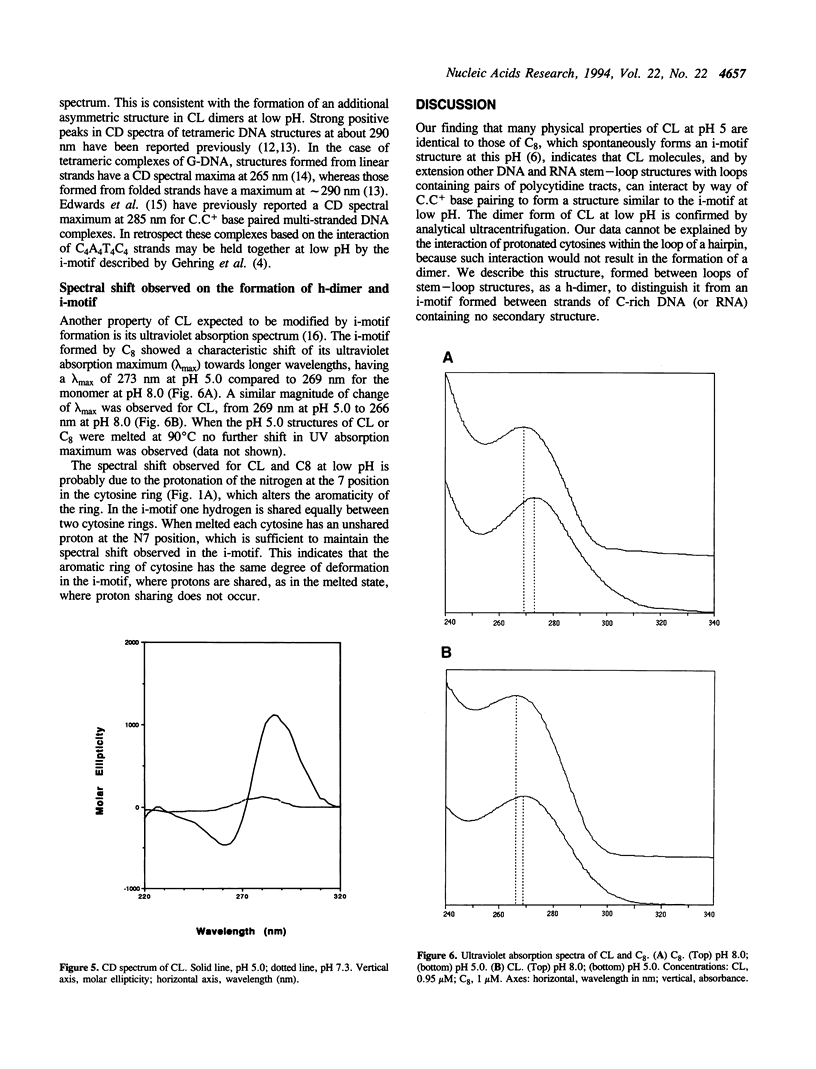
Thermal denaturation profiles of an oligodeoxynucleotide that forms a hairpin structure with a cytidine-rich loop show an unexpected transition at 60 degrees C at pH 5.0 but not at pH 8.0. Analytical ultracentrifugation shows that this transition reflects dimer formation via the interaction of loops from two molecules to form a novel structure termed the h-dimer. The dependence of this structure on low pH implies the formation of cytosine-protonated cytosine base pairs. NMR spectroscopy, thermal denaturation and ultraviolet absorption spectral analysis suggest a similarity to the i-motif structure recently proposed for the interaction of deoxycytidine oligomers. The use of hairpin loops to form i-motif-like structures may prove useful in searches for cognate proteins and possibly in the production of antibodies.
Full text
PDF



Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Edwards E. L., Patrick M. H., Ratliff R. L., Gray D. M. A.T and C.C+ base pairs can form simultaneously in a novel multistranded DNA complex. Biochemistry. 1990 Jan 23;29(3):828–836. doi: 10.1021/bi00455a033. [DOI] [PubMed] [Google Scholar]
- Gehring K., Leroy J. L., Guéron M. A tetrameric DNA structure with protonated cytosine.cytosine base pairs. Nature. 1993 Jun 10;363(6429):561–565. doi: 10.1038/363561a0. [DOI] [PubMed] [Google Scholar]
- Gutell R. R., Power A., Hertz G. Z., Putz E. J., Stormo G. D. Identifying constraints on the higher-order structure of RNA: continued development and application of comparative sequence analysis methods. Nucleic Acids Res. 1992 Nov 11;20(21):5785–5795. doi: 10.1093/nar/20.21.5785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardin C. C., Corregan M., Brown B. A., 2nd, Frederick L. N. Cytosine-cytosine+ base pairing stabilizes DNA quadruplexes and cytosine methylation greatly enhances the effect. Biochemistry. 1993 Jun 8;32(22):5870–5880. doi: 10.1021/bi00073a021. [DOI] [PubMed] [Google Scholar]
- Jaeger L., Michel F., Westhof E. Involvement of a GNRA tetraloop in long-range RNA tertiary interactions. J Mol Biol. 1994 Mar 11;236(5):1271–1276. doi: 10.1016/0022-2836(94)90055-8. [DOI] [PubMed] [Google Scholar]
- Jin R. Z., Breslauer K. J., Jones R. A., Gaffney B. L. Tetraplex formation of a guanine-containing nonameric DNA fragment. Science. 1990 Oct 26;250(4980):543–546. doi: 10.1126/science.2237404. [DOI] [PubMed] [Google Scholar]
- Jin R., Gaffney B. L., Wang C., Jones R. A., Breslauer K. J. Thermodynamics and structure of a DNA tetraplex: a spectroscopic and calorimetric study of the tetramolecular complexes of d(TG3T) and d(TG3T2G3T). Proc Natl Acad Sci U S A. 1992 Sep 15;89(18):8832–8836. doi: 10.1073/pnas.89.18.8832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang C., Zhang X., Ratliff R., Moyzis R., Rich A. Crystal structure of four-stranded Oxytricha telomeric DNA. Nature. 1992 Mar 12;356(6365):126–131. doi: 10.1038/356126a0. [DOI] [PubMed] [Google Scholar]
- Leroy J. L., Gehring K., Kettani A., Guéron M. Acid multimers of oligodeoxycytidine strands: stoichiometry, base-pair characterization, and proton exchange properties. Biochemistry. 1993 Jun 15;32(23):6019–6031. doi: 10.1021/bi00074a013. [DOI] [PubMed] [Google Scholar]
- Leroy J. L., Guéron M., Mergny J. L., Hélène C. Intramolecular folding of a fragment of the cytosine-rich strand of telomeric DNA into an i-motif. Nucleic Acids Res. 1994 May 11;22(9):1600–1606. doi: 10.1093/nar/22.9.1600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maher L. J., 3rd DNA triple-helix formation: an approach to artificial gene repressors? Bioessays. 1992 Dec;14(12):807–815. doi: 10.1002/bies.950141204. [DOI] [PubMed] [Google Scholar]
- Patel D. J. Nucleic acid architecture. Tetrads through interdigitation. Nature. 1993 Jun 10;363(6429):499–500. doi: 10.1038/363499a0. [DOI] [PubMed] [Google Scholar]
- Radhakrishnan I., Gao X., de los Santos C., Live D., Patel D. J. NMR structural studies of intramolecular (Y+)n.(R+)n(Y-)nDNA triplexes in solution: imino and amino proton and nitrogen markers of G.TA base triple formation. Biochemistry. 1991 Sep 17;30(37):9022–9030. doi: 10.1021/bi00101a016. [DOI] [PubMed] [Google Scholar]
- Revzin A., Neumann E., Katchalsky A. Metastable secondary structures in ribosomal RNA molecular hysteresis in the acid-base titration of Escherichia coli ribosomal RNA. J Mol Biol. 1973 Sep 5;79(1):95–114. doi: 10.1016/0022-2836(73)90272-6. [DOI] [PubMed] [Google Scholar]
- Scaria P. V., Shire S. J., Shafer R. H. Quadruplex structure of d(G3T4G3) stabilized by K+ or Na+ is an asymmetric hairpin dimer. Proc Natl Acad Sci U S A. 1992 Nov 1;89(21):10336–10340. doi: 10.1073/pnas.89.21.10336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith F. W., Feigon J. Quadruplex structure of Oxytricha telomeric DNA oligonucleotides. Nature. 1992 Mar 12;356(6365):164–168. doi: 10.1038/356164a0. [DOI] [PubMed] [Google Scholar]
- Wang Y., Patel D. J. Guanine residues in d(T2AG3) and d(T2G4) form parallel-stranded potassium cation stabilized G-quadruplexes with anti glycosidic torsion angles in solution. Biochemistry. 1992 Sep 8;31(35):8112–8119. doi: 10.1021/bi00150a002. [DOI] [PubMed] [Google Scholar]
- Woese C. R., Winker S., Gutell R. R. Architecture of ribosomal RNA: constraints on the sequence of "tetra-loops". Proc Natl Acad Sci U S A. 1990 Nov;87(21):8467–8471. doi: 10.1073/pnas.87.21.8467. [DOI] [PMC free article] [PubMed] [Google Scholar]



