SUMMARY
A cycle of palmitoylation/depalmitoylation of H-Ras mediates bidirectional trafficking between the Golgi apparatus and the plasma membrane but nothing is known about how this cycle is regulated. We show that the prolyl isomerase (PI) FKBP12 binds to H-Ras in a palmitoylation-dependent fashion and promotes depalmitoylation. A variety of inhibitors of the PI activity of FKBP12, including FK506, rapamycin and cycloheximide, increase steady-state palmitoylation. FK506 inhibits retrograde trafficking of H-Ras from the plasma membrane to the Golgi in a proline 179-dependent fashion, augments early GTP-loading of Ras in response to growth factors, and promotes H-Ras dependent neurite outgrowth from PC12 cells. These data demonstrate that FKBP12 regulates H-Ras trafficking by promoting depalmitoylation through cis-trans isomerization of a peptidyl-prolyl bond in proximity to the palmitoylated cysteines.
INTRODUCTION
Mammalian genomes harbor three Ras genes that encode four proteins: H-Ras, N-Ras, K-Ras4A and K-Ras4B. Each of these gene products requires post-translational modifications to target the protein to cellular membranes. All four proteins end in CAAX sequences that are modified by farnesylation, endoproteolysis and carboxyl methylation (Wright and Philips, 2006). Farnesyltransferase (FTase) modifies cytosolic Ras proteins with a 15-carbon farnesyl lipid and thereby directs the proteins to the cytosolic face of the endoplasmic reticulum (ER) where they are further processed by Ras converting enzyme 1 and isoprenylcysteine carboxyl methyltransferase. Three of the four Ras proteins are further processed at the Golgi apparatus where the protein acyl transferase DHHC9/GCP16 adds a palmitate to one or two cysteine residues in the hypervariable region immediately upstream of the CAAX sequence (Swarthout et al., 2005). Whereas farnesylation affords relatively weak affinity for membranes, palmitoylation markedly increases membrane affinity. Ras proteins palmitoylated and affinity trapped in Golgi membranes traffic to the plasma membrane (PM) via vesicular transport. Once associated with the inner leaflet of the PM, Ras proteins can become depalmitoylated and thereby released into the cytosol from whence they traffic by diffusion back to the Golgi for another round of palmitoylation (Goodwin et al., 2005; Rocks et al., 2005). Thus the palmitoylated isoforms of Ras cycle back and forth between the Golgi and PM. Because GTP-bound Ras proteins can engage effectors both at the PM and Golgi, as well as on vesicles, and because signaling outputs from these compartments differ (Chiu et al., 2002), the palmitoylation/depalmitoylation cycle modulates signaling.
Although it has long been appreciated that the half-life of palmitate is much shorter than that of the Ras protein (Magee et al., 1987), little is known about the regulation of palmitoylation/depalmitoylation of Ras. DHHC9/GCP16 has been identified as a relevant palmitoyl acyl transferase (Swarthout et al., 2005), but no mode of regulation of this enzyme has been reported. The identity of the thioesterase that specifically depalmitoylates Ras remains unknown and the lability of the thioester bond has led some Ras biologists to speculate that depalmitoylation may be non-enzymatic. Measurements of the half-life of palmitate on Ras vary widely (Baker et al., 2003; Laude and Prior, 2008; Magee et al., 1987) and have been reported to be shorter for constitutively active H-Ras12V, linking the palmitoylation/depalmitoylation cycle with the GTP/GDP cycle (Baker et al., 2003).
FK506 binding proteins (FKBPs), together with cyclophilins and parvulins, constitute the superfamily of prolyl isomerases (Lu et al., 2007). Although generally thought of as chaperones assisting protein folding by catalyzing the interconversion of cis and trans peptidyl-prolyl bonds, recent evidence suggests that prolyl isomerization, in some contexts, regulates signaling (Lu et al., 2007). The best characterized FKBP, FKBP12, has pharmacologic activity independent of prolyl isomerization; when FKBP12 binds to FK506, the two molecules form a tight complex with calcineurin and thereby inhibit the phosphatase activity that is required for the activation of NFAT. This mode of action explains the potent immunosuppressant activity of FK506 (Liu et al., 1991). FKBP12 also associates constitutively with the IP3 (Cameron et al., 1995), ryanodine (Wehrens et al., 2003) and TGFβ1 (Wang et al., 1996) receptors, suggesting other roles in signal transduction.
All three palmitoylated Ras proteins have prolines in proximity to the cysteines that are modified by palmitate. In this study we show that FKBP12 binds to Ras in a palmitoylation-dependent fashion and promotes depalmitoylation and retrograde trafficking of Ras in a manner dependent on a proline in the hypervariable region of Ras. These data demonstrate a new mechanism for regulating the palmitoylation/depalmitoylation cycle of Ras, reveal a new function of FKBP12 and suggest that Ras may mediate some of the pharmacologic effects of prolyl isomerase inhibitors such as FK506 and rapamycin.
RESULTS
FKBP12 Regulates Palmitoylation of H-Ras
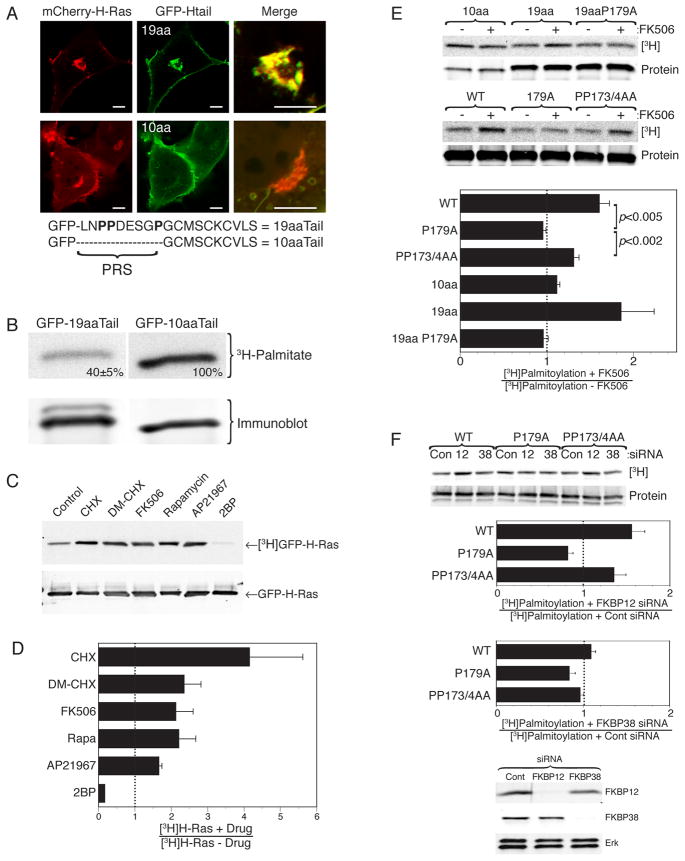
The last 10 amino acids of H-Ras have been characterized as the minimal PM targeting sequence (Choy et al., 1999; Hancock et al., 1991). Indeed, these 10 amino acids contain both the CAAX motif and the two additional cysteines that are sites for palmitoylation. However, we noticed that GFP extended with this sequence (GFP-H-Ras10aaTail) did not recapitulate the steady-state expression of GFP-tagged full-length H-Ras with respect to distribution between the PM and Golgi apparatus. Whereas H-Ras tagged at the N-terminus with fluorescent proteins revealed, in addition to expression on the PM, significant accumulation on the Golgi (Fig. 1A), GFP-H-Ras10aaTail showed much less expression on that organelle. In contrast, GFP-H-Ras19aaTail faithfully recapitulated the distribution of full-length H-Ras (Fig. 1A). Thus, the 9 amino acids upstream of the minimal PM targeting motif contain information required for proper steady-state distribution between organelles. Because transport of H-Ras from the Golgi to the PM is regulated by palmitoylation (Apolloni et al., 2000; Choy et al., 1999), we compared the steady-state level of palmitoylation of the 10 versus 19 amino acid targeting sequences. We found that GFP-H-Ras10aaTail is significantly more palmitoylated than is GFP-H-Ras19aaTail (Fig. 1B), consistent with more efficient transfer to and/or retention on the PM.
Figure 1. FKBP12 Regulates H-Ras Palmitoylation.
(A) COS-1 cells were co-transfected with mCherry-H-Ras and GFP extended either with the last 10 or 19 amino acids of H-Ras (GFP-Htail) and imaged alive with a confocal microscope. Bars indicate 2μm. A proline rich sequence (PRS) found only in the 19aa tail is shown below. (B-F) Metabolic labeling of COS-1 cells with [3H]palmitic acid. (B) Cells expressing GFP-19aaTail or GFP-10aaTail. Text indicates relative levels of palmitoylation after normalization for protein (mean ± SEM, n=3, p<0.0001). (C, D) Cells expressing GFP-H-Ras labeled in the presence of vehicle (DMSO), CHX (50 ng/ml), DM-CHX (10 μM), FK506 (1 μM), rapamycin (500 nM), AP21967 (2 μM) or 2-bromopalmitate (10 μM). (E) Cells expressing the indicated H-Ras or H-Ras tail constructs were treated with FK506 or vehicle. (F) Cells expressing GFP-H-Ras, GFP-H-RasP179A or GFP-H-RasPP173/4AA and pre-treated for 72 hrs with a control siRNA or siRNA directed to FKBP12 or FKBP38 (immunoblot below shows extent of knockdown). (C-F) Representative fluorograms and immunoblots are shown as are plots of pooled data where fluorogram values were normalized to protein expression as determined by Li-Cor Odyssey quantification of immunoblots. The ratio of the normalized values with/without drug or siRNA is plotted (mean ± SEM, n≥3) such that a value>1 indicates enhanced palmitoylation.
Inspection of the 9 amino acid sequence present in GFP-H-Ras19aaTail but not GFP-H-Ras10aaTail revealed that it is rich in proline (Fig. 1A). This led us to hypothesize that cis-trans isomerization of one or more of the peptidyl-proline bonds present in this sequence might affect H-Ras palmitoylation and thereby protein trafficking. Three families of prolyl isomerases (PIs), including the FK506 binding proteins (FKBPs), catalyze cis-trans isomerization. To test the hypothesis that prolyl isomerization by FKBPs regulates palmitoylation we studied the steady-state incorporation of [3H]palmitate into GFP-H-Ras in the presence or absence of various FKBP prolyl isomerase inhibitors (PIIs). Although cycloheximide (CHX) is best known for its ability to inhibit protein synthesis, it is also a potent inhibitor of the FKBP family of PIs (Christner et al., 1999). CHX markedly increased steady-state incorporation of [3H]palmitate into H-Ras (Fig. 1C, D). In contrast, puromycin, a translation inhibitor lacking PII activity, had no effect (not shown). The ability of CHX to block protein synthesis assures that the observed effect is related to the palmitoylation/depalmitoylation cycle rather than de novo synthesis of H-Ras. A derivative of CHX that does not inhibit protein synthesis but retains PII activity against FKBPs, DM-CHX (Edlich et al., 2006), also augmented H-Ras palmitoylation. Both FK506 and rapamycin bind to FKBP12 and inhibit its PI activity, although this activity is not believed to contribute to the ability of these drugs to inhibit calcineurin or mTor, respectively. We found that both FK506 and rapamycin augmented H-Ras palmitoylation to a level similar to that of DM-CHX (Fig. 1C, D), implicating FKBP12 as the relevant PI. A dose response with FK506 revealed no increased effect at concentrations above 1 μM (not shown). Importantly, a rapamycin analog that binds and inhibits the PI activity of FKBP12 but cannot bind to mTor, AP21967, also augmented palmitoylation of H-Ras (Fig. 1C, D). Thus, five chemically distinct compounds that share the property of inhibiting the PI activity of FKBP12 increased the steady-state palmitoylation of H-Ras, providing strong evidence that FKBP12 regulates this post-translational modification. The involvement of FKBP12 was confirmed by silencing its expression with siRNA, which had the same effect as FK506: augmented steady-state palmitoylation of H-Ras (Fig. 1F). Among the other FKBP family members, FKBP38 is the most sensitive to prolyl isomerase inhibiton by DM-CHX (Edlich et al., 2006). Silencing expression of FKBP38 had no effect on H-Ras palmitoylation (Fig. 1F) demonstrating that the activity of FKBP12 toward palmitoylated H-Ras was not a property of all FKBPs.
FK506 potentiated [3H]palmitate labeling of GFP-H-Ras19aaTail but not GFP-H-Ras10aaTail, confirming that the effect of the PII requires the proline-rich sequence (Fig. 1E). Among the three peptidyl-prolyl bonds in amino acids 172–179 of H-Ras, the G-P bond at position 178–179 is the most favorable for cis-trans isomerization since the side group preceding the proline is a hydrogen atom. To test the role of P179 in H-Ras palmitoylation we mutated this residue. The steady-state palmitoylation of neither full-length H-Ras with alanine substituted for P179 nor the equivalent modification of GFP-H-Ras19aaTail was affected by FK506, demonstrating that P179 is required (Fig. 1E). In contrast, FK506 augmented palmitoylation of GFP-H-RasPP173/4AA in which both of the other two prolines in amino acids 172–179 are substituted, demonstrating that proline 179 is the only one required for the action of FKBP12. GFP-H-RasP179A but not GFP-H-RasPP173/4AA was also resistant to the effects of knocking down FKBP12 (Fig. 1F), confirming the involvement of this PI in acting on the G-P bond at position 178–179. Together our data show that palmitoylation of H-Ras is regulated by the PI activity of FKBP12 directed toward the G-P bond at position 178–179.
FKBP12 Promotes Depalmitoylation of H-Ras
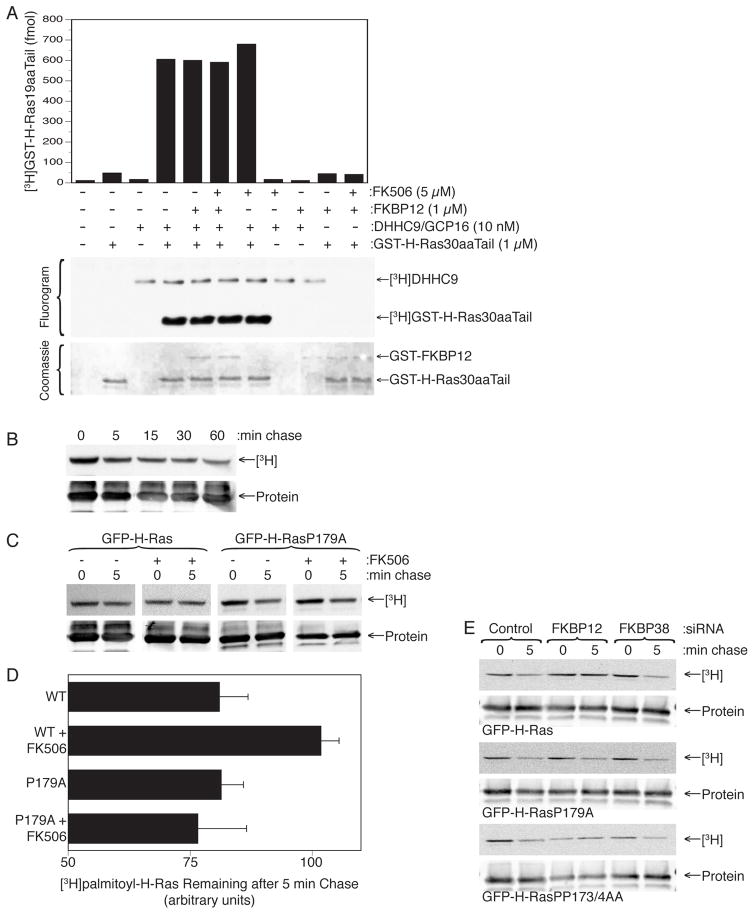
The increase in steady-state labeling of H-Ras with [3H]palmitate when FKBP12 is inhibited or silenced could result from an increase in the rate of palmitoylation, a decrease in the rate of depalmitoylation, or both. It follows that FKBP12 could be a negative regulator of palmitoylation, a positive regulator of depalmitoylation, or both. To investigate the effect of FKBP12 on palmitoylation we expressed in insect cells and affinity purified DHHC9/GCP16, the Golgi associated palmitoyl acyl transferase (PAT) that modifies H-Ras (Swarthout et al., 2005). When incubated in vitro with prenylated GST-H-Ras30aaTail produced in yeast and [3H]palmitoyl CoA, DHHC9/GCP16 readily transferred labeled palmitate to both itself and GST-H-Ras30aaTail (Fig. 2A). We found that neither FK506 nor up to 1 μM recombinant FKBP12 had an effect on the incorporation of [3H]palmitate into GST-H-Ras30aaTail (Fig. 2A). The same result was obtained when we substituted 6His-tagged full-length H-Ras for GST-H-Ras30aaTail. These data argue against FKBP12 as a negative regulator of palmitoylation.
Figure 2. FKBP12 Facilitates H-Ras Depalmitoylation.
(A) Palmitoylation of GST-H-Ras30aa tail in vitro using purified DHHC9/GPC16 and [3H]palmitoyl CoA in the presence or absence of FK506 or recombinant FKBP12. (B) [3H]palmitate pulse (1 mCi/ml × 5 min)–chase labeling of GFP-H-Ras expressed in COS-1 cells. (C, D) [3H]palmitate pulse–chase labeling as in (B) for the GFP-H-Ras and GFP-H-RasP179A, with or without FK506 (1 μM). A representative fluorogram and immunoblot are shown (C) and pooled results are plotted as percent 3H remaining after 5 min chase (D) (mean ± SEM, n=3). (E) [3H]palmitate pulse–chase labeling as in (B) in cells expressing the indicated H-Ras constuct and pretreated for 72 hrs with a control siRNA or siRNA directed to FKBP12 or FKBP38.
We attempted to develop an in vitro assay of H-Ras depalmitoylation analogous to the palmitoylation assay described above to determine the effects of FKBP12. We expressed GFP-H-Ras in COS-1 cells, metabolically labeled the cells with [3H]palmitate and immuno-affinity purified [3H]palmitoyl-GFP-H-Ras. We found that post-nuclear supernatants from HeLa S2 cells and mouse brain homogenates led to a very rapid, and near complete, loss of label from GFP-H-Ras but that the kinetics were so rapid that the reaction was essentially complete at the earliest time point we could measure. Separation of HeLa S2 post-nuclear supernatants into membrane and cytosolic fractions revealed that all of the depalmitoylating activity was in the membrane (Fig. S1). Neither recombinant FKBP12 nor FK506 affected the end-point loss of label, although effects on the rate of loss could not be measured.
To determine the effect of FKBP12 catalytic activity on H-Ras depalmitoylation in intact cells we performed pulse-chase [3H]palmitate labeling of H-Ras in COS-1 cells in the presence or absence of FK506. Following a 5 min pulse we found that more than 50% of the label was lost at the earliest chase time we could assay (5 min). After the first 5 min, the label was lost at a relatively slow rate over the next 60 min (Fig. 2B). Given the short half-life of the palmitate, we compared the [3H]palmitate lost from pulse-labeled H-Ras after a 5 min chase with and without drug. FK506 increased the amount of retained label after a 5 min chase by 33% (Fig. 2C, D). Similar results were obtained for oncogenic H-Ras12V (not shown). In contrast, FK506 had no effect on the half-life of [3H]palmitoyl-H-RasP179A (Fig. 2C, D). Thus FK506 increases the half-life of the palmitate on H-Ras in a P179 dependent fashion, consistent with a model whereby FKBP12 accelerates depalmitoylation of H-Ras. To confirm the effect of FKBP12 on depalmitoylation of H-Ras we silenced either FKBP12 or FKBP38 with siRNA and found that only knockdown of the former inhibited the loss of [3H]palmitate from H-Ras after a 5 min chase and that this effect was lost by substitution of proline 179 but not double substitution of prolines 172 and 173 (Fig. 2E). Thus FKBP12 promotes efficient depalmitoylation of H-Ras and this process requires proline 179.
FKBP12 Binds to H-Ras and N-Ras in a Palmitoylation-Dependent Fashion
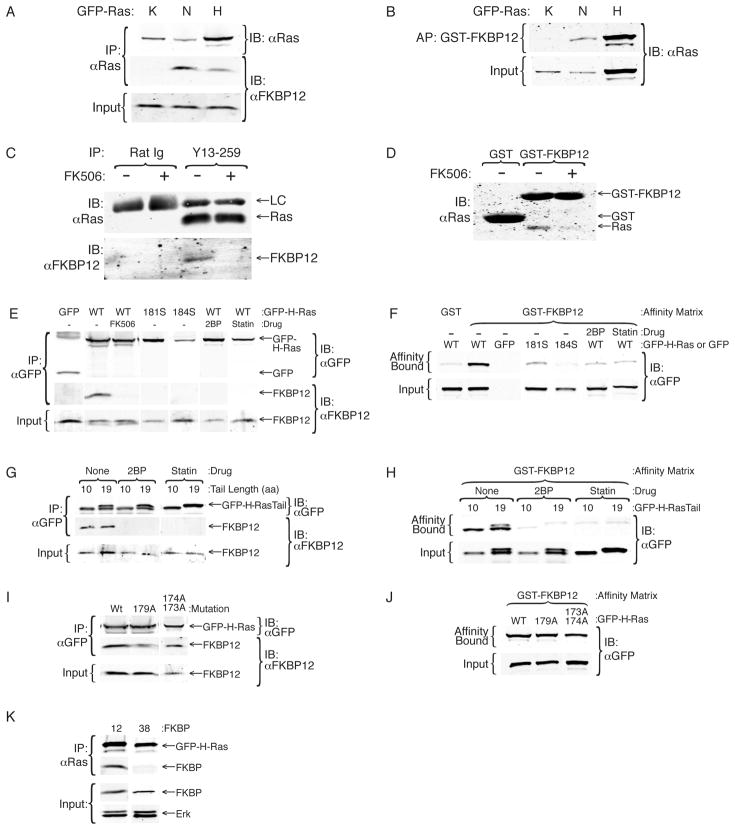
To determine if FKBP12 interacts with Ras we performed co-immunoprecipitations and found that endogenous FKBP12 bound GFP-H-Ras and GFP-N-Ras but not GFP-K-Ras (Fig. 3A). FKBP12 also bound to oncogenic H-Ras12V (not shown). The interaction was confirmed by affinity purification of GFP-H-Ras and GFP-N-Ras, but not GFP-K-Ras, with GST-FKBP12 (Fig. 3B). Brain extracts are relatively rich in H-Ras. The interaction between endogenous Ras and endogenous FKBP12 could be detected using detergent extracts of mouse brain (Fig. 3C). Endogenous Ras from detergent extracts of HeLa cell membranes interacted with GST-FKBP12 (Fig. 3D), confirming this result. Importantly, these interactions were blocked completely by FK506 (Fig 3C, D), suggesting that the drug competes with Ras for the same binding site.
Figure 3. FKBP12 Binds Palmitoylated Ras.
(A, B) Lysates of COS-1 cells expressing GFP-K, N, or H-Ras were immunoprecipitated with anti-Ras Y13-259 antibody and immunoblotted for Ras and FKBP12 (A) or affinity purified with GST-FKBP12 and immunoblotted for Ras (B). (C) Detergent extracts of neonatal mouse brains were immunoprecipitated with Y13-259 or a control rat Ig in the presence or absence of FK506 (1 μM) and precipitates were immunoblotted for endogenous FKBP12 and Ras. (D) Lysates of HeLa S2 cells were affinity purified with GST or GST-FKBP12 and immunoblotted for endogenous Ras. GST binds the secondary antibody non-specifically. (E, F) Lysates of COS-1 expressing GFP, GFP-H-Ras (WT) ± FK506 (1 μM), 2-bromopalmitate (2BP, 50 nM) or simvastatin (statin, 10 μM) or H-Ras palmitoylation-deficient mutants (181S or 184S) were immunoprecipitated with Y13-259 (E) or affinity purified with GST or GST-FKBP12 (F) and immunoblotted for Ras. (G, H) Lysates from COS-1 cells expressing GFP-H-Ras 10aa tail or 19aa tail ± 2BP or simvastatin as in (E, F) were immunoprecipitated with anti-GFP antibody (G) or affinity purified with GST-FKBP12 (H) and immunoblotted for GFP. (I, J) Lysates from COS-1 expressing GFP-H-Ras wild type or single or double proline mutants (179A or 173A/174A) were analyzed as in (G, H). (K) Lysates of COS-1 expressing GFP-H-Ras were immunoprecipitated with anti-GFP antibodies and immunoblotted for GFP and either FKBP12 or FKBP38.
The interaction of H-Ras and N-Ras, but not K-Ras, with FKBP12 suggested that palmitoylation is required for binding. To test this hypothesis we performed co-immunoprecipitation and GST-FKBP12 affinity purification with GFP-H-Ras or palmitoylation-deficient mutants thereof. Mutating either cysteine 181 or 184 to serine completely blocked co-immunoprecipitation of FKBP12 with GFP-H-Ras (Fig. 3E). Monopalmitoylated GFP-H-RasC184S and GFP-H-RasC181S also had markedly diminished affinity for GST-FKBP12 (Fig. 3F). Treatment of cells with 2-bromopalmitate (2BP) directly blocks palmitoylation of proteins. Statins that inhibit prenylation also block palmitoylation of H-Ras because farnesylation is a pre-requisite for palmitoylation. Treatment of cells with 2BP or simvastatin inhibited the interaction between H-Ras and FKBP12 (Fig. 3E, F), confirming that palmitoylation of H-Ras is required for the GTPase to interact with FKBP12. Similar results were obtained using GFP-N-Ras (data not shown). Co-immunoprecipitation and GST-FKBP12 affinity purification revealed that both GFP-H-Ras10aaTail and GFP-H-Ras19aaTail bound to FKBP12 (Fig. 3G, H). Treatment with 2BP or simvastatin blocked the interactions of both of these proteins with FKBP12 demonstrating that, as for full-length H-Ras, palmitoylation is required for the interaction (Fig. 3G, H). The binding of GFP-H-Ras10aaTail to FKBP12 demonstrates that the prolines in the C-terminal 19 amino acid sequence of H-Ras are not required for the interaction with FKBP12. This observation was confirmed by mutating either P179 or P173/174 in full length H-Ras (Fig. 4I, J). Thus, FKBP12 binds to the C-terminus of H-Ras in a proline-independent, palmitoylation-dependent fashion. The binding of palmitoylated H-Ras to FKBP12 did not reflect a general property of FKBPs because no such binding could be detected with FKBP38 (Fig. 3K).
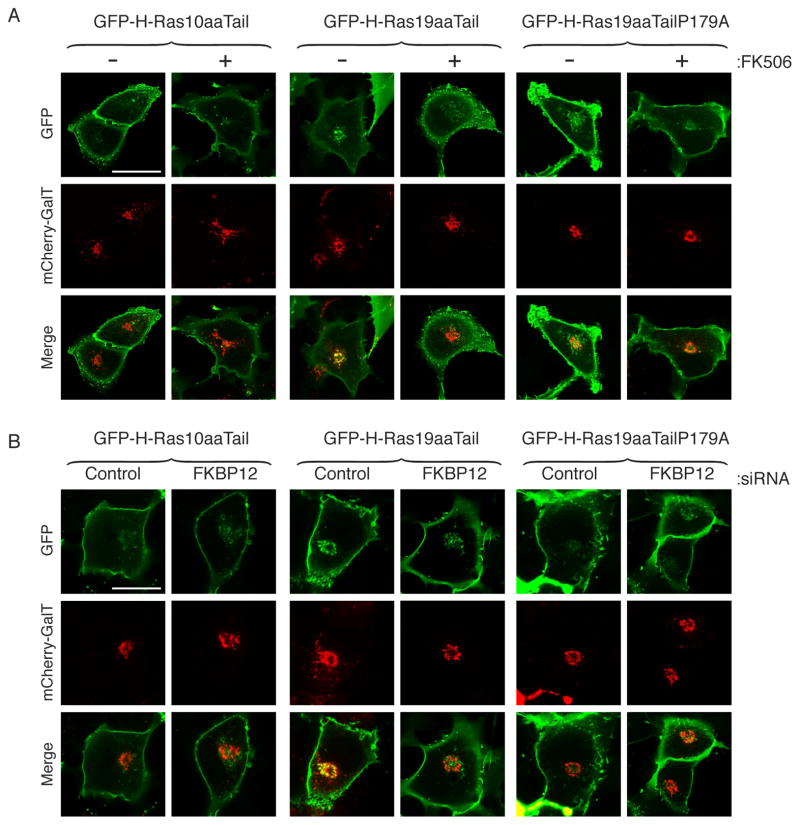
Figure 4. FKBP12 Regulated H-Ras Trafficking.
(A) COS-1 cells were co-transfected with mCherry-GalT (Golgi marker) and GFP extended with either the C-terminal 10 or 19 aa of H-Ras (GFP-H-Ras-10aaTail and GFP-H-Ras-19aaTail) or 19 aa in which the equivalent of proline 179 was changed to alanine (GFP-H-Ras-19aaTailP179A). Cells were imaged alive by confocal microscopy following overnight treatment with FK506 (1 μM) or vehicle control (DMSO). (B) COS-1 cells transfected and imaged as in (A) 3 days after transfection with siRNA directed against FKBP12 or a non-targeting control siRNA. Bar represents 10 μm. Each experiment was performed at least 3 times and 5 to 14 cells were imaged on each plate. The patterns shown are representative of 80–100% of the cells examined.
FKBP12 Regulates H-Ras Trafficking
To determine the effect of FKBP12 on H-Ras trafficking we studied the steady-state distribution between PM and Golgi of GFP-H-Ras10aaTail and GFP-H-Ras19aaTail, with or without treatment with FK506 or silencing of FKBP12 (Fig. 4). We used mCherry tagged galactosyl transferase (mCherry-GalT) as a marker of the Golgi. We found a paucity of GFP-H-Ras10aaTail on the Golgi and that FK506 did not affect the PM/Golgi distribution of this construct. In contrast, GFP-H-Ras19aaTail colocalized with mCherry-GalT on the Golgi. However, in cells that were treated with FK506 the distribution of GFP-H-Ras19aaTail changed to that of GFP-H-Ras10aaTail revealing markedly less expression on the Golgi (Fig. 4A). The FK506 induced changes in the distribution of the molecule were dependent on P179 because GFP-H-Ras19aaTailP179A was resistant to the effect of the drug (Fig. 4A). Identical results were obtained when, instead of inhibiting FKBP12 with FK506, the protein was knocked down with siRNA. Cells transfected with control siRNA revealed GFP-H-Ras19aaTail but not GFP-H-Ras10aaTail on the Golgi, but in cells transfected with siRNA targeting FKBP12 no construct, including GFP-H-Ras19aaTail, showed enrichment on the Golgi (Fig. 4B). These results indicate that FKBP12 is required for the normal distribution of H-Ras between PM and Golgi and that in its absence there is an enrichment of H-Ras on the PM at the expense of the Golgi. These imaging studies are consistent with the biochemical data described above implicating FKBP12 in promoting the depalmitoylation of H-Ras because steady-state expression of H-Ras on the Golgi is dependent on retrograde trafficking from the PM, which is regulated by depalmitoylation.
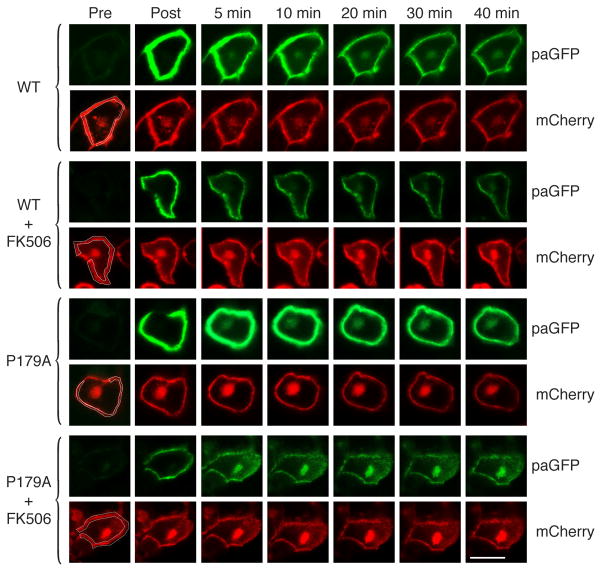
To analyze specifically retrograde trafficking of H-Ras from the PM to the Golgi we utilized photoactivatable GFP (paGFP) (Patterson and Lippincott-Schwartz, 2002; Rocks et al., 2005). We studied confluent MDCK cells because their semi-cuboidal geometry afforded an unambiguous region of PM well separated from the Golgi upon which paGFP-H-Ras could be activated with a 405 nm laser. We co-expressed mCherry-H-Ras such that both the PM and Golgi pools of H-Ras could be visualized throughout the experiment. We activated paGFP-H-Ras only on the PM and followed accumulation of the fluorescent paGFP-H-Ras on the Golgi over 40 min (Fig. 5). As predicted, although activated paGFP-H-Ras produced intense fluorescence at the PM immediately following activation, little if any fluorescence was observed on the Golgi. After 5 min paGFP-H-Ras was observed on the Golgi and the intensity increased up to 20 min. When the cells expressing paGFP-H-Ras and mCherry-H-Ras were pretreated with FK506, very little paGFP-H-Ras accumulated on the Golgi following activation at the PM. paGFP-H-RasP179A activated at the PM also accumulated on the Golgi by 5 min but was resistant to the effect of FK506. Thus, although a peptidyl-prolyl bond at position 178–179 is not required for retrograde trafficking, in wild-type H-Ras isomerization of this bond by FKBP12 regulates translocation from the PM to the Golgi.
Figure 5. FKBP12 Promotes Retrograde Trafficking of H-Ras from the PM to the Golgi.
Confluent monolayers of MDCK cells were co-transfected with mCherry H-Ras and H-Ras wt or P179A tagged at the N-terminus with a photoactivatable GFP (paGFP). Cells were imaged alive in the presence or absence of FK506 (1 μM) before and after photoactivation of paGFP-H-Ras exclusively at the PM compartment marked by mCherry-H-Ras (activation region outlined in white in pre-activation panels). Bar represents 10 μm. Images are representative of 13 to 15 experiments. Accumulation of photoactivated paGFP-H-Ras on the Golgi was observed as follows: WT 8/14; WT + FK506 1/14; P179A 10/13; P179A + FK506 12/15.
Since retrograde trafficking of H-Ras is regulated by depalmitoylation, our imaging data are concordant with our biochemical data and suggest a model whereby FKBP12 acting on P179 of H-Ras promotes depalmitoylation which allows release of H-Ras from the PM and eventual association with the Golgi where another round of palmitoylation can ensue.
The Effect of FKBP12 on Ras Signaling
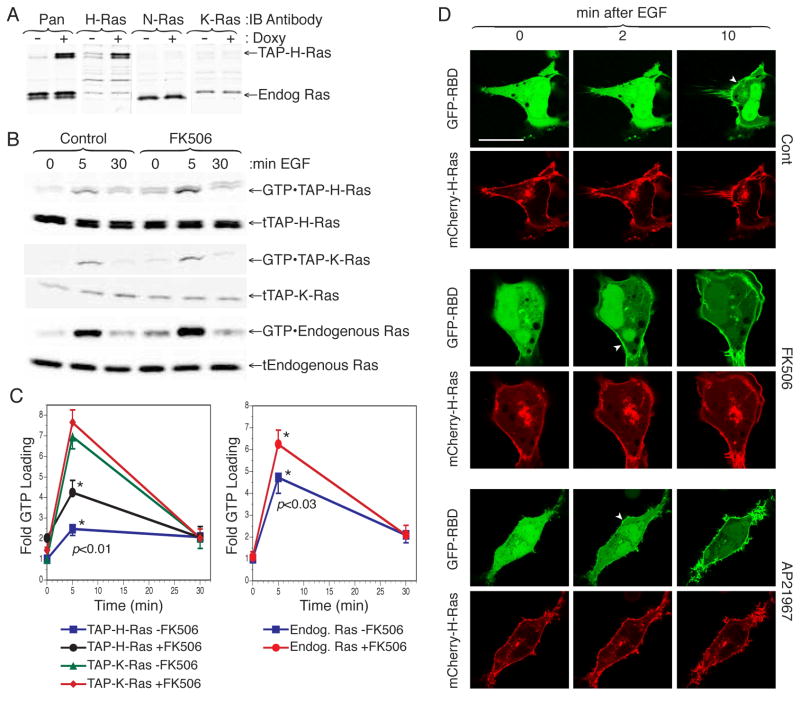
To determine if the action of FKBP12 on H-Ras could affect its signaling properties we determined the extent of GTP-loading on H-Ras following EGF stimulation with or without pretreatment with FKBP12 PI inhibitors. To accomplish this we generated HEK293 cell lines that express various tandem affinity peptide (TAP) tagged Ras proteins at endogenous levels in response to tetracycline (Fig. 6A). FK506 enhanced GTP loading at 5 min on endogenous Ras and TAP-H-Ras but not TAP-K-Ras (Fig. 6B, C). Like FK506, AP21967 augmented GTP loading on H-Ras (not shown), indicating that the FK506 effect was not mediated through calcineurin or mTor but rather through inhibition of the prolyl isomerases.
Figure 6. FKBP12 Limits H-Ras Activation at the PM.
(A) Lysates from HEK293 stably expressing a tetracycline inducible, tandem affinity peptide tagged H-Ras (TAP-H-Ras) were immunoblotted, before and after induction with doxycycline, using antibodies specific for H-Ras, N-Ras or K-Ras. (B) HEK293 expressing TAP-H-Ras or TAP-K-Ras were serum starved overnight in the presence or absence of FK506 (1 μM) and then stimulated with 10 ng/ml EGF for 5 or 30 minutes. GTP-bound Ras was affinity purified from cell lysates with GST-RBD. Ras was detected by immunoblot with a pan-Ras antibody from both GST-RBD pull downs (top panels) and lysates (bottom panels). (C) Quantification of GTP-Ras from (B) with results presented as fold change in GTP-bound Ras normalized to expression. Values plotted are mean ± SEM, n=4. (D) Spatiotemporal activation of H-Ras in live COS-1 cells co-transfected with H-Ras and GFP-RBD. Cells were serum starved, treated with FK506 (1 μM), AP21967 (2 μM) or vehicle and imaged alive before and after stimulation with EGF (50 ng/ml). Arrowheads indicate initial recruitment of GFP-RBD to the PM. Bar represents 10 μm. The experiment shown is representative of eight.
We have previously shown that the Ras binding domain (RBD) of Raf-1 fused to GFP can be used as an in vivo probe for Ras activation (Chiu et al., 2002). To confirm the results of the GST-RBD pull down assay (Fig. 6B, C) by single cell analysis, we studied recruitment of GFP-RBD to membrane associated H-Ras in response to physiologic concentrations of EGF (50 ng/ml). Whereas 10 min were required to see unambiguous recruitment of GFP-RBD to the PM of untreated cells, Ras activation was apparent on this compartment by 2 min in cells pretreated with either FK506 or AP21967 (Fig 6D). Thus, FKBP12 prolyl isomerase inhibitors accelerated the steady state accumulation of GTP-bound H-Ras on the PM, consistent with their ability to block depalmitoylation and retrograde trafficking to the Golgi.
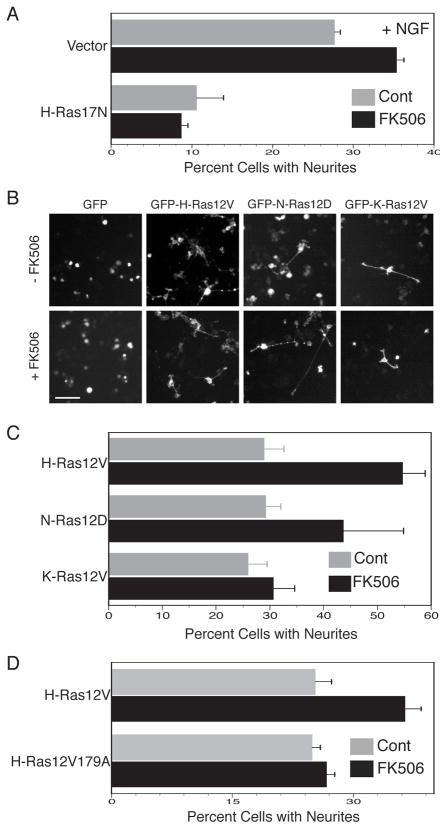
FK506 has been reported to promote differentiation of PC12 cells in response to nerve growth factor (Lyons et al., 1994) but the mechanism of action has never been determined. We verified this effect of FK506 and employed a dominant negative form of Ras, H-Ras17N, to block Ras signaling (Fig. 7A). We found that, as expected (Szeberenyi et al., 1990), expression of H-Ras17N dramatically inhibited NGF stimulated neurite formation. Importantly, H-Ras17N completely eliminated the augmented neurite formation induced by FK506 (Fig. 7A), suggesting that the mechanism of action of FK506 in this process involves Ras, concordant with our observation that FK506 augments GTP-loading of H-Ras (Fig. 6).
Figure 7. FKBP12 Restricts H-Ras Signaling.
(A) PC12 cells were co-transfected with H-Ras17N or vector and GFP and then treated with NGF (50 ng/ml) with or without FK506 (1 μM) or vehicle control (DMSO). After 4 days GFP positive cells were scored for neurite processes with lengths 1.5 times the diameter of the cell soma. Results shown are mean±SEM, n=3, p<0.001 for vector ± FK506. (B) PC12 cells were transiently transfected with either GFP alone or GFP-H-Ras12V, GFP-N-Ras12D, or GFP-K-Ras12V. After 3 days in the presence of FK506 (1 μM) or vehicle control (DMSO), GFP positive cells were scored for neurites. Bar represents 50 μm. (C) Results from (B) are plotted as percent of cells with neurite outgrowth (mean ± SEM, n=4). (D) PC12 cells were transfected with GFP-H-Ras12V or GFP-H-Ras12V179A and treated with or without FK506. Results shown are mean±SEM, n=4.
Expression of activated Ras proteins induces PC12 cell differentiation in the absence of growth factor stimulation (Bar-Sagi and Feramisco, 1985). We therefore sought to determine if the effect of FK506 is specific to palmitoylated isoforms of Ras. PC12 cells expressing GFP alone did not form neurites and FK506 had no effect on these cells. In contrast, expression of GFP-tagged forms of H-Ras12V, N-Ras12D and K-Ras12V all induced differentiation of PC12 cells as determined by neurite outgrowth (Fig. 7B, C). Treatment with FK506 significantly enhanced the efficiency of differentiation in cells expressing GFP-H-Ras12V and GFP-N-Ras12D but not GFP-Kras12V (Fig. 7B, C). The effect of FK506 on H-Ras12V induced PC12 cell differentiation depended on proline 179 since FK506 was without effect on the differentiation induced by H-Ras12V179A (Fig. 7D). The data are consistent with enhanced signaling of palmitoylated Ras in PC12 cells when the PI activity of FKBP12 is inhibited with FK506. This effect can be attributed to increased palmitoylation and a shift in the steady-state distribution of the protein toward accumulation on the PM. The data support the idea that the action of FKBP12 on H-Ras plays a role not only in subcellular trafficking of this GTPase, but also in its signal output.
DISCUSSION
Palmitoylation of H-Ras is required for its association with the PM. This is a consequence both of increased affinity for phospholipid bilayers and entry into specific membrane trafficking pathways. It has long been appreciated that protein S-palmitoylation is a reversible modification (Smotrys and Linder, 2004) and that palmitate turns over on acylated proteins, including Ras (Magee et al., 1987). More recently, the palmitoylation/depalmitoylation cycle of H-Ras was shown to be causally linked to bidirectional trafficking between the Golgi and PM (Goodwin et al., 2005; Rocks et al., 2005). What remains unknown is how the palmitoylation/depalmitoylation cycle of H-Ras is regulated. Our data show that FKBP12 regulates the palmitoylation/depalmitoylation cycle of H-Ras by binding to the GTPase in a palmitoylation-dependent fashion and promoting depalmitoylation through a mechanism that requires the prolyl-isomerase activity of FKBP12 and proline 179 of H-Ras.
The half-life of palmitate has been reported to be anywhere from 20 min (Magee et al., 1987) to 2.4 hrs (Baker et al., 2003). In contrast, our pulse/chase data suggest that the loss of palmitate from H-Ras is biphasic with a rapid initial phase followed by much slower loss (Fig. 2B). Half of the label was lost in the initial, rapid phase that had a t1/2 of less than 5 min. The discrepancy between our results and those previously reported might be explained by the significantly shorter pulse time we employed and the avoidance of CHX, which we now know affects depalmitoylation. In the older studies “pulse” labeling with [3H]palmitate ranged from 2–4 hrs. Two factors call into question the utility of such long pulse times. To become a substrate for protein acyl transferases, [3H]palmitate must not only be transported into the cell but also conjugated with coenzyme A. Long pulse times allow cellular pools of [3H]palmitoyl-CoA to reach equilibrium making an efficient chase untenable. In addition, [3H]palmitate released from labeled cellular proteins and lipids can be recycled to [3H]palmitoyl-CoA such that a large equilibrium pool of labeled protein serves as a reservoir of [3H]palmitoyl-CoA, confounding the ability to efficiently chase with unlabeled palmitate. We minimized these obstacles by labeling with a true “pulse” of only 5 min and compensating by significantly increasing the concentration of [3H]palmitate. The more rapid turnover of [3H]palmitate that we observed can be more easily reconciled with H-Ras trafficking data in live cells than can the older measurements; fluorescence recovery after photobleaching and photoactivation studies have shown that the t1/2 of H-Ras recovery on, or delivery to, the Golgi is less than 10 min (Goodwin et al., 2005; Rocks et al., 2005).
Although the last ten amino acids of H-Ras that include both palmitoylation sites was established almost two decades ago as the minimal PM targeting sequence (Hancock et al., 1991), several recent studies have reported signaling and trafficking information in the portion of the hypervariable region immediately upstream of this sequence. Rotblat reported that three separable domains contributed to H-Ras association with PM microdomains: the lipid anchor (aa 179–189), the hypervariable linker region (hvr = aa 166–179) and the GTP-binding domain (aa 1–165) (Rotblat et al., 2004). Whereas the lipid anchor associated with lipid rafts, the hvr afforded affinity for non-raft membranes. Laude and Prior showed that the composition of the hvr of H-Ras affected both the distribution of the protein between the PM and Golgi and steady-state palmitoylation (Laude and Prior, 2008).
Although FKBP12 is best known for its ability to bind to and inhibit calcineurin in the presence of FK506, physical interactions with several receptors have led to the idea that FKBP12 plays a role in signaling. FKBP12 binds and regulates the activity of TGFβ1R (Wang et al., 1996), IP3R (Cameron et al., 1995), and the ryanodine receptor (Wehrens et al., 2003). Our data provide evidence for an additional role in signaling downstream of receptors at the level of Ras.
Our observation that the binding of FKBP12 to a substrate for prolyl isomerization does not depend on the proline is consistent with binding data for TGFβ1R (Wang et al., 1996). Our discovery that palmitoylation is required for H-Ras binding to FKBP12 is consistent with the crystal structure of the isomerase which reveals a hydrophobic groove. This groove serves as the binding surface for both peptide substrates and FK506, consistent with the observation that FK506 acts to inhibit PI activity by competing with substrates for binding (Van Duyne et al., 1991). The ability of FK506 to compete for H-Ras binding confirms the substrate pocket as the binding site for H-Ras. However, this model presents a conundrum: how can the acyl chains of H-Ras simultaneously insert into the phospholipid bilayer and associate with the hydrophobic pocket of FKBP12? One possibility is that FKBP12 acts as a cytosolic chaperone that can extract palmitoylated Ras proteins from membranes and render them soluble in much the same way that RhoGDI acts on geranylgeranylated Rho proteins (Michaelson et al., 2001).
The evidence for the reversibility of S-acylation of H-Ras is compelling but the mechanism of depalmitoylation remains unclear. Several thioesterases can catalyze the hydrolysis of the thioester bonds of H-Ras in vitro (Smotrys and Linder, 2004), but which, if any, of these plays a physiologic role in H-Ras trafficking and signaling has not been determined. One of these, APT1, was first purified from rat liver cytosol and shown to have activity in vitro toward both G protein α subunits and H-Ras, although the efficiency was six fold higher for the Gα (Duncan and Gilman, 1998) and the S. cerevisiae ortholog acts on the G protein α subunit Gpa1p but not Ras2p (Duncan and Gilman, 2002). Besides the apparent substrate specificity favoring G protein α subunits over H-Ras, the other problem with considering APT1 as the relevant thioesterase is the fact that, whereas APT1 is a soluble, cytosolic enzyme, its putative substrate is associated with membranes in an orientation that makes the thioester bond inaccessible. However, APT1 itself has been reported to be palmitoylated (Yang et al.), suggesting a mechanism whereby a pool of the enzyme might associate with the membrane and thereby come into proximity with membrane-associated substrates. Our observation that all of the in vitro thioesterase activity in HeLa cell homogenates is in the membrane fraction argues that, if APT1 is the relevant thioesterase, it is the acylated pool that is active. One parsimonious corollary that follows from a model in which FKBP12 serves as a chaperone for palmitoylated Ras proteins is that the H-Ras/FKPB12 complex would be expected to render the thioester bond accessible to thioesterases like APT1. A role for FKBP12 in presenting the thioester bond to a membrane-associated thioesterase is plausible considering that the farnesyl modification will afford some membrane affinity, even when the acyl chains are disengaged from the membrane.
The ability of FKBP12 to extract palmitoylated Ras proteins from membranes might be enough to explain its role in promoting depalmitoylation. However, our data strongly suggest that the PI activity of FKBP12 is also important. GFP extended with either 10 or 19 of the C-terminal amino acids of H-Ras bound equally well to FKBP12, but the steady-state palmitoylation of only the latter construct containing prolines was affected by FK506. The ability of FK506 to augment palmitoylation of H-Ras, to affect PM to Golgi trafficking and to stimulate H-Ras12V induced PC12 cell differentiation was dependent on proline 179. This suggests that cis-trans isomerization about the G-P bond at position 178–179 promotes hydrolysis of the thioester bonds at positions 181 and 184. One model that incorporates this observation posits that the thioesterase that acts on H-Ras associated with FKBP12 works more efficiently when the peptidyl-prolyl bond at position 178–179 is in a cis conformation.
Although prolyl isomerization is most often thought of in the context of folding of nascent proteins, the concept of prolyl isomerization operating like other post-translational modifications to mediate information flow down signaling pathways is attractive. The best evidence for such regulation comes from Pin1, a phosphoprotein-directed PI in the parvulin family that has been shown to act on multiple regulators of the cell cycle, including the Ras/MAPK pathway at the level of Raf-1 (Lu et al., 2007). Cis-trans isomerization about G-P bonds catalyzed by CypA has been shown to regulate the Ikt kinase (Brazin et al., 2002) and the adaptor protein Crk (Nicholson and Lu, 2007). In each of these examples, prolyl isomerization can be thought of as a molecular timer regulated by PIs (Lu et al., 2007). The dwell time of palmitoylated Ras on the inner leaflet of the PM is regulated by depalmitoylation. Our data are consistent with a model in which cis-trans isomerization of the G-P bond at position 178–179 acts as a molecular timer to trigger depalmitoylation and thereby limit the time of residence of H-Ras at the PM.
EXPERIMENTAL PROCEDURES
Metabolic Labeling
For steady-state monitoring of H-Ras palmitoylation, COS-1 cells were transfected with GFP-H-Ras using Lipofectamine 2000 according to the manufacturer’s instructions. The cells were then incubated overnight in Labeling Media (DMEM, 0.2 mCi/ml [3H]palmitic acid, 10% dialyzed FBS, 5 mM sodium pyruvate, 3.6 mg/ml fatty acid free BSA). Pharmacologic agents were used at the following concentrations: 1 μM FK506; 50 ng/ml CHX; 10 μM DM-CHX; 500 nM rapamycin; 2 μM AP21967 (ARIAD Pharmaceuticals, Cambridge, MA); 25 μM 2-bromopalmitate. Cells were harvested in RIPA buffer (20 mM Tris, pH 7.5, 137 mM NaCl, 2 mM EDTA, 0.5% sodium deoxycholate, 0.1% SDS, 1% Triton X-100, 10% glycerol, and protease inhibitors). For knockdown experiments, pooled siRNAs directed against FKBP12 or a control non-targeting pool (Thermo-Fisher Scientific, Waltham, MA) were transfected using Dharmafect Reagent according to the manufacturer’s protocol three days prior to metabolic labeling or imaging. GFP-H-Ras was immunoprecipitated from RIPA extracts of post-nuclear supernatants with Y13-259AC agarose-conjugated antibody (Santa Cruz Biotechnology, Santa Cruz, CA). Beads were washed in RIPA, eluted with 2X Laemmli sample buffer containing 5 mM DTT, and incubated at 70°C for 3min. Eluates were analyzed by 14% SDS-PAGE. Protein transferred to PVDF membranes were blocked with milk and then incubated with rabbit anti-GFP (Invitrogen, Carlsbad, CA) or Ras10 pan-Ras monoclonal (EMD Chemicals, Darmstadt Germany) antibodies at 1:5,000, followed by goat anti-mouse or rabbit IR800 infrared fluorophore conjugated secondary antibodies (Li-Cor, Lincoln NE) at 1:20,000. Blots were scanned and GFP Ras bands quantified using a Li-Cor Odyssey. Membranes were then dried and exposed to BioMax MS film in a BioMax Transcreen LE folder at −80°C for 1–3 days. 3H fluorograms were quantified using an Epson V700 transparency scanner and ImageQuant analysis software (GE Healthcare, Piscataway, NJ) and the values were normalized to protein levels determined by immunoblot and Li-Cor scan.
In vitro Palmitoylation and Pulse Chase Depalmitoylation Assay
In vitro palmitoylation was performed as previously described (Jennings et al., 2009). To monitor depalmitoylation, COS-1 cells were incubated in Labeling Media (DMEM, 10% FBS) supplemented with 1 mCi/ml [3H]palmitate for 5 min. Cells were then washed twice and incubated in Chase Media (same as Labeling Media but with 200 μM palmitic acid) for up to 60 min before [3H]palmitoyl-GFP-H-Ras was immunoprecipated and analyzed as described above.
Co-Immunoprecipitation and GST-FKBP12 Affinity Capture
Confluent 10 cm dishes of COS-1 cells were transfected with GFP-Ras constructs using Lipofectamine 2000. The following day, cells were lysed in RIPA buffer and GFP-Ras was immunoprecipitated with either Y13-259AC or anti-GFP antibodies. PVDF membranes were probed with antibodies directed against either GFP or Ras at 1:5000 and FKBP12 (Thermo Fisher, Rockford, IL) at 1:500. Primary antibodies were detected with IR fluorophore conjugated secondary antibodies and Li-Cor Odyssey scanning. For affinity purification GST-FKBP12 conjugated glutathione sepharose was incubated with clarified COS-1 RIPA lysates of cells expressing GFP-Ras. Neonatal mouse brains were homogenized in hypotonic Homogenization Buffer (10 mM Tris, pH 7.5, 5 mM MgCl2, 2 mM EDTA, 2 mM EGTA) by Dounce and post-nuclear supernatants were extracted by adding 1% Triton X-100. Endogenous Ras was immunoprecipitated with Y13-259AC. HeLa S2 cells were disrupted by nitrogen cavitation in Relaxation Buffer (10 mM HEPES, pH 7.3, 100 mM KCl, 3 mM NaCl, 3.5 mM MgCl2) and membranes prepared from post-nuclear supernatants by centrifugation at 100,000 x g were extracted with 1% Triton X-100 in Relaxation Buffer. Extracts were applied to GST-FKBP12-conjugated glutathione sepharose and affinity purified proteins were analyzed by immunoblot as described above.
Confocal Microscopy and Photoactivation
COS-1 cells expressing GFP extended with various H-Ras C-terminal peptides and mCherry tagged H-Ras or GalT were imaged with a Zeiss 510 laser scanning confocal microscope using a 63X, NA 1.4 objective and a humidified metabolic chamber maintained at 37°C, 5% CO2 (Pecon GmbH, Erbach, Germany). To monitor dynamic trafficking of Ras, confluent MDCK cells were transfected with paGFP-H-Ras and mCherry-H-Ras in a 10:1 ratio. To photoactivate paGFP, a region of interest determined by mCherry-H-Ras at the PM was activated with 25 scans of a 405 nM 30 mW laser at 100% transmission. Images were collected at 2 min intervals before and after photoactivation with the detector pinhole at maximum diameter and laser power at a minimum to prevent photobleaching.
Ras Activation
The coding regions of H-Ras and K-Ras cDNAs were cloned into pNTAP vectors (Agilent Technologies, La Jolla, CA) that place dual affinity peptides at the N-termini of the fusion proteins. The coding region of TAP-Ras was then amplified by PCR and cloned into pcDNA4/TO which was then applied to the Flp-In™ T-REx™ System (Invitrogen, Carlsbad, CA) to generate lines of 293 cells that express TAP-tagged H-Ras and K-Ras from a Tet inducible promoter. These cells were serum starved overnight in DMEM with 0.1% serum and TAP-Ras was induced with 2 μg/ml doxycycline to achieve levels of TAP-Ras similar to endogenous protein. Cells were treated with or without 1 μM FK506 and stimulated with 10 ng/ml EGF and lysed in Magnesium Lysis Buffer (25 mM HEPES pH 7.5, 125 mM NaCl, 10 mM MgCl2, 1 mM EDTA, 1% IGEPAL CA-630, 10% glycerol). Clarified lysates were incubated for 45 min with 10 μg GST-RBD glutathione sepharose beads. Affinity bound GTP-Ras was detected by immunoblot. To monitor H-Ras activation in live cells, COS-1 cells expressing mCherry-H-Ras and GFP-RBD were serum starved and confocal images were acquired every 2 min before and after stimulation with 50 ng/ml EGF.
Neurite Outgrowth
PC12 cells were grown in DMEM supplemented with 10% horse serum and 5% FBS and then transfected with constitutively active isoforms of GFP-Ras using Lipofectamine 2000. After 3 days in the presence or absence of 1 μM FK506, GFP expressing cells were scored for extension of neurites longer than 1.5 times the cell soma. For NGF studies the PC12 cells were co-transfected with pEGFP and H-Ras17N or vector and maintained in DMEM supplemented with 0.1% horse serum and 0.05% FBS with or without FK506 (1 μM) ± NGF (50 ng/ml) and scored for neurites after 4 days.
Supplementary Material
Acknowledgments
This work was supported by NIH grants GM55279 (MRP), CA116034 (MRP), CA118495 (MRP) and GM51466 (MEL), by an award from the Jeffrey Rosenzweig Foundation (MRP), and by a predoctoral fellowship from the AHA Midwest Affiliate (BCJ). We thank Mattias Weiwad and Gunter Fischer (MPF for Enzymology of Protein Folding, Halle/Saale, Germany) for the kind gift of DM-CHX. We thank ARIAD Pharmaceuticals, Cambridge, MA for supplying AP21967.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Apolloni A, Prior IA, Lindsay M, Parton RG, Hancock JF. H-ras but not K-ras traffics to the plasma membrane through the exocytic pathway. Mol Cell Biol. 2000;20:2475–2487. doi: 10.1128/mcb.20.7.2475-2487.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker TL, Zheng H, Walker J, Coloff JL, Buss JE. Distinct rates of palmitate turnover on membrane-bound cellular and oncogenic H-ras. J Biol Chem. 2003;278:19292–19300. doi: 10.1074/jbc.M206956200. [DOI] [PubMed] [Google Scholar]
- Bar-Sagi D, Feramisco JR. Microinjection of the ras oncogene protein into PC12 cells induces morphological differentiation. Cell. 1985;42:841–848. doi: 10.1016/0092-8674(85)90280-6. [DOI] [PubMed] [Google Scholar]
- Brazin KN, Mallis RJ, Fulton DB, Andreotti AH. Regulation of the tyrosine kinase Itk by the peptidyl-prolyl isomerase cyclophilin A. Proc Natl Acad Sci U S A. 2002;99:1899–1904. doi: 10.1073/pnas.042529199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cameron AM, Steiner JP, Sabatini DM, Kaplin AI, Walensky LD, Snyder SH. Immunophilin FK506 binding protein associated with inositol 1,4,5-trisphosphate receptor modulates calcium flux. Proc Natl Acad Sci U S A. 1995;92:1784–1788. doi: 10.1073/pnas.92.5.1784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiu VK, Bivona T, Hach A, Sajous JB, Silletti J, Wiener H, Johnson RL, Cox AD, Philips MR. Ras signalling on the endoplasmic reticulum and the Golgi. Nat Cell Biol. 2002;4:343–350. doi: 10.1038/ncb783. [DOI] [PubMed] [Google Scholar]
- Choy E, Chiu VK, Silletti J, Feoktistov M, Morimoto T, Michaelson D, Ivanov IE, Philips MR. Endomembrane trafficking of ras: the CAAX motif targets proteins to the ER and Golgi. Cell. 1999;98:69–80. doi: 10.1016/S0092-8674(00)80607-8. [DOI] [PubMed] [Google Scholar]
- Christner C, Wyrwa R, Marsch S, Kullertz G, Thiericke R, Grabley S, Schumann D, Fischer G. Synthesis and cytotoxic evaluation of cycloheximide derivatives as potential inhibitors of FKBP12 with neuroregenerative properties. J Med Chem. 1999;42:3615–3622. doi: 10.1021/jm991038t. [DOI] [PubMed] [Google Scholar]
- Duncan JA, Gilman AG. A cytoplasmic acyl-protein thioesterase that removes palmitate from G protein alpha subunits and p21(RAS) J Biol Chem. 1998;273:15830–15837. doi: 10.1074/jbc.273.25.15830. [DOI] [PubMed] [Google Scholar]
- Duncan JA, Gilman AG. Characterization of Saccharomyces cerevisiae acyl-protein thioesterase 1, the enzyme responsible for G protein alpha subunit deacylation in vivo. J Biol Chem. 2002;277:31740–31752. doi: 10.1074/jbc.M202505200. [DOI] [PubMed] [Google Scholar]
- Edlich F, Weiwad M, Wildemann D, Jarczowski F, Kilka S, Moutty MC, Jahreis G, Lucke C, Schmidt W, Striggow F, et al. The specific FKBP38 inhibitor N-(N′,N′-dimethylcarboxamidomethyl)cycloheximide has potent neuroprotective and neurotrophic properties in brain ischemia. J Biol Chem. 2006;281:14961–14970. doi: 10.1074/jbc.M600452200. [DOI] [PubMed] [Google Scholar]
- Goodwin JS, Drake KR, Rogers C, Wright L, Lippincott-Schwartz J, Philips MR, Kenworthy AK. Depalmitoylated Ras traffics to and from the Golgi complex via a nonvesicular pathway. J Cell Biol. 2005;170:261–272. doi: 10.1083/jcb.200502063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hancock JF, Cadwallader K, Paterson H, Marshall CJ. A CAAX or a CAAL motif and a second signal are sufficient for plasma membrane targeting of ras proteins. EMBO Journal. 1991;10:4033–4039. doi: 10.1002/j.1460-2075.1991.tb04979.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jennings BC, Nadolski MJ, Ling Y, Baker MB, Harrison ML, Deschenes RJ, Linder ME. 2-Bromopalmitate and 2-(2-hydroxy-5-nitro-benzylidene)-benzo[b]thiophen-3-one inhibit DHHC-mediated palmitoylation in vitro. J Lipid Res. 2009;50:233–242. doi: 10.1194/jlr.M800270-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laude AJ, Prior IA. Palmitoylation and localisation of RAS isoforms are modulated by the hypervariable linker domain. J Cell Sci. 2008;121:421–427. doi: 10.1242/jcs.020107. [DOI] [PubMed] [Google Scholar]
- Liu J, Farmer JD, Jr, Lane WS, Friedman J, Weissman I, Schreiber SL. Calcineurin is a common target of cyclophilin-cyclosporin A and FKBP-FK506 complexes. Cell. 1991;66:807–815. doi: 10.1016/0092-8674(91)90124-h. [DOI] [PubMed] [Google Scholar]
- Lu KP, Finn G, Lee TH, Nicholson LK. Prolyl cis-trans isomerization as a molecular timer. Nat Chem Biol. 2007;3:619–629. doi: 10.1038/nchembio.2007.35. [DOI] [PubMed] [Google Scholar]
- Lyons WE, George EB, Dawson TM, Steiner JP, Snyder SH. Immunosuppressant FK506 promotes neurite outgrowth in cultures of PC12 cells and sensory ganglia. Proc Natl Acad Sci U S A. 1994;91:3191–3195. doi: 10.1073/pnas.91.8.3191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magee AI, Gutierrez L, McKay IA, Marshall CJ, Hall A. Dynamic fatty acylation of p21N-ras. Embo J. 1987;6:3353–3357. doi: 10.1002/j.1460-2075.1987.tb02656.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michaelson D, Silletti J, Murphy G, D’Eustachio P, Rush M, Philips MR. Differential Localization of Rho GTPases in Live Cells. Regulation by hypervariable regions and rhogdi binding. J Cell Biol. 2001;152:111–126. doi: 10.1083/jcb.152.1.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicholson LK, Lu KP. Prolyl cis-trans Isomerization as a molecular timer in Crk signaling. Mol Cell. 2007;25:483–485. doi: 10.1016/j.molcel.2007.02.005. [DOI] [PubMed] [Google Scholar]
- Patterson GH, Lippincott-Schwartz J. A photoactivatable GFP for selective photolabeling of proteins and cells. Science. 2002;297:1873–1877. doi: 10.1126/science.1074952. [DOI] [PubMed] [Google Scholar]
- Rocks O, Peyker A, Kahms M, Verveer PJ, Koerner C, Lumbierres M, Kuhlmann J, Waldmann H, Wittinghofer A, Bastiaens PI. An Acylation Cycle Regulates Localization and Activity of Palmitoylated Ras Isoforms. Science. 2005;307:1746–1752. doi: 10.1126/science.1105654. [DOI] [PubMed] [Google Scholar]
- Rotblat B, Prior IA, Muncke C, Parton RG, Kloog Y, Henis YI, Hancock JF. Three separable domains regulate GTP-dependent association of H-ras with the plasma membrane. Mol Cell Biol. 2004;24:6799–6810. doi: 10.1128/MCB.24.15.6799-6810.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smotrys JE, Linder ME. Palmitoylation of intracellular signaling proteins: regulation and function. Annu Rev Biochem. 2004;73:559–587. doi: 10.1146/annurev.biochem.73.011303.073954. [DOI] [PubMed] [Google Scholar]
- Swarthout JT, Lobo S, Farh L, Croke MR, Greentree WK, Deschenes RJ, Linder ME. DHHC9 and GCP16 constitute a human protein fatty acyltransferase with specificity for H- and N-Ras. J Biol Chem. 2005;280:31141–31148. doi: 10.1074/jbc.M504113200. [DOI] [PubMed] [Google Scholar]
- Szeberenyi J, Cai H, Cooper GM. Effect of a dominant inhibitory Ha-ras mutation on neuronal differentiation of PC12 cells. Mol Cell Biol. 1990;10:5324–5332. doi: 10.1128/mcb.10.10.5324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Duyne GD, Standaert RF, Karplus PA, Schreiber SL, Clardy J. Atomic structure of FKBP-FK506, an immunophilin-immunosuppressant complex. Science. 1991;252:839–842. doi: 10.1126/science.1709302. [DOI] [PubMed] [Google Scholar]
- Wang T, Li BY, Danielson PD, Shah PC, Rockwell S, Lechleider RJ, Martin J, Manganaro T, Donahoe PK. The immunophilin FKBP12 functions as a common inhibitor of the TGF beta family type I receptors. Cell. 1996;86:435–444. doi: 10.1016/s0092-8674(00)80116-6. [DOI] [PubMed] [Google Scholar]
- Wehrens XH, Lehnart SE, Huang F, Vest JA, Reiken SR, Mohler PJ, Sun J, Guatimosim S, Song LS, Rosemblit N, et al. FKBP12.6 deficiency and defective calcium release channel (ryanodine receptor) function linked to exercise-induced sudden cardiac death. Cell. 2003;113:829–840. doi: 10.1016/s0092-8674(03)00434-3. [DOI] [PubMed] [Google Scholar]
- Wright LP, Philips MR. Thematic review series: lipid posttranslational modifications. CAAX modification and membrane targeting of Ras. J Lipid Res. 2006;47:883–891. doi: 10.1194/jlr.R600004-JLR200. [DOI] [PubMed] [Google Scholar]
- Yang W, Di Vizio D, Kirchner M, Steen H, Freeman MR. Proteome scale characterization of human S-acylated proteins in lipid raft-enriched and non-raft membranes. Mol Cell Proteomics. 2010;9:54–70. doi: 10.1074/mcp.M800448-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.