Abstract
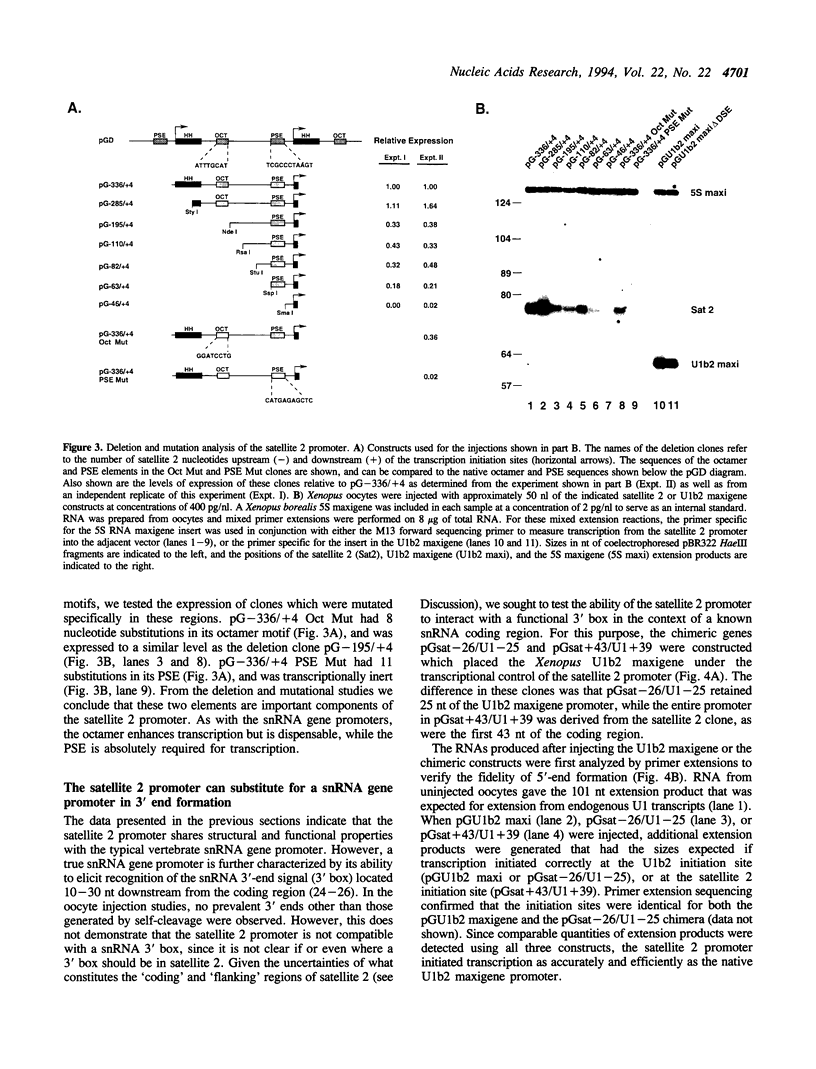
The transcriptional promoter of satellite 2 from the eastern newt, Notophthalmus viridescens, was analyzed by assaying the activity of deleted or mutated satellite 2 clones in Xenopus laevis oocytes. Two elements in the promoter were found to be important for transcription. These elements have sequences that are similar to the sequences of the octamer and the proximal sequence element of vertebrate snRNA genes transcribed by RNA polymerase II. Furthermore, the organization of these elements and their respective roles in transcription are the same as their organization and roles in the snRNA genes. To further investigate the relationship between the satellite 2 and snRNA gene promoters, the ability of the satellite 2 promoter to drive transcription of a true snRNA gene was tested. The satellite 2 promoter initiated transcription of the Xenopus U1b2 snRNA gene as efficiently as the native U1b2 promoter, and the 3' ends of the resulting U1b2 transcripts were accurately formed. This latter result confirms that the satellite 2 promoter is a functional analog of the snRNA promoter, since 3'-end formation of snRNA genes transcribed by RNA polymerase II requires that transcription be initiated from a compatible promoter. The structural and functional similarities between the satellite 2 and the snRNA gene promoters suggest that these elements are evolutionarily related. These findings were used to extend a previously proposed model concerning the nature and derivation of satellite 2.
Full text
PDF





Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bogenhagen D. F., Brown D. D. Nucleotide sequences in Xenopus 5S DNA required for transcription termination. Cell. 1981 Apr;24(1):261–270. doi: 10.1016/0092-8674(81)90522-5. [DOI] [PubMed] [Google Scholar]
- Ciliberto G., Buckland R., Cortese R., Philipson L. Transcription signals in embryonic Xenopus laevis U1 RNA genes. EMBO J. 1985 Jun;4(6):1537–1543. doi: 10.1002/j.1460-2075.1985.tb03814.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cremisi F., Scarabino D., Carluccio M. A., Salvadori P., Barsacchi G. A newt ribozyme: a catalytic activity in search of a function. Proc Natl Acad Sci U S A. 1992 Mar 1;89(5):1651–1655. doi: 10.1073/pnas.89.5.1651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deininger P. L., Batzer M. A., Hutchison C. A., 3rd, Edgell M. H. Master genes in mammalian repetitive DNA amplification. Trends Genet. 1992 Sep;8(9):307–311. doi: 10.1016/0168-9525(92)90262-3. [DOI] [PubMed] [Google Scholar]
- Epstein L. M., Coats S. R. Tissue-specific permutations of self-cleaving newt satellite-2 transcripts. Gene. 1991 Nov 15;107(2):213–218. doi: 10.1016/0378-1119(91)90321-2. [DOI] [PubMed] [Google Scholar]
- Epstein L. M., Gall J. G. Self-cleaving transcripts of satellite DNA from the newt. Cell. 1987 Feb 13;48(3):535–543. doi: 10.1016/0092-8674(87)90204-2. [DOI] [PubMed] [Google Scholar]
- Epstein L. M., Mahon K. A., Gall J. G. Transcription of a satellite DNA in the newt. J Cell Biol. 1986 Oct;103(4):1137–1144. doi: 10.1083/jcb.103.4.1137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Epstein L. M., Pabón-Peña L. M. Alternative modes of self-cleavage by newt satellite 2 transcripts. Nucleic Acids Res. 1991 Apr 11;19(7):1699–1705. doi: 10.1093/nar/19.7.1699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green B., Pabón-Peña L. M., Graham T. A., Peach S. E., Coats S. R., Epstein L. M. Conserved sequence and functional domains in satellite 2 from three families of salamanders. Mol Biol Evol. 1993 Jul;10(4):732–750. doi: 10.1093/oxfordjournals.molbev.a040041. [DOI] [PubMed] [Google Scholar]
- Guerrier-Takada C., Gardiner K., Marsh T., Pace N., Altman S. The RNA moiety of ribonuclease P is the catalytic subunit of the enzyme. Cell. 1983 Dec;35(3 Pt 2):849–857. doi: 10.1016/0092-8674(83)90117-4. [DOI] [PubMed] [Google Scholar]
- Haseloff J., Gerlach W. L. Simple RNA enzymes with new and highly specific endoribonuclease activities. Nature. 1988 Aug 18;334(6183):585–591. doi: 10.1038/334585a0. [DOI] [PubMed] [Google Scholar]
- Hernandez N., Lucito R. Elements required for transcription initiation of the human U2 snRNA gene coincide with elements required for snRNA 3' end formation. EMBO J. 1988 Oct;7(10):3125–3134. doi: 10.1002/j.1460-2075.1988.tb03179.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernandez N., Weiner A. M. Formation of the 3' end of U1 snRNA requires compatible snRNA promoter elements. Cell. 1986 Oct 24;47(2):249–258. doi: 10.1016/0092-8674(86)90447-2. [DOI] [PubMed] [Google Scholar]
- Kazmaier M., Tebb G., Mattaj I. W. Functional characterization of X. laevis U5 snRNA genes. EMBO J. 1987 Oct;6(10):3071–3078. doi: 10.1002/j.1460-2075.1987.tb02614.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiss T., Marshallsay C., Filipowicz W. Alteration of the RNA polymerase specificity of U3 snRNA genes during evolution and in vitro. Cell. 1991 May 3;65(3):517–526. doi: 10.1016/0092-8674(91)90469-f. [DOI] [PubMed] [Google Scholar]
- Krol A., Lund E., Dahlberg J. E. The two embryonic U1 RNA genes of Xenopus laevis have both common and gene-specific transcription signals. EMBO J. 1985 Jun;4(6):1529–1535. doi: 10.1002/j.1460-2075.1985.tb03813.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunkel T. A., Roberts J. D., Zakour R. A. Rapid and efficient site-specific mutagenesis without phenotypic selection. Methods Enzymol. 1987;154:367–382. doi: 10.1016/0076-6879(87)54085-x. [DOI] [PubMed] [Google Scholar]
- Leeflang E. P., Liu W. M., Hashimoto C., Choudary P. V., Schmid C. W. Phylogenetic evidence for multiple Alu source genes. J Mol Evol. 1992 Jul;35(1):7–16. doi: 10.1007/BF00160256. [DOI] [PubMed] [Google Scholar]
- Lund E., Bostock C. J., Dahlberg J. E. The transcription of Xenopus laevis embryonic U1 snRNA genes changes when oocytes mature into eggs. Genes Dev. 1987 Mar;1(1):47–56. doi: 10.1101/gad.1.1.47. [DOI] [PubMed] [Google Scholar]
- Mattaj I. W., Lienhard S., Jiricny J., De Robertis E. M. An enhancer-like sequence within the Xenopus U2 gene promoter facilitates the formation of stable transcription complexes. Nature. 1985 Jul 11;316(6024):163–167. doi: 10.1038/316163a0. [DOI] [PubMed] [Google Scholar]
- Murgo S., Krol A., Carbon P. Sequence, organization and transcriptional analysis of a gene encoding a U1 snRNA from the axolotl, Ambystoma mexicanum. Gene. 1991 Mar 15;99(2):163–170. doi: 10.1016/0378-1119(91)90123-s. [DOI] [PubMed] [Google Scholar]
- Neuman de Vegvar H. E., Dahlberg J. E. Nucleocytoplasmic transport and processing of small nuclear RNA precursors. Mol Cell Biol. 1990 Jul;10(7):3365–3375. doi: 10.1128/mcb.10.7.3365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pabón-Peña L. M., Zhang Y., Epstein L. M. Newt satellite 2 transcripts self-cleave by using an extended hammerhead structure. Mol Cell Biol. 1991 Dec;11(12):6109–6115. doi: 10.1128/mcb.11.12.6109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roebuck K. A., Szeto D. P., Green K. P., Fan Q. N., Stumph W. E. Octamer and SPH motifs in the U1 enhancer cooperate to activate U1 RNA gene expression. Mol Cell Biol. 1990 Jan;10(1):341–352. doi: 10.1128/mcb.10.1.341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanger F., Coulson A. R., Barrell B. G., Smith A. J., Roe B. A. Cloning in single-stranded bacteriophage as an aid to rapid DNA sequencing. J Mol Biol. 1980 Oct 25;143(2):161–178. doi: 10.1016/0022-2836(80)90196-5. [DOI] [PubMed] [Google Scholar]
- Savino R., Hitti Y., Gerbi S. A. Genes for Xenopus laevis U3 small nuclear RNA. Nucleic Acids Res. 1992 Oct 25;20(20):5435–5442. doi: 10.1093/nar/20.20.5435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shatkin A. J. Capping of eucaryotic mRNAs. Cell. 1976 Dec;9(4 Pt 2):645–653. doi: 10.1016/0092-8674(76)90128-8. [DOI] [PubMed] [Google Scholar]
- Shinshi H., Miwa M., Sugimura T. Enzyme cleaving the 5'-terminal methylated blocked structure of messenger RNA. FEBS Lett. 1976 Jun 1;65(2):254–257. doi: 10.1016/0014-5793(76)80492-9. [DOI] [PubMed] [Google Scholar]
- Symons R. H. Self-cleavage of RNA in the replication of small pathogens of plants and animals. Trends Biochem Sci. 1989 Nov;14(11):445–450. doi: 10.1016/0968-0004(89)90103-5. [DOI] [PubMed] [Google Scholar]
- Uhlenbeck O. C. A small catalytic oligoribonucleotide. Nature. 1987 Aug 13;328(6131):596–600. doi: 10.1038/328596a0. [DOI] [PubMed] [Google Scholar]
- Ullu E., Tschudi C. Alu sequences are processed 7SL RNA genes. Nature. 1984 Nov 8;312(5990):171–172. doi: 10.1038/312171a0. [DOI] [PubMed] [Google Scholar]
- Weiner A. M., Denison R. A. Either gene amplification or gene conversion may maintain the homogeneity of the multigene family encoding human U1 small nuclear RNA. Cold Spring Harb Symp Quant Biol. 1983;47(Pt 2):1141–1149. doi: 10.1101/sqb.1983.047.01.129. [DOI] [PubMed] [Google Scholar]
- Yang H., Moss M. L., Lund E., Dahlberg J. E. Nuclear processing of the 3'-terminal nucleotides of pre-U1 RNA in Xenopus laevis oocytes. Mol Cell Biol. 1992 Apr;12(4):1553–1560. doi: 10.1128/mcb.12.4.1553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Vegvar H. E., Lund E., Dahlberg J. E. 3' end formation of U1 snRNA precursors is coupled to transcription from snRNA promoters. Cell. 1986 Oct 24;47(2):259–266. doi: 10.1016/0092-8674(86)90448-4. [DOI] [PubMed] [Google Scholar]