Abstract
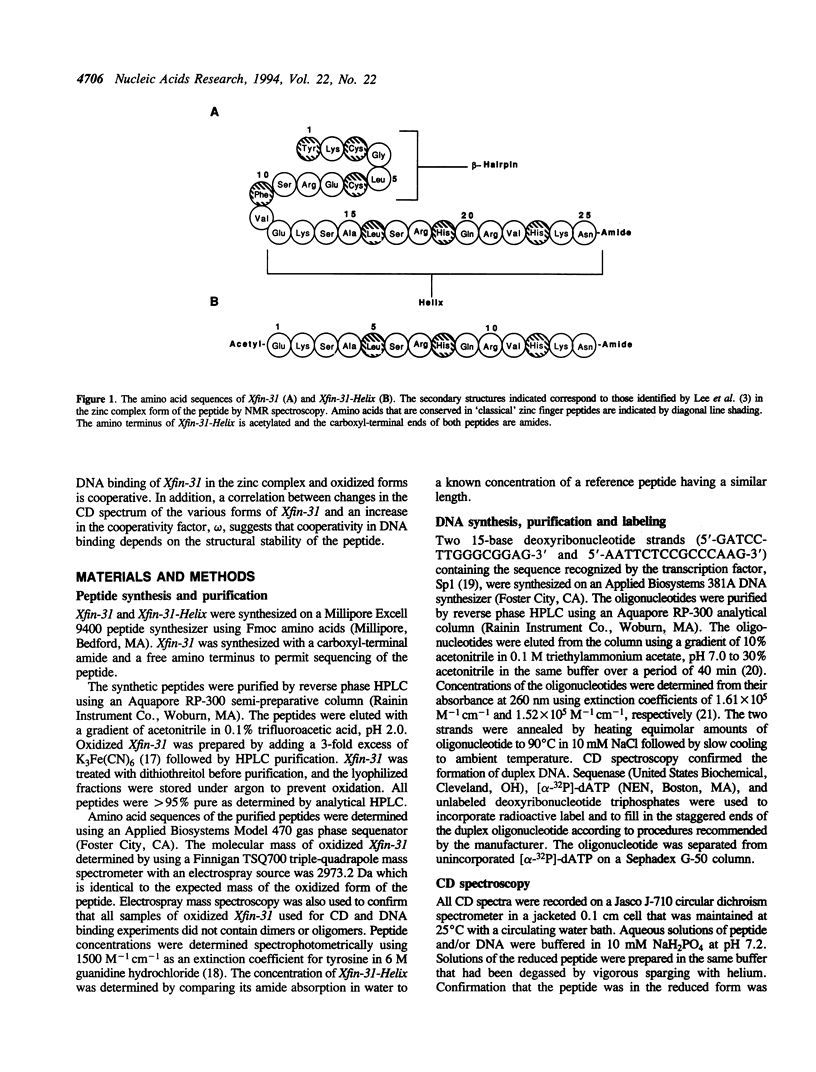
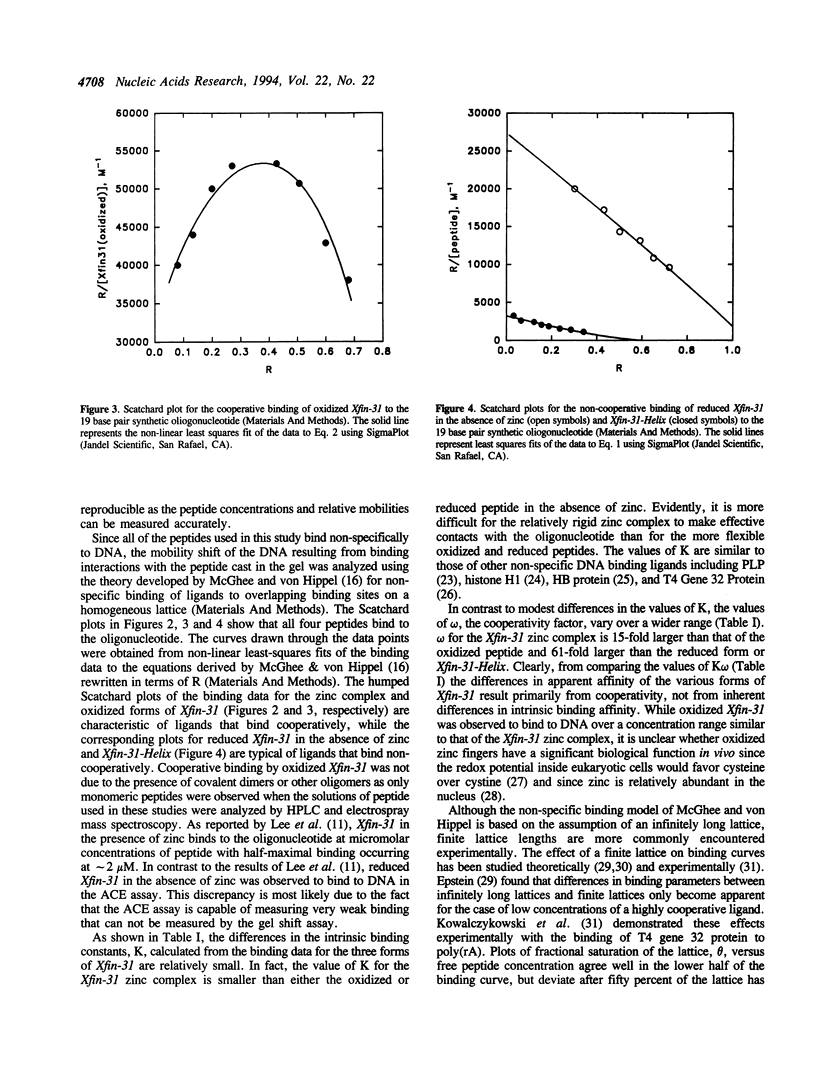
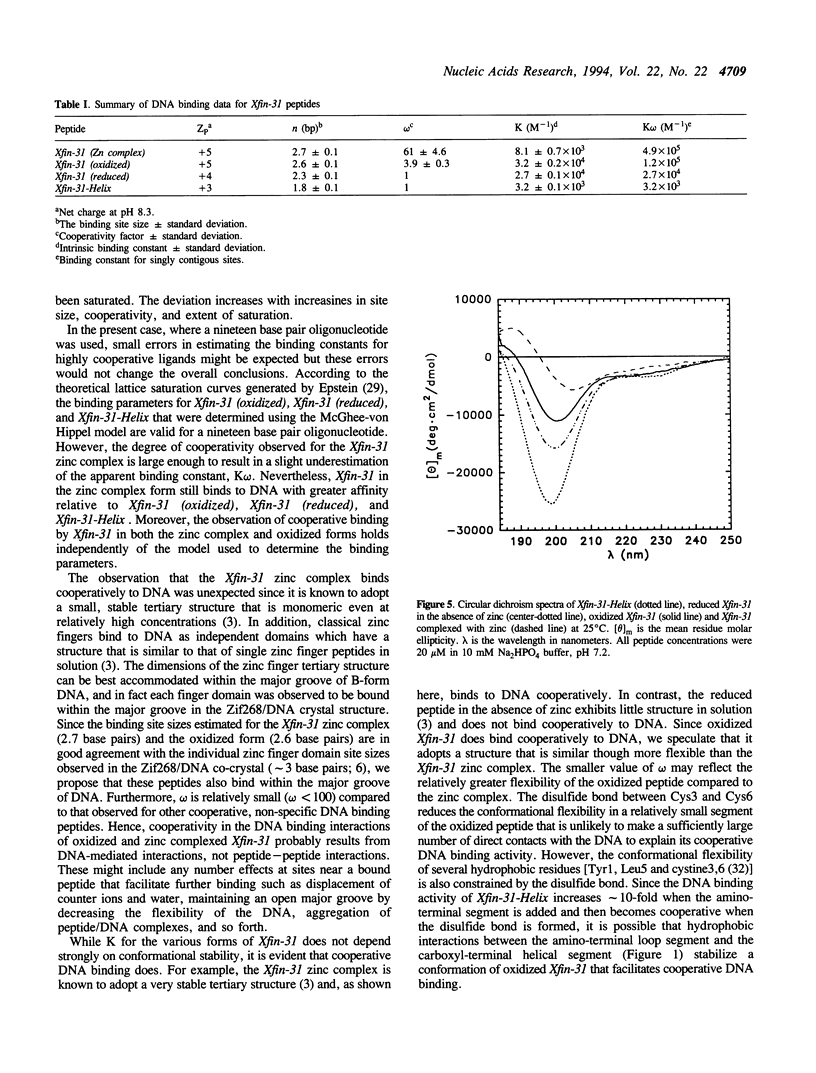
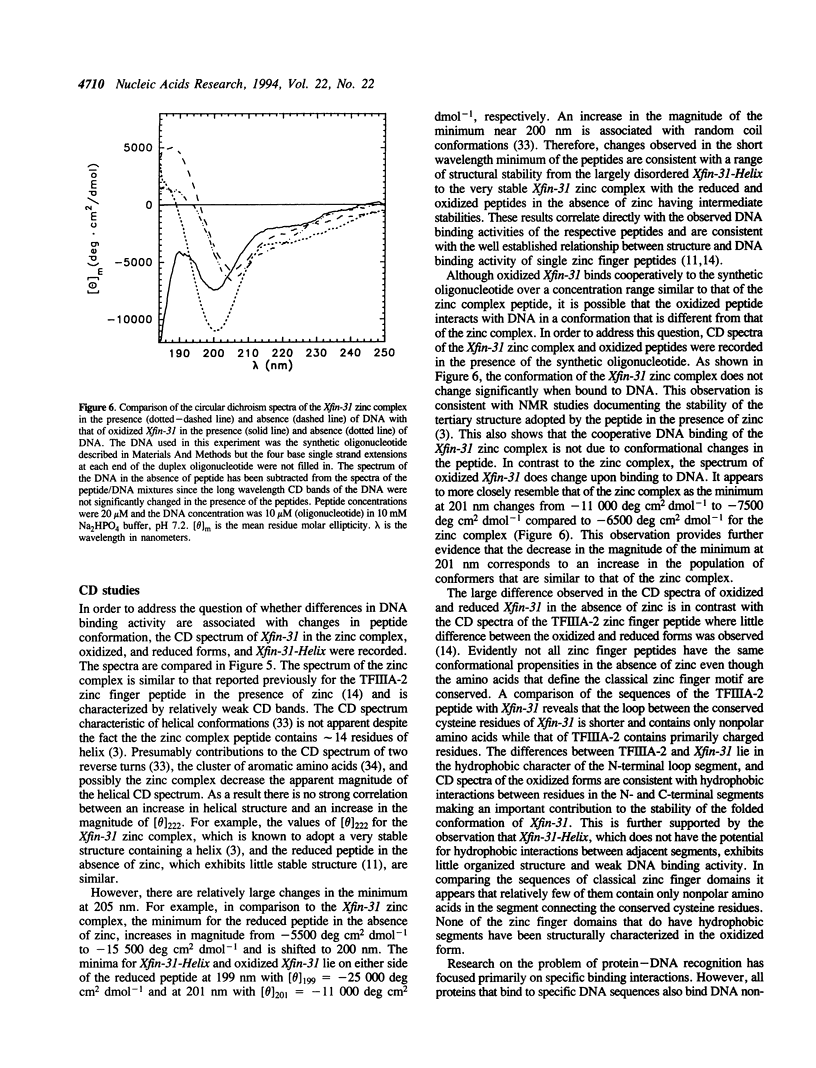
The DNA binding and structural properties of Xfin-31 (Lee, M.S., Gippert, G.P., Soman, K.V., Case, D.A. and Wright, P.E., 1989, Science 245, 635-637), a twenty five amino acid zinc finger peptide, in the reduced, oxidized and zinc complex forms, as well as the fourteen residue helical segment of the zinc finger (residues 12-25) have been compared using affinity coelectrophoresis (ACE) and circular dichroism (CD) spectroscopy. The zinc complex and oxidized peptides bind cooperatively to DNA although the cooperativity factor, omega, is more than 15-fold greater for the zinc complex. The reduced peptide in the absence of zinc and the helical segment do not bind cooperatively (omega = 1). Hence, the binding constant for singly contiguous sites (K omega) ranges over 100-fold for the various peptides even though the intrinsic binding constants (K) are similar. An increase in binding order and affinity for the other forms of Xfin-31 is correlated with an increasing similarity of the CD spectrum to that of the Xfin-31 zinc complex. The surprising DNA binding activity of the oxidized peptide may result from hydrophobic interactions between the amino-terminal loop formed by the Cys3-Cys6 disulfide bond and conserved hydrophobic residues in the carboxyl-terminal segment. Xfin-31 may be a particularly useful model for studying several poorly understood aspects of cooperative, non-specific DNA binding since it is small, has a stable, well-defined structure, and structures of zinc fingers bound to DNA have been determined.
Full text
PDF


Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Carr M. D., Pastore A., Gausepohl H., Frank R., Roesch P. NMR and molecular dynamics studies of the mKr2 'zinc finger'. Eur J Biochem. 1990 Mar 10;188(2):455–461. doi: 10.1111/j.1432-1033.1990.tb15423.x. [DOI] [PubMed] [Google Scholar]
- Diakun G. P., Fairall L., Klug A. EXAFS study of the zinc-binding sites in the protein transcription factor IIIA. Nature. 1986 Dec 18;324(6098):698–699. doi: 10.1038/324698a0. [DOI] [PubMed] [Google Scholar]
- Edelhoch H. Spectroscopic determination of tryptophan and tyrosine in proteins. Biochemistry. 1967 Jul;6(7):1948–1954. doi: 10.1021/bi00859a010. [DOI] [PubMed] [Google Scholar]
- Epstein I. R. Cooperative and non-cooperative binding of large ligands to a finite one-dimensional lattice. A model for ligand-oligonucleotide interactions. Biophys Chem. 1978 Sep;8(4):327–339. doi: 10.1016/0301-4622(78)80015-5. [DOI] [PubMed] [Google Scholar]
- Frankel A. D., Berg J. M., Pabo C. O. Metal-dependent folding of a single zinc finger from transcription factor IIIA. Proc Natl Acad Sci U S A. 1987 Jul;84(14):4841–4845. doi: 10.1073/pnas.84.14.4841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gidoni D., Dynan W. S., Tjian R. Multiple specific contacts between a mammalian transcription factor and its cognate promoters. 1984 Nov 29-Dec 5Nature. 312(5993):409–413. doi: 10.1038/312409a0. [DOI] [PubMed] [Google Scholar]
- Hwang C., Sinskey A. J., Lodish H. F. Oxidized redox state of glutathione in the endoplasmic reticulum. Science. 1992 Sep 11;257(5076):1496–1502. doi: 10.1126/science.1523409. [DOI] [PubMed] [Google Scholar]
- Klevit R. E., Herriott J. R., Horvath S. J. Solution structure of a zinc finger domain of yeast ADR1. Proteins. 1990;7(3):215–226. doi: 10.1002/prot.340070303. [DOI] [PubMed] [Google Scholar]
- Kowalczykowski S. C., Paul L. S., Lonberg N., Newport J. W., McSwiggen J. A., von Hippel P. H. Cooperative and noncooperative binding of protein ligands to nucleic acid lattices: experimental approaches to the determination of thermodynamic parameters. Biochemistry. 1986 Mar 25;25(6):1226–1240. doi: 10.1021/bi00354a006. [DOI] [PubMed] [Google Scholar]
- Lee M. S., Gippert G. P., Soman K. V., Case D. A., Wright P. E. Three-dimensional solution structure of a single zinc finger DNA-binding domain. Science. 1989 Aug 11;245(4918):635–637. doi: 10.1126/science.2503871. [DOI] [PubMed] [Google Scholar]
- Lee M. S., Gottesfeld J. M., Wright P. E. Zinc is required for folding and binding of a single zinc finger to DNA. FEBS Lett. 1991 Feb 25;279(2):289–294. doi: 10.1016/0014-5793(91)80170-8. [DOI] [PubMed] [Google Scholar]
- Libertini L. J., Ausió J., van Holde K. E., Small E. W. Highly cooperative binding to DNA by a histone-like, sperm-specific protein from Spisula solidissima. Biopolymers. 1988 Sep;27(9):1459–1477. doi: 10.1002/bip.360270911. [DOI] [PubMed] [Google Scholar]
- Lim W. A., Sauer R. T., Lander A. D. Analysis of DNA-protein interactions by affinity coelectrophoresis. Methods Enzymol. 1991;208:196–210. doi: 10.1016/0076-6879(91)08014-9. [DOI] [PubMed] [Google Scholar]
- Lohman T. M., Kowalczykowski S. C. Kinetics and mechanism of the association of the bacteriophage T4 gene 32 (helix destabilizing) protein with single-stranded nucleic acids. Evidence for protein translocation. J Mol Biol. 1981 Oct 15;152(1):67–109. doi: 10.1016/0022-2836(81)90096-6. [DOI] [PubMed] [Google Scholar]
- Manning M. C., Woody R. W. Theoretical study of the contribution of aromatic side chains to the circular dichroism of basic bovine pancreatic trypsin inhibitor. Biochemistry. 1989 Oct 17;28(21):8609–8613. doi: 10.1021/bi00447a051. [DOI] [PubMed] [Google Scholar]
- McGhee J. D., von Hippel P. H. Theoretical aspects of DNA-protein interactions: co-operative and non-co-operative binding of large ligands to a one-dimensional homogeneous lattice. J Mol Biol. 1974 Jun 25;86(2):469–489. doi: 10.1016/0022-2836(74)90031-x. [DOI] [PubMed] [Google Scholar]
- Michael S. F., Kilfoil V. J., Schmidt M. H., Amann B. T., Berg J. M. Metal binding and folding properties of a minimalist Cys2His2 zinc finger peptide. Proc Natl Acad Sci U S A. 1992 Jun 1;89(11):4796–4800. doi: 10.1073/pnas.89.11.4796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagai K., Nakaseko Y., Nasmyth K., Rhodes D. Zinc-finger motifs expressed in E. coli and folded in vitro direct specific binding to DNA. Nature. 1988 Mar 17;332(6161):284–286. doi: 10.1038/332284a0. [DOI] [PubMed] [Google Scholar]
- Omichinski J. G., Clore G. M., Appella E., Sakaguchi K., Gronenborn A. M. High-resolution three-dimensional structure of a single zinc finger from a human enhancer binding protein in solution. Biochemistry. 1990 Oct 9;29(40):9324–9334. doi: 10.1021/bi00492a004. [DOI] [PubMed] [Google Scholar]
- Omichinski J. G., Clore G. M., Robien M., Sakaguchi K., Appella E., Gronenborn A. M. High-resolution solution structure of the double Cys2His2 zinc finger from the human enhancer binding protein MBP-1. Biochemistry. 1992 Apr 28;31(16):3907–3917. doi: 10.1021/bi00131a004. [DOI] [PubMed] [Google Scholar]
- Pavletich N. P., Pabo C. O. Zinc finger-DNA recognition: crystal structure of a Zif268-DNA complex at 2.1 A. Science. 1991 May 10;252(5007):809–817. doi: 10.1126/science.2028256. [DOI] [PubMed] [Google Scholar]
- Párraga G., Horvath S. J., Eisen A., Taylor W. E., Hood L., Young E. T., Klevit R. E. Zinc-dependent structure of a single-finger domain of yeast ADR1. Science. 1988 Sep 16;241(4872):1489–1492. doi: 10.1126/science.3047872. [DOI] [PubMed] [Google Scholar]
- Record M. T., Jr, Lohman M. L., De Haseth P. Ion effects on ligand-nucleic acid interactions. J Mol Biol. 1976 Oct 25;107(2):145–158. doi: 10.1016/s0022-2836(76)80023-x. [DOI] [PubMed] [Google Scholar]
- Sakaguchi K., Appella E., Omichinski J. G., Clore G. M., Gronenborn A. M. Specific DNA binding to a major histocompatibility complex enhancer sequence by a synthetic 57-residue double zinc finger peptide from a human enhancer binding protein. J Biol Chem. 1991 Apr 15;266(11):7306–7311. [PubMed] [Google Scholar]
- Saunders A. J., Young G. B., Pielak G. J. Polarity of disulfide bonds. Protein Sci. 1993 Jul;2(7):1183–1184. doi: 10.1002/pro.5560020713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- South T. L., Summers M. F. Zinc fingers. Adv Inorg Biochem. 1990;8:199–248. [PubMed] [Google Scholar]
- THIERS R. E., VALLEE B. L. Distribution of metals in subcellular fractions of rat liver. J Biol Chem. 1957 Jun;226(2):911–920. [PubMed] [Google Scholar]
- Wallace R. B., Miyada C. G. Oligonucleotide probes for the screening of recombinant DNA libraries. Methods Enzymol. 1987;152:432–442. doi: 10.1016/0076-6879(87)52050-x. [DOI] [PubMed] [Google Scholar]
- Watanabe F. Cooperative interaction of histone H1 with DNA. Nucleic Acids Res. 1986 Apr 25;14(8):3573–3585. doi: 10.1093/nar/14.8.3573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe F., Stankowski S., Schwarz G. Interaction of the HB protein of Bacillus globigii with nucleic acids. Analysis of the binding to DNA and polynucleotides. Eur J Biochem. 1984 Apr 2;140(1):215–219. doi: 10.1111/j.1432-1033.1984.tb08089.x. [DOI] [PubMed] [Google Scholar]
- von Hippel P. H. Protein-DNA recognition: new perspectives and underlying themes. Science. 1994 Feb 11;263(5148):769–770. doi: 10.1126/science.8303292. [DOI] [PubMed] [Google Scholar]


