Summary
Most bacteria surround themselves with a peptidoglycan (PG) exoskeleton synthesized by polysaccharide polymerases called penicillin-binding proteins (PBPs). Because they are the targets of penicillin and related antibiotics, the structure and biochemical functions of the PBPs have been extensively studied. Despite this, we still know surprisingly little about how these enzymes build the PG layer in vivo. Here, we identify the Escherichia coli outer membrane lipoproteins LpoA and LpoB as essential PBP cofactors. We show that LpoA and LpoB form specific trans-envelope complexes with their cognate PBP and are critical for PBP function in vivo. We further show that LpoB promotes PG synthesis by its partner PBP in vitro and that it likely does so by stimulating glycan chain polymerization. Overall, our results indicate that PBP accessory proteins play a central role in PG biogenesis and, like the PBPs they work with, these factors are attractive targets for antibiotic development.
Introduction
To fortify their cytoplasmic membrane and protect it from osmotic rupture, most bacteria surround themselves with a peptidoglycan (PG) exoskeleton. This tough polysaccharide layer is constructed from long glycan chains crosslinked to one another via attached peptides to form a continuous matrix that envelops the cell (Fig. 1A). Because bacteria are encased within this polymeric shell, their growth and morphogenesis is intimately linked to PG synthesis and remodeling; PG expansion is essential for growth, and the shape of the organism is defined by the PG network (Margolin, 2009).
Figure 1. Cell wall structure and assembly.
A. Schematic of bacterial cells with the cell wall (PG layer) in green. Gram-negative cells have a relatively thin PG layer surrounded by an additional (outer) membrane. Above the cells is a schematic detailing the structure of PG, which continues in all directions to envelop the cell (green arrows). M, N-acetylmuramic acid; G, N-acetylglucosamine. Dots represent the attached peptides. B. Overview of PG assembly. A generic multi-protein complex (grey) containing a class A PBP (purple) is shown. For simplicity, the other PG assembly factors thought to participate in the final stages of PG construction are not specifically labeled. See text for details. PGT, peptidoglycan glycosyltransferase domain; TP, transpeptidase domain.
The PG synthesis pathway is of tremendous practical importance as it is the target of many of our most effective antibiotics, notably penicillin and related β-lactams. Since its discovery over eighty years ago, penicillin has served as a key probe of the PG assembly process as well as a widely used therapeutic. It covalently modifies and inhibits PG synthases called high - molecular-weight penicillin-binding proteins (HMW-PBPs) (Sauvage et al., 2008), which henceforth will be referred to simply as PBPs. Bacteria typically encode two varieties of PBPs: class A and class B (Sauvage et al., 2008). Both types are integral membrane proteins with relatively large domains facing the cell exterior. Class A PBPs are thought to be the primary cellular PG synthases since they are bifunctional and have both peptidoglycan glycosyl transferase (PGT) and transpeptidase (TP) domains capable of polymerizing the glycan strands of PG and crosslinking them, respectively (Fig. 1A and 1B). Class B PBPs are mono-functional and only have TP activity.
Although purified bifunctional PBPs can polymerize and crosslink PG in vitro from lipid-II substrate (Fig.1B) (Bertsche et al., 2005; Barrett et al., 2007), the PBPs alone are insufficient for the proper assembly of the cell-shaped PG network in vivo. To build a dynamic, uniformly-shaped PG mesh that grows in step with the rest of the cell, PBP activity must be spatially and temporally controlled (Margolin, 2009). Adding to the complexity, rod-shaped bacteria like Escherichia coli and Bacillus subtilis must properly switch between different modes of PG growth: elongation of the cylindrical portion of the rod and synthesis of the hemispherical polar caps during division. It is therefore not surprising that, besides the PBPs, an array of additional proteins have been implicated in PG assembly (den Blaauwen et al., 2008; Margolin, 2009). Recent work indicates that many of these factors are organized into multi-enzyme PG synthesizing complexes by cytoskeletal polymers of FtsZ (tubulin-like) and MreB (actin-like), which are thought to direct their activity to appropriate subcellular locations (Fig. 1B). Studies from a number of laboratories have identified likely components of these complexes, including: (i) many, if not all, of the enzymes required for lipid-II synthesis (MurA-MurG), (ii) both classes of synthetic PBPs, (iii) various PG hydrolases, and (iv) several essential integral membrane proteins of unknown function, such as SEDS-domain proteins like RodA or FtsW and the elongation specific factors MreC and MreD (den Blaauwen et al., 2008; Margolin, 2009; White et al., 2010). Many questions regarding the function and composition of these PBP-containing, multi-enzyme complexes remain to be addressed. One of the most fundamental, however, is whether such complexes promote PG synthesis simply by providing the PBPs with access to substrate via lipid-II synthesis and flipping (Fig. 1B), or whether they also contain critical accessory factors that facilitate and/or regulate PBP activity by affecting lipid-II utilization or the incorporation of nascent PG into the existing network.
Here, we report the discovery of protein cofactors essential for the in vivo function of the bifunctional PBPs. They were identified using directed genetic screens in the model gram-negative bacterium, E. coli. We have designated them LpoA (YraM) and LpoB (YcfM) for lipoprotein activators of bifunctional PBP activity from the outer membrane. We demonstrate that they directly affect PBP activity through the formation of specific trans-envelope Lpo-PBP complexes. In addition, we show that LpoB promotes PG synthesis by its partner PBP in vitro and that it does so by stimulating glycan chain polymerization. Overall, our results indicate that PBP accessory proteins play a central role in PG biogenesis and, like the PBPs they work with, these factors are attractive targets for antibiotic development. An accompanying report from Typas et al. (Typas et al., 2010) describes the independent discovery of the Lpo factors using large-scale phenotyping and proteomic approaches.
Results
Rationale for the synthetic lethal screens
E. coli, like many bacteria, encodes multiple class A PBPs: PBP1a, PBP1b, and PBP1c (Sauvage et al., 2008). Prior genetic studies indicated that cells remain viable if either PBP1a or PBP1b is inactivated, but not if they both are (Yousif et al., 1985; Kato et al., 1985). This suggests that there are at least two major PG synthesizing complexes in the cell, one containing PBP1a and the other PBP1b, an idea supported by crosslinking studies in Haemophilus influenzae and immunoprecipitation studies in E. coli (Charpentier et al., 2002; Alaedini and Day, 1999). Based on this, we reasoned that we could identify critical components of these multi-enzyme PBP complexes by performing a screen for mutations synthetically lethal with the loss of either PBP1a or PBP1b. For example, if factor X is an important component of a PBP1a synthetic complex, one might expect mutations that inactivate factor X to be just as lethal when PBP1b is depleted as the inactivation of PBP1a is. Therefore, a screen for mutants synthetically lethal with the loss of PBP1b should yield factor X mutants as well as ponA (PBP1a) mutants. The converse is also true. Important components of the PBP1b synthetic complex should be identified by a screen for mutants synthetically lethal with the loss of PBP1a.
Screening for mutants synthetically lethal with the loss of PBP1a or PBP1b
Our synthetic lethal screen employed methodology we used previously to identify new cell division factors and regulators of division site placement (Bernhardt and de Boer, 2004; Bernhardt and de Boer, 2005). To apply the screen to the PBPs, we initially searched for mutants synthetically lethal with the loss of PBP1b function (slb mutants). Our parental strain for the screen had chromosomal deletions removing both lacZ and ponB (PBP1b). The strain also harbored an unstable plasmid containing both the ponB and lacZ genes under control of the inducible lactose promoter (Plac). Since PBP1b is not essential, cells frequently lost the plasmid when they were grown on non-selective media containing the Plac inducer, IPTG, and the LacZ indicator, X-gal. They formed either white (LacZ-) colonies or blue-sectored colonies resulting from plasmid loss before or during colony formation, respectively (Fig. 2A and 2B). To find slb mutants, we mutagenized the parental strain with a transposon and plated the resulting library on indicator medium to look for rare solid-blue colonies (Fig. 2A and 2B). Mutants forming such colonies presumably could not lose the plasmid because the transposon insertion rendered PBP1b essential. To confirm this, we tested the slb candidates for growth with or without IPTG. Mutants truly dependent on PBP1b for growth should require the presence of IPTG to induce ponB expression from the plasmid.
Figure 2. Synthetic lethal screens and terminal phenotypes.
A-B. Colonies of transposon mutants used for the slb mutant screen grown on an indicator plate (LB-IPTG-Xgal) for 2 days at 30°C. The arrow points to a rare solid-blue colony that retained the unstable plasmid. The boxed region in (A) is enlarged in (B). C. Schematics indicating the approximate locations of the transposon insertions in the lpo genes. Triangles represent transposon insertion points (green: transcription of the KanR cassette is in the same direction as the target gene; red: transcription is in the opposite direction). D. Plating defects of representative slb and sla mutants isolated in the screens. Cells of TU122/pTU110 [ΔponB/Plac∷ponB lacZ] (top) or TU121/pCB1 [ΔponA/Plac∷gfp-ponA lacZ] (bottom) and their derivatives with the indicated transposon insertions were grown overnight at 30°C. Culture densities were normalized, 10-fold serial dilutions were prepared for each, and 5 μl of each dilution was spotted onto LB with or without IPTG as indicated. E. Cells of TU121(attλTB309) [ΔponA(Para∷ponA)] (top) or MM11 [Para∷ponB] (bottom) and their derivatives were grown overnight at 37°C. Serial dilutions were prepared as in (D) and dilutions were spotted onto the indicated media. F-G. Strains used in (E) were grown in LB-arabinose at 37°C to an OD600 = 0.6-1.1. They were then pelleted, washed three times with LB, and resuspended in LB-glucose at an OD600 = 0.08 or 0.02 for TU121(attλTB309) (F) or MM11 derivatives (G), respectively. Cell growth following subculture (t = 0) was then monitored by regular OD600 measurements. Please also see data in Figure S2 and S3.
We screened a total of 30,000 colonies and isolated 16 slb mutants. Less than half of these (5/16) were completely dependent on IPTG for growth. Three of the IPTG-dependent mutants had transposon insertions that mapped to the gene for PBP1a (ponA), indicating that the screen worked as expected (Fig. 2D). Two of the remaining IPTG-dependent mutants had transposons that mapped within lpoA (yraM) (Fig. 2C and 2D). The lpoA reading frame codes for a 678 amino acid protein with an LppC domain that is well conserved among the γ-proteobacteria but has an unknown function (Finn et al., 2008; Vijayalakshmi et al., 2008). LpoA is also predicted to have a lipoprotein signal sequence for targeting it to the outer membrane (see below). Besides the ponA and lpoA alleles, most of the remaining slb isolates showed a synthetically sick phenotype in combination with a PBP1b defect. They all had transposon insertions that mapped to genes encoding factors previously implicated in cell division and will be described as part of a separate report.
We also performed the converse screen for mutants synthetically lethal with the loss of PBP1a function (sla mutants). The screen design was identical to the one described above except that the parental strain was deleted for the ponA (PBP1a) gene and gfp-ponA was encoded on the unstable plasmid. A total of 41 sla mutants displaying IPTG-dependent growth were isolated. The majority (34/41) were found to have transposon insertions disrupting the gene coding for PBP1b (Fig. 2D), and 7/41 sla mutants had transposon insertions disrupting the gene coding for LpoB (YcfM) (Fig. 2C and 2D). LpoB is a 213 amino acid protein of previously unknown function that, like LpoA, is predicted to have a lipoprotein signal sequence for targeting it to the outer membrane (see below). Since the bias in the isolation of ponB mutants might have prevented us from identifying additional sla loci, we repeated the sla screen in a strain containing a second copy of ponB. In this case, only one mutant was isolated and it contained a transposon insertion in lpoB. We therefore conclude that ponB and lpoB are likely to be the only sla loci in the genome.
Specificity of the synthetically lethal combinations and their terminal phenotypes
To test the specificity of the synthetic lethal combinations, we constructed strains TU121(attλTB309) [ΔponA(Para∷ponA)] and MM11 [Para∷ponB] in which the production of either PBP1a or PBP1b was controlled by the arabinose promoter, respectively. In the case of TU121 (attλTB309), the native ponA locus was deleted and a second copy of ponA under arabinose promoter control was integrated at the λ att site. For MM11, the native ponB promoter was replaced with the arabinose promoter. Growth of TU121(attλTB309) [ΔponA(Para∷ponA)] derivatives lacking PBP1b or LpoB was severely inhibited on media without arabinose (Fig. 2E). Growth of the corresponding LpoA- strain was unaffected (Fig. 2E). Similarly, MM11 [Para∷ponB] derivatives defective for PBP1a or LpoA failed to grow without arabinose supplementation, while the corresponding LpoB- strain showed robust growth (Fig. 2E). Thus, LpoA is specifically required in the absence of PBP1b and LpoB is specifically required in the absence of PBP1a.
The terminal phenotype of PBP1a depletion in the absence of PBP1b is cell lysis, and vice versa (Fig. 2F and 2G) (Yousif et al., 1985). To determine if cells lacking specific Lpo-PBP1 combinations also lyse, we followed the growth and morphology of strains lacking Lpo factors after initiating PBP1 depletion. When PBP1a was depleted, cells lacking PBP1b or LpoB displayed a dramatic lysis phenotype. About three generations following the initiation of PBP1a depletion, the optical density of the mutant cultures began to decline rapidly (Fig. 2F). This coincided with the appearance of cell ghosts and lysing cells with membrane blebs in the cultures (Fig. S1). The only difference between the PBP1b- and LpoB- cells upon PBP1a depletion was that the LpoB- cells consistently began lysing about 10 minutes later than PBP1b- cells (Fig. 2F). Similar results were obtained when we compared the effect of PBP1b depletion on cells defective for PBP1a or LpoA. In both cases, the terminal phenotype was lysis (Fig. 2G). As on plates, growth of PBP1a- LpoA- or PBP1b- LpoB- cells in liquid was normal (data not shown). Importantly, the synthetic lethal phenotypes resulting from the lpoA or lpoB deletions were corrected by their expression in trans, indicating that the phenotypes observed were not due to an adverse effect of the deletions on the expression of nearby genes (Fig. S2). In addition, Bocillin labeling and immunoblotting indicated that the defects observed for the LpoA/B- mutants were not due to a decrease in PBP1 protein levels (Fig. S3).
Although PBP1a and PBP1b appear to be largely redundant, the phenotypes of cells lacking individual PBP1s are not identical. Loss of PBP1b leads to a hypersensitivity to β-lactam antibiotics that is not observed for PBP1a- mutants (Schmidt et al., 1981; Yousif et al., 1985). In addition, inhibition of the class B PBPs, PBP2 with mecillinam or PBP3 with cephalexin, normally leads to the formation of spherical or filamentous cells, respectively, that are slow to lyse (Spratt and Pardee, 1975). Similar treatments of mutants lacking PBP1b, however, rapidly induce cell lysis (Schmidt et al., 1981; Garcia del Portillo and de Pedro, 1990). Although the reasons for the aberrant β-lactam phenotypes of PBP1b mutants are not known, we used these observations to further investigate the equivalence of LpoB and PBP1b defects. We found that LpoB defective mutants indeed shared a β-lactam hypersensitivity phenotype with PBP1b- mutants (Fig. S4A). Moreover, LpoB- cells also lysed rapidly when treated with mecillinam or cephalexin (Fig. S4B-D). Thus, a LpoB- mutant has all of the hallmarks of a PBP1b- defect.
At least one Lpo factor is required for growth
Our results thus far suggest that LpoA is important for PBP1a function and that LpoB is important for PBP1b function. If this is true, then the combined loss of LpoA and LpoB should phenocopy the lysis and lethality observed when PBP1a and PBP1b are simultaneously inactivated. Accordingly, when LpoA was depleted in the absence of LpoB, cells lysed and failed to grow on media lacking inducer for lpoA expression (Fig. 3A and 3B).
Figure 3. Lpo factors are essential for growth and PBP1 function.
A. Cells of MM22(attHKMM10) [ΔlpoA(Plac∷lpoA-gfp)] and its derivatives were grown overnight in LB-IPTG (50μM) at 37°C. Serial dilutions were prepared as in Figure 2, and spotted onto the indicated solid media. Identical results were obtained when LpoB was depleted in the absence of LpoA (data not shown). B. Cultures of cells from (A) were grown to an OD600 of 0.3-0.4 in LB-IPTG (50μM) at 37°C. The cells were then pelleted, washed three times with LB, and resuspended in LB-glucose at an OD600 = 0.02. Cell growth following subculture (t = 0) was then monitored by regular OD600 measurements. C-D. Cells of MM13 [Para∷ponB ΔlpoA] (C) or CB4(attλTB309) [ΔponA ΔlpoB(Para∷ponA)] (D) containing the low-copy plasmids pTB284 [Plac-con∷gfp], pCB62 [Plac-con∷ponA], or pCB72 [Plac-con∷ponB] were grown to an OD600 of 0.5-0.7 in LB-Ara-Spc (C) or M9-Ara-Spc (D) at 37°C. They were then washed, diluted into LB-IPTG (1mM), and growth at 37°C was followed as in (B). E-F. To determine the extent of PBP1a or PBP1b overproduction in the cultures from (C) and (D), respectively, extracts were prepared from cells harvested at times indicated by the arrows above the growth curves (C-D). Immunoblot analysis was then performed to determine the levels of PBP1a (E) or PBP1b (F) in strains MM13/pCB62 or CB4(attλTB309)/pCB72, respectively, relative to corresponding control strains harboring pTB284. Numbers above lanes indicate the amount of total protein loaded. G. Cultures of TB28 [WT], MM13, or CB4(attλTB309) containing the aforementioned plasmids were diluted, plated on the indicated media containing Spc, and incubated overnight at 37°C. Plasmids pCB62 [Plac-con∷ponA] and pCB72 [Plac-con∷ponB] possess native ribosome binding sites and 5′UTRs for ponA and ponB, respectively. pCB72 likely has a higher basal level of expression than pCB62 since it can correct the PBP1B- LpoA- phenotype of MM13 without IPTG induction. Plac-con is a synthetic lac promoter with consensus -35 and -10 elements. Please also see data in Figure S1 and S4.
The Lpo factors are essential for PBP1 function
To determine whether or not the Lpo proteins are essential for PBP1 function, we tested the ability of PBP1a/b overproduction to suppress the synthetic lethal phenotypes associated with a LpoA/B- defect. Overproduction of PBP1a by approximately 4 fold from plasmid pCB62 [Plac-con∷ponA] was not sufficient to suppress the Slb phenotype of MM13 [Para∷ponB ΔlpoA] cells upon PBP1b depletion in liquid or solid medium (Fig. 3C, 3E, and 3G). Only a minor delay in the timing of lysis was observed for MM13 cells harboring pCB62 relative to pTB284 [Plac-con∷gfp] (Fig. 3C). Similarly, an approximately 8 fold overproduction of PBP1b from pCB72 [Plac-con∷ponB] failed to rescue the lytic phenotype and plating defects of CB4(attλTB309) [ΔponA ΔlpoB(Para∷ponA)] cells depleted of PBP1a (Fig. 3D, 3F, and 3G). We conclude that the Lpo proteins are essential for the in vivo function of their cognate PBP as opposed to factors that simply stimulate PBP activity but are not critical for their function.
Lpo factors localize throughout the outer membrane
Based on the sequence of their signal peptides, LpoA and LpoB are both predicted to be outer membrane lipoproteins (Fig. S5A). To test this, we used a cytological assay for lipoprotein transport developed by Pugsley and co-workers (Lewenza et al., 2006). In this assay, cells expressing a fluorescent protein fused to a lipoprotein or lipobox-containing signal sequence are visualized following osmotic shock. This treatment induces the formation of plasmolysis bays where the inner membrane retracts from the rest of the cell envelope. Thus, a fluorescently labeled lipoprotein that is retained in the inner membrane will localize to the invaginations of the plasmolysis bays whereas an outer membrane lipoprotein will retain a peripheral localization pattern.
Before osmotic shock, GFP fusions to LpoA and LpoB displayed a patchy, non-uniform peripheral localization pattern (Fig. S6E and S6G), indicating that they are broadly distributed throughout the envelope. In unconstricted cells, GFP-LpoA and GFP-LpoB retained their peripheral distribution following osmotic shock, suggesting that they are indeed outer membrane proteins (Fig. 4A and 4D). This was confirmed using membrane fractionation experiments (Fig. S5B-E). In dividing cells, the GFP-LpoA and GFP-LpoB signals became enriched at the septum following osmotic shock (Fig. S6F and S6H). This was also observed for GFP-PBP1b, but not GFP-PBP1a (Fig. S6A-D). It is not clear why plasmolysis leads to the enrichment of these fusion proteins at the septum, but this may reflect some propensity of the native proteins to be recruited to the division apparatus (Bertsche et al., 2006; Typas et al., 2010).
Figure 4. LpoA and LpoB are outer membrane lipoproteins.
A-F. Cytological assay of membrane localization. Cells of MM13 [Para∷ponB ΔlpoA] (A-C) or CB4(attλTB309) [ΔponA ΔlpoB(Para∷ponA)] (D-F) harboring the integrated expression constructs (A) attHKMM10 [Plac∷lpoA-gfp], (B) attHKCB42 [Plac∷ssdsbA-lpoA-gfp], (C) attHKMM50 [Plac∷lpoA(D+2D+3)-gfp], (D) attHKCB28 [Plac∷lpoB-gfp], (E) attHKCB41 [Plac∷ssdsbA-lpoB-gfp], or (F) attHKMM51 [Plac∷lpoB(D+2E+3)-gfp] were grown at 30°C to mid-log in M9-arabinose supplemented with 100 μM IPTG. The cells were then plasmolyzed and visualized using GFP (panels 1) and phase contrast (panels 2) optics. Arrows highlight clear examples of protein localization (outer membrane, A and D; periplasm B and E; inner membrane, C and F). Bar equals 2 microns. G-H. Functionality of signal sequence mutants. Cultures of cells from (A-F) were diluted and plated on the indicated media as described in Figure 2. Please also see data in Figure S5 and S6
To determine the importance of outer membrane localization and/or lipidation for Lpo factor function, we generated LpoA-GFP and LpoB-GFP variants with D+2 D/E+3 substitutions in their signal sequences to cause their retention in the inner membrane (Fig. S5A), as well as variants in which the entire lipoprotein signal sequence was replaced with that of DsbA, a secreted periplasmic protein. All variants displayed the expected localization pattern following osmotic shock (Fig. 4A-F). Fusions with the DsbA signal sequence appeared to fill the periplasm of the plasmolysis bays (Fig. 4B and 4E), and those with the D+2 D/E+3 substitutions appeared to co-localize with invaginations of the inner membrane where it retracted from the envelope (Fig. 4C and 4F). We tested the ability of these variants to support the function of their cognate PBP1 by assaying whether or not they could correct the synthetic lethal phenotypes of Lpo/PBP1 mutant combinations. As shown in Figure 4G, LpoA variants with a DsbA signal peptide or the D+2 D+3 double substitution were not active, suggesting that LpoA must be targeted to the outer membrane to promote PBP1a function. Immunoblot analysis showed that all of the LpoA variants were produced at levels similar to the wild-type protein (data not shown), indicating that the observed defects were not due to reduced protein accumulation. In contrast to the results with LpoA, all of the LpoB variants were functional (Fig. 4H), suggesting that LpoB does not need to be lipidated or transported to the outer membrane to support PBP1b function.
Specific interaction of the Lpo factors with their cognate PBP
Overall, our results suggest that the Lpo factors are promoting the in vivo activity of the PBP1s. To test whether they do so directly, we purified untagged versions of PBP1a and PBP1b, and 6xHis tagged versions of LpoA (H-LpoA) and LpoB (H-LpoB) lacking their signal sequences and lipid modification signals. We then used a pull-down assay to investigate potential PBP1-Lpo interactions. When PBP1a (4μM) was incubated with H-LpoA (4μM), it was found in the eluate from Ni-NTA beads following two wash steps (Fig. 5A). Purified FtsZ (4μM) was included in the binding reactions as a negative control. It was not retained on the beads, indicating that the washes were effective. In contrast to the results with PBP1a, only a very small amount of PBP1b co-purified with H-LpoA (Fig. 5B). Conversely, when PBP1a (4μM) or PBP1b (4μM) were incubated with H-LpoB (4μM), PBP1b was specifically retained on NiNTA beads (Fig. 5C and 5D). Thus, as implied by the genetic results, LpoA specifically associates with PBP1a, while LpoB specifically associates with PBP1b.
Figure 5. LpoA and LpoB specifically interact with their cognate PBP.
A-D. H-LpoA (A-B) or H-LpoB (C-D) was incubated with PBP1a (A and C) or PBP1b (B and D) for 60 min at room temperature in binding buffer [20mM Tris (pH 7.4), 0.1% Triton-X-100, and either 300mM or 150mM NaCl for A-B or C-D, respectively]. Ni-NTA resin (Qiagen) was then added to each reaction and they were further incubated for 2 hr at 4°C with rotation. The resin was pelleted by centrifugation, washed twice with binding buffer containing 20mM imadazole, and the proteins retained on the resin were eluted with sample buffer containing EDTA (100 mM). Proteins in the initial reaction (input), initial supernatant (UB), wash supernatants (W1 and W2), and eluate were separated on a 12% SDS polyacrylamide gel and stained with Coomassie Brilliant Blue. FtsZ was included in each reaction as a non-specific control. All proteins were present in the initial binding reaction at a concentration of 4 μM. Positions of molecular weight markers (numbers in kDa) are given to the left of each gel.
Similar amounts of Lpo and PBP1 proteins appeared to elute from the Ni-NTA resin, suggesting that the PBP1-Lpo stoichiometries in the isolated complexes were close to 1:1. Accordingly, the cellular copy numbers of the Lpo factors measured using semi-quantitative immunoblotting mirrored those determined for their cognate PBPs. About 500 molecules per cell each of LpoA and PBP1a were detected while PBP1b and LpoB were found to have copy numbers of about 1000 and 2300 molecules per cell, respectively. The in vivo PBP1:Lpo stoichiometry is therefore in the range of 1:1 to 1:2. The measured PBP1a/b levels were 2-4 times higher than those determined previously using radiolabeled penicillin as a probe (Dougherty et al., 1996), but were consistent with past measurements of PBP1b levels by immunoblotting (Den Blaauwen and Nanninga, 1990). Radiolabeled penicillin thus appears to underestimate PBP levels when used for copy number determinations.
Lpo factors promote PBP activity in vivo and in vitro
To assess the effect of the Lpo proteins on PG assembly, we compared the chemical composition of wild-type PG with PG isolated from PBP1a- PBP1b- or LpoA- LpoB- cells just before they lysed (Table S1). For the analysis, purified PG was digested with a muramidase (mutanolysin) that cleaves the β-1,4 linkages between the N-acetylmuramic acid (M) and N-acetylglucosamine (G) sugars of PG (Fig. 1A). The resulting mixtures of muropeptides, primarily consisting of monomeric G-M disaccharides with attached peptides or dimeric G-M disaccharides connected by crosslinked peptides, were separated by HPLC and the component muropeptides quantitated. The absence of either the PBP1s or the Lpo proteins caused a strikingly similar alteration in relative PG composition. PG from the mutants showed a decrease in peptide crosslinking, from 22% in the wild-type control down to 18-19% (Table S1). In addition, a dramatic increase (4-7 fold) in pentapeptide-containing muropeptides was observed in the PG isolated from the mutants (Table S1). Since the terminal D-Ala is cleaved from pentapeptides as part the PBP crosslinking reaction, the observed increase in pentapeptides in the mutant PG preparations coupled with the reduction in crosslinking is consistent with a failure in the efficient incorporation of new PG material into the existing cell wall network.
Next, we examined the effect of LpoB on PG synthesis in vitro using ether-permeabilized (EP) cells. In the latter stages of the PG synthesis pathway, lipid-II (Fig.1B) is made from the activated sugar precursors UDP-M-pentapeptide (UDP-M-pep5) and UDP-G. PG synthesis can be reconstituted and measured in ether-permeabilized (EP) cells by supplying them with purified UDP-M-pep5 and UDP-[14C]G and monitoring the incorporation of label into the detergent-insoluble PG fraction (Mirelman et al., 1976). PBP1b appears to be the major PBP responsible for PG synthesis in this system because EP cells from PBP1b- mutants are almost completely defective in label incorporation whereas the absence of PBP1a has little to no effect (Fig. 6A) (Tamaki et al., 1977). EP cells thus provide a convenient system for following PBP1b activity in a cellular context. To determine if LpoB is required for PBP1b activity in this system, we prepared EP cells from a LpoB- mutant and tested their ability to synthesize PG. LpoB- EP cells behaved identically to those prepared from a PBP1b- mutant and were dramatically reduced in their ability to incorporate UDP-[14C]G into the detergent-insoluble PG fraction (Fig. 6A). Remarkably, the addition of purified untagged LpoB completely restored the ability of LpoB- EP cells to synthesize PG (Fig. 6A), indicating that the PG synthesis defect was due solely to the absence of LpoB. As expected based on prior results with PBP1a- mutants, a LpoA defect did not affect PG synthesis in EP cells (Fig. 6A).
Figure 6. LpoB activates PBP1b PGT activity and affects polymer length.
A. PG synthesis in EP cells. EP cells from the indicated strains were incubated with or without LpoB (0.5-4 μM, as indicated). Reactions were initiated with the addition of UDP-M-pentapeptide (4 nmol) and UDP-[14C]G. After 60 min they were boiled in 4% SDS and filtered. Labeled PG retained on the filter was quantified by liquid scintillation counting. B-C. PBP PGT activity was measured by the incorporation of lipid-II into peptidoglycan in the presence of penicillin G. [14C]G-labeled lipid-II (4-8 μM) was incubated with or without LpoA or LpoB (50 nM) prior to the addition of PBP1a or PBP1b (50 nM), which initiated the reaction. At the indicated time points, reactions were quenched and analyzed for remaining substrate and PG product by paper chromatography. Results of single experiments are shown. They are representative of multiple trials (see Supplementary Information). D. Glycan chains generated in reactions similar to those in (B-C) were separated on an acrylamide gel (9%) and visualized using a phosphorimager. Lipid-II substrate was present at 4 μM in each reaction and protein amounts are indicated above the gel lanes.
To further investigate LpoB function, we monitored its effect on the PGT activity of PBP1b in a purified system using radiolabled lipid-II substrate. LpoB specifically enhanced the initial rate of PBP1b transglycosylation by an average of 1.5× (Fig. 6B). However, it did not affect the activity of PBP1a, nor did LpoA affect the PGT activity of either PBP (Fig. 6B-C). Although the effect of LpoB on the rate of PBP1b PGT activity was relatively modest, it specifically affected the length of glycan chains produced by PBP1b. Much shorter polymers were produced in the presence of LpoB than those formed in its absence (Fig. 6D). We propose that this indicates that LpoB stimulates the initiation of glycan strand formation (see Discussion). Overall, we conclude that LpoB is absolutely required for PBP1b function in a cellular context and is capable of directly and specifically modulating PBP1b PGT activity in a purified system.
Discussion
Despite the fundamental role of PG assembly in bacterial growth and its long history as an effective antibiotic target, we are only just beginning to uncover the molecular mechanisms underlying this complex process. Over the years, a great deal of effort has focused on the structural and biochemical characterization of the PBPs, the major PG synthases. This has led to important insights into the mechanisms by which these enzymes catalyze PG polymerization and crosslinking, how they are inhibited by antibiotic molecules, and why certain variants confer antibiotic resistance (Sauvage et al., 2008). While much still remains to be learned about the biochemical function of the PBPs, progress in this area has significantly outpaced our knowledge of the in vivo function of the PBPs and how they cooperate with other cellular factors to build the cell-shaped PG mesh. One of the principal reasons for this is that functional redundancy among PG assembly factors has limited the effectiveness of genetic analysis. Here, we turned this redundancy into an advantage by employing synthetic lethal screens to uncover the E. coli lipoproteins LpoA and LpoB as the first set of essential PBP cofactors. Our results suggest that, like other polymerases, the PBPs may generally require accessory factors to augment or regulate their activity. Moving forward, these factors will surely play a significant role in advancing both our understanding of PBP function in vivo and PBP enzymology in vitro.
The outer membrane and PG biogenesis
Although PG precursor biosynthesis occurs inside the cell, the final stages of PG assembly take place at the cell surface (Fig. 1B). This presents an interesting challenge for bacteria, which must somehow coordinate PG assembly outside the cytoplasmic membrane with processes that control growth and division on the inside. Recent results indicate that this coordination is, at least in part, mediated by the MreB and FtsZ cytoskeletons (Margolin, 2009).
Because they surround their PG layer with a second (outer) membrane, gram-negative bacteria face the additional challenge of coordinating PG synthesis with outer membrane assembly. The discovery of LpoA and LpoB as outer membrane activators of the PBPs suggests that they may play an important role in coupling outer membrane biogenesis and PG synthesis. While further work is required to test this possibility, it highlights the potential for the PG synthetic machinery receiving regulatory input from the outer membrane as well as cytoskeletal elements in the cytoplasm. An equally attractive possibility is that the outer membrane localization of the Lpo factors might facilitate a “template” function for the existing PG matrix. The glycan strands of PG are thought to be oriented perpendicular to the long-axis of the cell (Gan et al., 2008) (Fig. S7). Adjacent “tracks” of glycan strands are therefore likely to restrict the lateral diffusion of trans-envelope complexes formed between the Lpo proteins in the outer membrane and the PBPs in the inner membrane. We envision that this arrangement would force the insertion of new PG material along a consistent path directed by the “tracks” in the existing structure (Fig. S7). Thus, as implied by earlier work (Schwarz and Leutgeb, 1971; Goodell and Schwarz, 1975), the PG layer itself may collaborate with cytoskeletal elements to provide a robust mechanism for shape-maintenance and the regular expansion of the PG network. Importantly, the Lpo factors may represent just one set of potential connections between the PBPs and the outer membrane. Indeed, it was shown previously that PBP1b is connected to the outer membrane through its interactions with the bridging protein MipA that, in turn, associates with the outer membrane PG hydrolase MltA (Vollmer et al., 1999). In contrast to LpoB, neither MipA or MltA are required for PBP1b activity. However, the ability of these and potentially other proteins to connect PBP1b to the outer membrane may be one reason why an outer membrane localization is not absolutely required for LpoB to support PBP1b function.
Biochemical activities of LpoA and LpoB
In addition to promoting PBP1b activity in EP cells and stimulating PBP1b PGT activity in vitro, LpoB also caused PBP1b to produce dramatically shorter glycan chains. A wide range of glycan strand lengths has been detected in purified cellular PG, but essentially nothing is known about how strand length is determined in vivo and whether or not it is regulated. The observation that LpoB affects the length distribution of glycan chains produced by PBP1b suggests the attractive possibility that it regulates PBP1b product length by stimulating the termination of glycan synthesis. However, such a role is difficult to reconcile with the observation that LpoB is essential for PG synthesis by PBP1b in EP cells. We therefore favor a model in which LpoB stimulates the initiation of glycan chain synthesis to explain its effect on product length distribution in PBP1b PGT assays. This model is based on the previous observation that the length distribution of polymers produced by a variety of purified PBPs was unaffected by the PBP:substrate ratio (Wang et al., 2008). Surprisingly, even in reactions with a 1:1 PBP:substrate ratio, where at most only a few turnovers would be expected, long glycan strands with a size distribution typical of the PBP being assayed were produced. This suggests that the initiation step of glycan chain synthesis is rate-limiting and that the subsequent elongation process is rapid and processive (Wang et al., 2008). Thus, consistent with our observations, activation of glycan strand initiation by LpoB would be predicted to result in a greater number of elongating PBP1b polymerases in PGT reactions such that lipid-II is exhausted before longer polymer lengths can be achieved. Because LpoB is not absolutely required for PBP1b activity in a purified system, a subset of PBP1b molecules in PGT reactions lacking LpoB must be capable of initiating glycan strand formation. Initiation may be tightly controlled in the context of multi-protein complexes formed in vivo, however, leading to a strict LpoB dependence for PBP1b PGT activity in the cell. Further work will be required to investigate the potential role of LpoB in glycan chain initiation, and whether, in such a capacity, it serves as critical PBP1b regulator that helps coordinate its PGT activity with the activity of other components of the PG or outer membrane assembly machinery.
While our results clearly show that LpoA is essential for PBP1a function in vivo, we have yet to detect a biochemical activity that sheds light on the specific role of LpoA in the reactions catalyzed by PBP1a. One potential reason for this is that lipidation and outer membrane localization of LpoA was shown to be essential for its activity in vivo and the protein we purified was not lipidated. Interestingly, the structure of the C-terminal domain of H. influenzae LpoA was recently solved and was found to share similarities with periplasmic solute binding proteins like maltose binding protein (Vijayalakshmi et al., 2008). The closest structural relative to LpoA is E. coli LivJ (PBD ID# 1z15), a branched-chain amino acid binding protein (Vijayalakshmi et al., 2008). This suggests that LpoA may associate with the peptide moieties of PG to facilitate crosslinking of nascent material into the PG matrix by PBP1a. Consistent with this idea, in the accompanying report from Typas et al. (Typas et al., 2010), the authors present evidence that LpoA greatly enhances the formation of crosslinked PG by purified PBP1a.
A specific role for PBP1b-LpoB complexes in cell division?
At least two distinct multi-enzyme complexes are though to be necessary for proper PG biogenesis in rod shaped cells: one organized by MreB for cell elongation and the other organized by the division protein FtsZ (den Blaauwen et al., 2008). Although they are individually dispensable for growth, at least one type of PBP1 protein is thought to be required for the activity of each of these complexes. It therefore seems likely that PBP1a and PBP1b, along with their associated Lpo activators, are both capable of productively interfacing with the MreB- and FtsZ-directed PG synthesizing machineries. Consistent with this possibility, in normal, unplasmolyzed cells, GFP fusions to the PBP1s and the Lpo factors were found to localize in patches and foci that were broadly distributed around the cell periphery (Fig. 4 and S6). In addition, immunofluorescence localization of PBP1b performed previously also found it to be present in many foci throughout the cell cylinder, although a modest enrichment at the division sites of constricting cells was also reported (Bertsche et al., 2006).
While PBP1a and PBP1b appear to be largely interchangeable in their capacities to support growth, it is likely that they display some level of functional specialization in wild-type cells. Accordingly, PBP1b has been shown to interact with the division-specific, class B PBP, PBP3, and the division factor FtsN (Bertsche et al., 2006; Muller et al., 2007). This suggests that PBP1b, along with LpoB, may play a specific role in the division process that PBP1a-LpoA sub-complexes are unable to perform. Interestingly, a number of mutants with lesions in genes coding for factors involved in septal PG biogenesis were isolated using our screen for cells with a Slb phenotype. Rather than identifying additional co-factors needed for PBP1a activity like LpoA, we favor the interpretation that these division mutants have a Slb phenotype because PBP1b- cells are partially defective for cell division and are thus sensitized to further insults to the division machinery. A similar class of division mutants was not recovered in the corresponding screen for sla mutants, nor did the alleles identified in the Slb screen display synthetic phenotypes with the absence of PBP1a when this was tested directly (data not shown). The phenotype of the PBP1b-/division mutant combinations in each case was a high frequency of cell lysis, suggesting that the absence of PBP1b combined with a defect in one of these division factors often results in catastrophic failures in septal PG assembly. Further analysis of these mutants and their connection to PBP1b function should allow us to elucidate how PBP1b participates in the construction of the polar PG caps, and whether or not this role is unique to PBP1b-LpoB containing complexes.
Lpo factor conservation and the prospect for additional PBP accessory proteins
LpoA is broadly conserved among the γ-proteobacteria while LpoB is primarily restricted to the enterobacteriaceae family within this class (Finn et al., 2008). We therefore suspect that organisms belonging to bacterial classes other than the γ-proteobacteria are likely to possess PBP accessory proteins distinct from LpoA and LpoB that also promote the function of their PBPs. Some of these factors may play critical roles in the PG assembly process itself, while others may help coordinate PG synthesis with additional envelope components like the outer membranes of other proteobacteria or the teichoic acids of gram-positive organisms.
We anticipate that several of the many PBP-interacting proteins identified previously will also prove to be PBP accessory proteins that directly assist the PBPs in PG assembly. Prime candidates are the PG hydrolases. These enzymes break bonds in the PG meshwork and have long been thought necessary for providing space in the existing matrix for the insertion of new material. Although this possibility remains attractive on a theoretical basis, experimental support for it is still lacking. Addressing the role of these and other PBP-interacting factors in PBP function represents an important challenge for future work.
PBP accessory factors and antibiotic development
The discovery of penicillin and its ability to inhibit the PBPs to induce bacteriolysis ushered in the antibiotic age of clinical medicine. Since then, the PBPs have proven to be one of the most important drug targets ever identified. Sadly, antibiotic resistance continues to erode the effectiveness of our current cache of antibacterial therapies, including penicillin derivatives and other β-lactams (Taubes, 2008). Regardless of their precise biochemical function, the discovery of accessory factors essential for the in vivo function of the PBPs suggests new avenues for antibiotic development to help combat drug-resistant bacteria. The observation that LpoA is essential in H. influenzae (Wong and Akerley, 2003) suggests that direct targeting of PBP accessory proteins might be effective in some circumstances. Additionally, further study of the mechanisms by which these accessory factors influence PBP activity is likely to reveal novel ways to block PBP function for therapeutic purposes.
Materials and Methods
Media, bacterial strains, and plasmids
Cells were grown in LB (1% tryptone, 0.5% yeast extract, 0.5% NaCl) or minimal M9 medium supplemented with 0.2% casamino acids and 0.2% sugar (arabinose; Ara, glucose; Glu, or maltose; Malt). The bacterial strains and plasmids used in this study are listed in Tables S2 and S3, respectively, and a detailed description of their construction is given in Supplementary Information.
Synthetic lethal screens
Screens for mutants with a Sla or Slb phenotype were performed as described previously (Bernhardt and de Boer, 2004). Detailed methods are presented in Supplementary Information.
Fluorescence microscopy and cytological assay for membrane localization
Fluorescence microscopy was performed essentially as described previously (Uehara et al., 2009). The cytological assay for membrane localization of lipoproteins was performed as described previously (Lewenza et al., 2006). Please see Supplementary Information for details about growth conditions and sample preparation methods used for specific experiments.
Protein purification and biochemical assays
Detailed protein purification procedures are presented in Supplementary Information, as are methods used for PG chemical analysis and biochemical reactions.
Supplementary Material
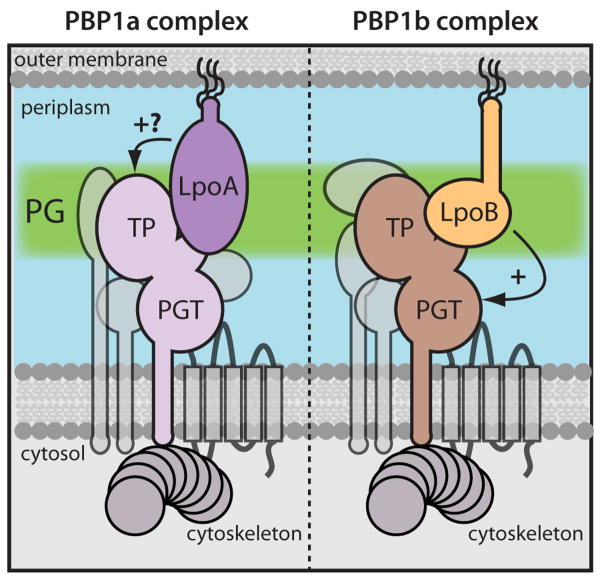
Figure 7. Model for Lpo protein function.
Shown are schematic diagrams of putative PBP-containing complexes in E. coli drawn as in Figure 1. The partial redundancy of PBP1a (lavender) and PBP1b (brown) suggest that they form part of independent PG synthesizing (sub)complexes that can substitute for one another. LpoA is an essential component of the PBP1a complex (left) that potentially stimulates the transpeptidase activity of this PBP (see text for details). LpoB is an essential component of the PBP1b complex and activates its PGT activity. Please also see model in Figure S7.
Acknowledgments
The authors would like to thank D. Rudner, M. Waldor, and members of our laboratories for critical reading of the manuscript. We would also like to thank A. Typas, C. Gross, W. Vollmer and co-workers for communicating their results in advance of publication. Special thanks to K. Suefuji and J. T. Park for generously providing the UDP-M-pep5. This work was supported by the Massachusetts Life Science Center (T.G.B), the Burroughs Wellcome Fund (T.G.B), and the National Institutes of Health (R01 AI083365-01A1 for T.G.B, GM066174 for D.E.K., and GM076710 for D.E.K and S.W.). T.G.B. holds a Career Award in the Biomedical Sciences from the Burroughs Wellcome Fund. C.P.B. was supported in part by a fellowship from FRSQ.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Alaedini A, Day RA. Identification of two penicillin-binding multienzyme complexes in Haemophilus influenzae. Biochem Biophys Res Commun. 1999;264:191–95. doi: 10.1006/bbrc.1999.1509. [DOI] [PubMed] [Google Scholar]
- Barrett D, Zhang Y, Kahne D, Sliz P, Walker S. Crystal structure of a peptidoglycan glycosyltransferase suggests a model for processive glycan chain synthesis. Proceedings of the National Academy of Sciences. 2007;104:5348–353. doi: 10.1073/pnas.0701160104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernhardt TG, de Boer PA. Screening for synthetic lethal mutants in Escherichia coli and identification of EnvC (YibP) as a periplasmic septal ring factor with murein hydrolase activity. Mol Microbiol. 2004;52:1255–269. doi: 10.1111/j.1365-2958.2004.04063.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernhardt TG, de Boer PA. SlmA, a nucleoid-associated, FtsZ binding protein required for blocking septal ring assembly over Chromosomes in E. coli. Mol Cell. 2005;18:555–564. doi: 10.1016/j.molcel.2005.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertsche U, Breukink E, Kast T, Vollmer W. In vitro murein peptidoglycan synthesis by dimers of the bifunctional transglycosylase-transpeptidase PBP1B from Escherichia coli. J Biol Chem. 2005;280:38096–8101. doi: 10.1074/jbc.M508646200. [DOI] [PubMed] [Google Scholar]
- Bertsche U, Kast T, Wolf B, Fraipont C, Aarsman ME, Kannenberg K, von Rechenberg M, Nguyen-Distèche M, den Blaauwen T, et al. Interaction between two murein (peptidoglycan) synthases, PBP3 and PBP1B, in Escherichia coli. Mol Microbiol. 2006;61:675–690. doi: 10.1111/j.1365-2958.2006.05280.x. [DOI] [PubMed] [Google Scholar]
- Charpentier X, Chalut C, Rémy MH, Masson JM. Penicillin-binding proteins 1a and 1b form independent dimers in Escherichia coli. J Bacteriol. 2002;184:3749–752. doi: 10.1128/JB.184.13.3749-3752.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Den Blaauwen T, Nanninga N. Topology of penicillin-binding protein 1b of Escherichia coli and topography of four antigenic determinants studied by immunocolabeling electron microscopy. J Bacteriol. 1990;172:71–79. doi: 10.1128/jb.172.1.71-79.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- den Blaauwen T, de Pedro MA, Nguyen-Distèche M, Ayala JA. Morphogenesis of rod-shaped sacculi. FEMS Microbiol Rev. 2008;32:321–344. doi: 10.1111/j.1574-6976.2007.00090.x. [DOI] [PubMed] [Google Scholar]
- Dougherty TJ, Kennedy K, Kessler RE, Pucci MJ. Direct quantitation of the number of individual penicillin-binding proteins per cell in Escherichia coli. J Bacteriol. 1996;178:6110–15. doi: 10.1128/jb.178.21.6110-6115.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finn RD, Tate J, Mistry J, Coggill PC, Sammut SJ, Hotz HR, Ceric G, Forslund K, Eddy SR, et al. The Pfam protein families database. Nucleic Acids Res. 2008;36:D281–88. doi: 10.1093/nar/gkm960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gan L, Chen S, Jensen GJ. Molecular organization of Gram-negative peptidoglycan. Proc Natl Acad Sci USA. 2008;105:18953–57. doi: 10.1073/pnas.0808035105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- García del Portillo F, de Pedro MA. Differential effect of mutational impairment of penicillin-binding proteins 1A and 1B on Escherichia coli strains harboring thermosensitive mutations in the cell division genes ftsA, ftsQ, ftsZ, and pbpB. J Bacteriol. 1990;172:5863–870. doi: 10.1128/jb.172.10.5863-5870.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodell EW, Schwarz U. Sphere-rod morphogenesis of Escherichia coli. J Gen Microbiol. 1975;86:201–09. doi: 10.1099/00221287-86-2-201. [DOI] [PubMed] [Google Scholar]
- Kato J, Suzuki H, Hirota Y. Dispensability of either penicillin-binding protein-1a or -1b involved in the essential process for cell elongation in Escherichia coli. Mol Gen Genet. 1985;200:272–77. doi: 10.1007/BF00425435. [DOI] [PubMed] [Google Scholar]
- Lewenza S, Vidal-Ingigliardi D, Pugsley AP. Direct visualization of red fluorescent lipoproteins indicates conservation of the membrane sorting rules in the family Enterobacteriaceae. J Bacteriol. 2006;188:3516–524. doi: 10.1128/JB.188.10.3516-3524.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margolin W. Sculpting the bacterial cell. Curr Biol. 2009;19:R812–822. doi: 10.1016/j.cub.2009.06.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mirelman D, Yashouv-Gan Y, Schwarz U. Peptidoglycan biosynthesis in a thermosensitive division mutant of Escherichia coli. Biochemistry. 1976;15:1781–790. doi: 10.1021/bi00654a001. [DOI] [PubMed] [Google Scholar]
- Müller P, Ewers C, Bertsche U, Anstett M, Kallis T, Breukink E, Fraipont C, Terrak M, Nguyen-Distèche M, Vollmer W. The essential cell division protein FtsN interacts with the murein (peptidoglycan) synthase PBP1B in Escherichia coli. J Biol Chem. 2007;282:36394–6402. doi: 10.1074/jbc.M706390200. [DOI] [PubMed] [Google Scholar]
- Sauvage E, Kerff F, Terrak M, Ayala JA, Charlier P. The penicillin-binding proteins: structure and role in peptidoglycan biosynthesis. FEMS Microbiol Rev. 2008;32:234–258. doi: 10.1111/j.1574-6976.2008.00105.x. [DOI] [PubMed] [Google Scholar]
- Schmidt LS, Botta G, Park JT. Effects of furazlocillin, a beta-lactam antibiotic which binds selectively to penicillin-binding protein 3, on Escherichia coli mutants deficient in other penicillin-binding proteins. J Bacteriol. 1981;145:632–37. doi: 10.1128/jb.145.1.632-637.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarz U, Leutgeb W. Morphogenetic aspects of murein structure and biosynthesis. J Bacteriol. 1971;106:588–595. doi: 10.1128/jb.106.2.588-595.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spratt BG, Pardee AB. Penicillin-binding proteins and cell shape in E. coli. Nature. 1975;254:516–17. doi: 10.1038/254516a0. [DOI] [PubMed] [Google Scholar]
- Tamaki S, Nakajima S, Matsuhashi M. Thermosensitive mutation in Escherichia coli simultaneously causing defects in penicillin-binding protein-1Bs and in enzyme activity for peptidoglycan synthesis in vitro. Proc Natl Acad Sci USA. 1977;74:5472–76. doi: 10.1073/pnas.74.12.5472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taubes G. The bacteria fight back. The bacteria fight back. Science. 2008;321:356–361. doi: 10.1126/science.321.5887.356. [DOI] [PubMed] [Google Scholar]
- Typas A, Banzhaf M, van den Berg van Saparoea B, Verheul J, Biboy J, Nichols RJ, Zietek M, Beilharz K, Kannenberg K et al. Regulation of peptidoglycan synthesis by outer membrane proteins. Cell. 2010 doi: 10.1016/j.cell.2010.11.038. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uehara T, Dinh T, Bernhardt TG. LytM-domain factors are required for daughter cell separation and rapid ampicillin-induced lysis in Escherichia coli. J Bacteriol. 2009;191:5094–5107. doi: 10.1128/JB.00505-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vijayalakshmi J, Akerley BJ, Saper MA. Structure of YraM, a protein essential for growth of Haemophilus influenzae. Proteins. 2008;73:204–217. doi: 10.1002/prot.22033. [DOI] [PubMed] [Google Scholar]
- Vollmer W, von Rechenberg M, Höltje JV. Demonstration of molecular interactions between the murein polymerase PBP1B, the lytic transglycosylase MltA, and the scaffolding protein MipA of Escherichia coli. J Biol Chem. 1999;274:6726–6734. doi: 10.1074/jbc.274.10.6726. [DOI] [PubMed] [Google Scholar]
- Wang G, Klein MG, Tokonzaba E, Zhang Y, Holden LG, Chen XS. The structure of a DnaB-family replicative helicase and its interactions with primase. Nat Struct Mol Biol. 2008;15:94–100. doi: 10.1038/nsmb1356. [DOI] [PubMed] [Google Scholar]
- White CL, Kitich A, Gober JW. Positioning Cell Wall Synthetic Complexes by the Bacterial Morphogenetic Proteins MreB and MreD. Mol Microbiol. 2010;76:613–633. doi: 10.1111/j.1365-2958.2010.07108.x. [DOI] [PubMed] [Google Scholar]
- Wong SM, Akerley BJ. Inducible expression system and marker-linked mutagenesis approach for functional genomics of Haemophilus influenzae. Gene. 2003;316:177–186. doi: 10.1016/s0378-1119(03)00762-5. [DOI] [PubMed] [Google Scholar]
- Yousif SY, Broome-Smith JK, Spratt BG. Lysis of Escherichia coli by beta-lactam antibiotics: deletion analysis of the role of penicillin-binding proteins 1A and 1B. J Gen Microbiol. 1985;131:2839–845. doi: 10.1099/00221287-131-10-2839. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.