Abstract
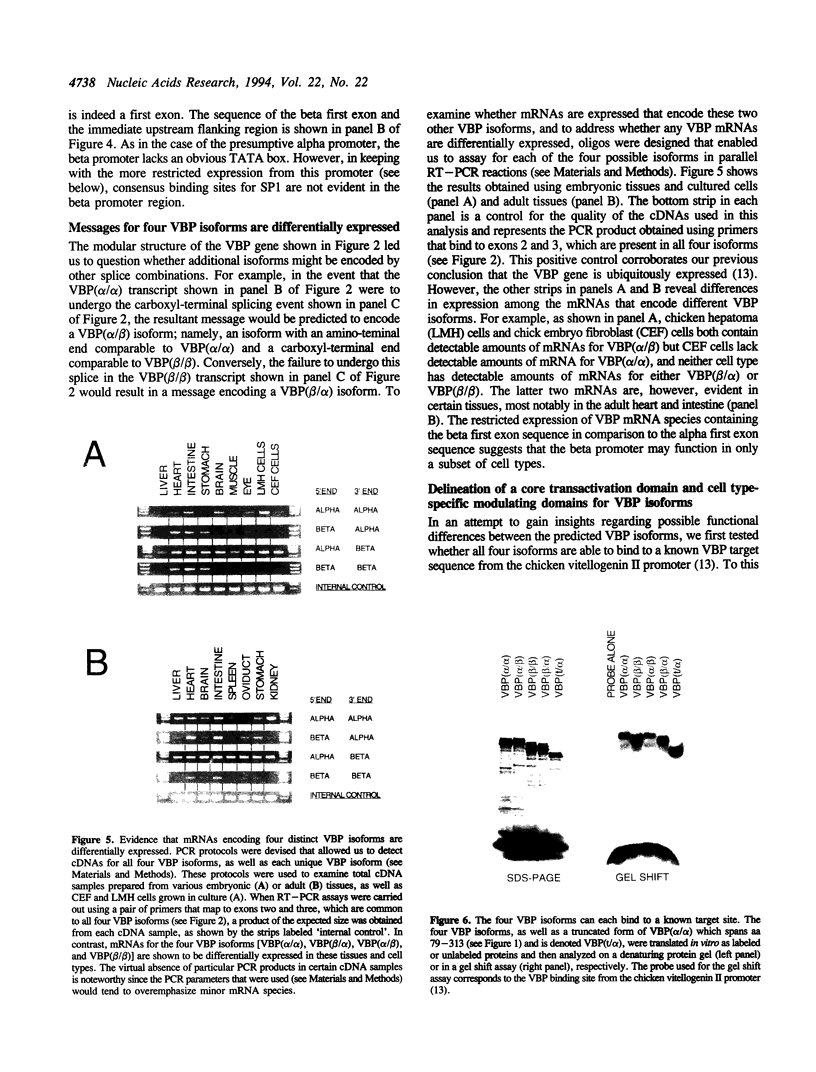
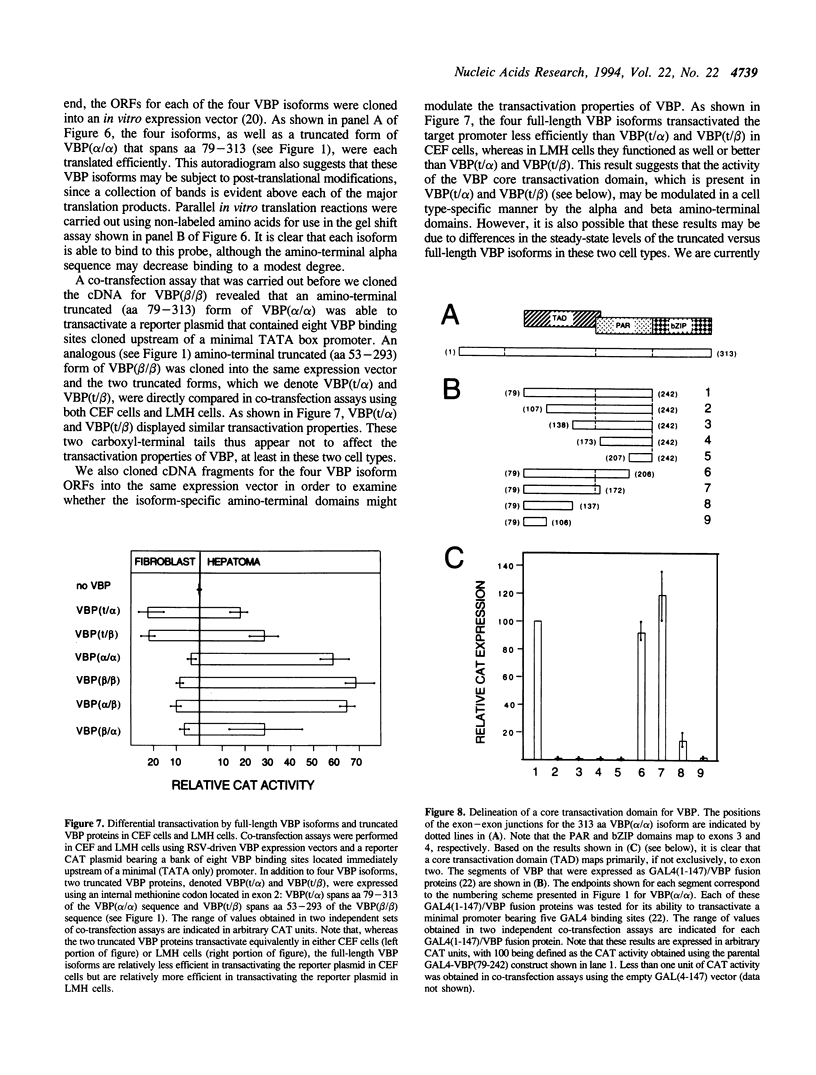
We previously isolated a set of overlapping cDNA clones that encoded a unique open reading frame for the chicken VBP transcription factor. We now report the isolation of a cDNA clone that encodes a complete open reading frame for a VBP isoform that differs from the previously reported sequence at both the amino-terminal and carboxyl-terminal ends. An analysis of the VBP gene revealed that the two different amino-terminal sequences map to alternative first exons and that the two different carboxyl-terminal sequences reflect an optional splicing event which can occur only on transcripts that are polyadenylated at the more distal of two polyadenylation sites. An RT-PCR analysis further revealed that a total of four VBP isoforms are encoded by the combinatorial use of these two splicing options. The mRNAs for these four isoforms are differentially expressed in different tissues and cell types. We provide evidence that one function of the amino-terminal domains is to impose cell type specificity on a core transactivation domain that is present in all four isoforms. Since it is known that VBP can heterodimerize with other members of the PAR subfamily of bZIP factors, our evidence for four VBP isoforms greatly expands the number of complexes that may be used to effect transcriptional regulation through PAR-factor binding sites.
Full text
PDF





Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Baichwal V. R., Park A., Tjian R. The cell-type-specific activator region of c-Jun juxtaposes constitutive and negatively regulated domains. Genes Dev. 1992 Aug;6(8):1493–1502. doi: 10.1101/gad.6.8.1493. [DOI] [PubMed] [Google Scholar]
- Beckmann H., Su L. K., Kadesch T. TFE3: a helix-loop-helix protein that activates transcription through the immunoglobulin enhancer muE3 motif. Genes Dev. 1990 Feb;4(2):167–179. doi: 10.1101/gad.4.2.167. [DOI] [PubMed] [Google Scholar]
- Binder R., MacDonald C. C., Burch J. B., Lazier C. B., Williams D. L. Expression of endogenous and transfected apolipoprotein II and vitellogenin II genes in an estrogen responsive chicken liver cell line. Mol Endocrinol. 1990 Feb;4(2):201–208. doi: 10.1210/mend-4-2-201. [DOI] [PubMed] [Google Scholar]
- Bradford M. M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976 May 7;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- Brent R., Ptashne M. A eukaryotic transcriptional activator bearing the DNA specificity of a prokaryotic repressor. Cell. 1985 Dec;43(3 Pt 2):729–736. doi: 10.1016/0092-8674(85)90246-6. [DOI] [PubMed] [Google Scholar]
- Chen E. Y., Seeburg P. H. Supercoil sequencing: a fast and simple method for sequencing plasmid DNA. DNA. 1985 Apr;4(2):165–170. doi: 10.1089/dna.1985.4.165. [DOI] [PubMed] [Google Scholar]
- Delmas V., Laoide B. M., Masquilier D., de Groot R. P., Foulkes N. S., Sassone-Corsi P. Alternative usage of initiation codons in mRNA encoding the cAMP-responsive-element modulator generates regulators with opposite functions. Proc Natl Acad Sci U S A. 1992 May 15;89(10):4226–4230. doi: 10.1073/pnas.89.10.4226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Descombes P., Schibler U. A liver-enriched transcriptional activator protein, LAP, and a transcriptional inhibitory protein, LIP, are translated from the same mRNA. Cell. 1991 Nov 1;67(3):569–579. doi: 10.1016/0092-8674(91)90531-3. [DOI] [PubMed] [Google Scholar]
- Drolet D. W., Scully K. M., Simmons D. M., Wegner M., Chu K. T., Swanson L. W., Rosenfeld M. G. TEF, a transcription factor expressed specifically in the anterior pituitary during embryogenesis, defines a new class of leucine zipper proteins. Genes Dev. 1991 Oct;5(10):1739–1753. doi: 10.1101/gad.5.10.1739. [DOI] [PubMed] [Google Scholar]
- Foulkes N. S., Mellström B., Benusiglio E., Sassone-Corsi P. Developmental switch of CREM function during spermatogenesis: from antagonist to activator. Nature. 1992 Jan 2;355(6355):80–84. doi: 10.1038/355080a0. [DOI] [PubMed] [Google Scholar]
- Foulkes N. S., Sassone-Corsi P. More is better: activators and repressors from the same gene. Cell. 1992 Feb 7;68(3):411–414. doi: 10.1016/0092-8674(92)90178-f. [DOI] [PubMed] [Google Scholar]
- Frankel A. D., Kim P. S. Modular structure of transcription factors: implications for gene regulation. Cell. 1991 May 31;65(5):717–719. doi: 10.1016/0092-8674(91)90378-c. [DOI] [PubMed] [Google Scholar]
- Gorman C. M., Moffat L. F., Howard B. H. Recombinant genomes which express chloramphenicol acetyltransferase in mammalian cells. Mol Cell Biol. 1982 Sep;2(9):1044–1051. doi: 10.1128/mcb.2.9.1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham F. L., van der Eb A. J. A new technique for the assay of infectivity of human adenovirus 5 DNA. Virology. 1973 Apr;52(2):456–467. doi: 10.1016/0042-6822(73)90341-3. [DOI] [PubMed] [Google Scholar]
- Hunger S. P., Ohyashiki K., Toyama K., Cleary M. L. Hlf, a novel hepatic bZIP protein, shows altered DNA-binding properties following fusion to E2A in t(17;19) acute lymphoblastic leukemia. Genes Dev. 1992 Sep;6(9):1608–1620. doi: 10.1101/gad.6.9.1608. [DOI] [PubMed] [Google Scholar]
- Inaba T., Roberts W. M., Shapiro L. H., Jolly K. W., Raimondi S. C., Smith S. D., Look A. T. Fusion of the leucine zipper gene HLF to the E2A gene in human acute B-lineage leukemia. Science. 1992 Jul 24;257(5069):531–534. doi: 10.1126/science.1386162. [DOI] [PubMed] [Google Scholar]
- Iyer S. V., Davis D. L., Seal S. N., Burch J. B. Chicken vitellogenin gene-binding protein, a leucine zipper transcription factor that binds to an important control element in the chicken vitellogenin II promoter, is related to rat DBP. Mol Cell Biol. 1991 Oct;11(10):4863–4875. doi: 10.1128/mcb.11.10.4863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson P. F., McKnight S. L. Eukaryotic transcriptional regulatory proteins. Annu Rev Biochem. 1989;58:799–839. doi: 10.1146/annurev.bi.58.070189.004055. [DOI] [PubMed] [Google Scholar]
- Kawaguchi T., Nomura K., Hirayama Y., Kitagawa T. Establishment and characterization of a chicken hepatocellular carcinoma cell line, LMH. Cancer Res. 1987 Aug 15;47(16):4460–4464. [PubMed] [Google Scholar]
- Koenig R. J., Lazar M. A., Hodin R. A., Brent G. A., Larsen P. R., Chin W. W., Moore D. D. Inhibition of thyroid hormone action by a non-hormone binding c-erbA protein generated by alternative mRNA splicing. Nature. 1989 Feb 16;337(6208):659–661. doi: 10.1038/337659a0. [DOI] [PubMed] [Google Scholar]
- Kozak M. Compilation and analysis of sequences upstream from the translational start site in eukaryotic mRNAs. Nucleic Acids Res. 1984 Jan 25;12(2):857–872. doi: 10.1093/nar/12.2.857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landschulz W. H., Johnson P. F., Adashi E. Y., Graves B. J., McKnight S. L. Isolation of a recombinant copy of the gene encoding C/EBP. Genes Dev. 1988 Jul;2(7):786–800. doi: 10.1101/gad.2.7.786. [DOI] [PubMed] [Google Scholar]
- Lillie J. W., Green M. R. Transcription activation by the adenovirus E1a protein. Nature. 1989 Mar 2;338(6210):39–44. doi: 10.1038/338039a0. [DOI] [PubMed] [Google Scholar]
- Mitchell P. J., Tjian R. Transcriptional regulation in mammalian cells by sequence-specific DNA binding proteins. Science. 1989 Jul 28;245(4916):371–378. doi: 10.1126/science.2667136. [DOI] [PubMed] [Google Scholar]
- Mueller C. R., Maire P., Schibler U. DBP, a liver-enriched transcriptional activator, is expressed late in ontogeny and its tissue specificity is determined posttranscriptionally. Cell. 1990 Apr 20;61(2):279–291. doi: 10.1016/0092-8674(90)90808-r. [DOI] [PubMed] [Google Scholar]
- Mumberg D., Lucibello F. C., Schuermann M., Müller R. Alternative splicing of fosB transcripts results in differentially expressed mRNAs encoding functionally antagonistic proteins. Genes Dev. 1991 Jul;5(7):1212–1223. doi: 10.1101/gad.5.7.1212. [DOI] [PubMed] [Google Scholar]
- Pabo C. O., Sauer R. T. Transcription factors: structural families and principles of DNA recognition. Annu Rev Biochem. 1992;61:1053–1095. doi: 10.1146/annurev.bi.61.070192.005201. [DOI] [PubMed] [Google Scholar]
- Pei D. Q., Shih C. H. Transcriptional activation and repression by cellular DNA-binding protein C/EBP. J Virol. 1990 Apr;64(4):1517–1522. doi: 10.1128/jvi.64.4.1517-1522.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson M. L., Perry R. P. The regulated production of mu m and mu s mRNA is dependent on the relative efficiencies of mu s poly(A) site usage and the c mu 4-to-M1 splice. Mol Cell Biol. 1989 Feb;9(2):726–738. doi: 10.1128/mcb.9.2.726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaffner W. How do different transcription factors binding the same DNA sequence sort out their jobs? Trends Genet. 1989 Feb;5(2):37–39. doi: 10.1016/0168-9525(89)90017-6. [DOI] [PubMed] [Google Scholar]
- Weber B. L., Westin E. H., Clarke M. F. Differentiation of mouse erythroleukemia cells enhanced by alternatively spliced c-myb mRNA. Science. 1990 Sep 14;249(4974):1291–1293. doi: 10.1126/science.2205003. [DOI] [PubMed] [Google Scholar]
- Wuarin J., Schibler U. Expression of the liver-enriched transcriptional activator protein DBP follows a stringent circadian rhythm. Cell. 1990 Dec 21;63(6):1257–1266. doi: 10.1016/0092-8674(90)90421-a. [DOI] [PubMed] [Google Scholar]
- Yamamoto K. K., Gonzalez G. A., Menzel P., Rivier J., Montminy M. R. Characterization of a bipartite activator domain in transcription factor CREB. Cell. 1990 Feb 23;60(4):611–617. doi: 10.1016/0092-8674(90)90664-z. [DOI] [PubMed] [Google Scholar]