Abstract
Protein microarrays provide an efficient way to identify and quantify protein–protein interactions in high throughput. One drawback of this technique is that proteins show a broad range of physicochemical properties and are often difficult to produce recombinantly. To circumvent these problems, we have focused on families of protein interaction domains. Here we provide protocols for constructing microarrays of protein interaction domains in individual wells of 96-well microtiter plates, and for quantifying domain–peptide interactions in high throughput using fluorescently labeled synthetic peptides. As specific examples, we will describe the construction of microarrays of virtually every human Src homology 2 (SH2) and phosphotyrosine binding (PTB) domain, as well as microarrays of mouse PDZ domains, all produced recombinantly in Escherichia coli. For domains that mediate high-affinity interactions, such as SH2 and PTB domains, equilibrium dissociation constants (KDs) for their peptide ligands can be measured directly on arrays by obtaining saturation binding curves. For weaker binding domains, such as PDZ domains, arrays are best used to identify candidate interactions, which are then retested and quantified by fluorescence polarization. Overall, protein domain microarrays provide the ability to rapidly identify and quantify protein–ligand interactions with minimal sample consumption. Because entire domain families can be interrogated simultaneously, they provide a powerful way to assess binding selectivity on a proteome-wide scale and provide an unbiased perspective on the connectivity of protein–protein interaction networks.
INTRODUCTION
Over the past decade, a variety of functional proteomics methods have been used to uncover protein–protein interactions in high throughput. These include the yeast two-hybrid assay1,2, the protein complementation assay3, affinity purification coupled with mass spectrometry4,5, peptide phage display6, oriented peptide library screening7 and protein microarrays8. Both the yeast two-hybrid assay and the protein complementation assay entail co–expressing a `bait' and `prey' protein in the same cell. If the two proteins interact, a reporter gene is expressed or a reporter protein becomes active. Because these assays are conducted within living cells, no biochemical purifications steps are required. As such, these methods allow one to assess very large numbers of potential interactions extremely efficiently and have been used to conduct genome-wide investigations9–15. One drawback of these techniques, however, is that they do not permit precise control over the assay conditions. The integrity of the proteins cannot be assessed and neither their concentrations nor their posttranslational modification states can be easily manipulated. Likewise, coupling affinity purification with tandem mass spectrometry provides a powerful way to identify protein–protein interactions on a genome-wide scale4,5,16, but does not afford control over the assay conditions or the states of the interacting proteins. In these experiments, an epitope-tagged target protein is expressed in a cell of interest, where it interacts with endogenous proteins or protein complexes. Following affinity purification of the bait protein, interaction partners are identified by tandem mass spectrometry. Although this technique is well suited to identifying physiologically relevant multiprotein complexes, the type and state of the cell has a large role in defining the outcome. Interaction partners that are highly expressed are likely to be identified, whereas those that are not expressed or are present at low concentrations will be missed.
In contrast to these cell-based methods, phage display and oriented peptide library screens are performed in vitro. In a typical phage display experiment, a library of peptide-displaying bacteriophage is passed over a solid support that has been coated with a protein of interest. Phage-displaying cognate peptides are isolated and subsequently amplified. Multiple rounds of selection are usually performed to enrich the library in high-affinity binders, and DNA sequencing of the selected phage reveals the binding preferences of the target protein17–20. In an oriented peptide library experiment, a library of peptides is chemically synthesized and passed over a column displaying the target protein. Peptides captured by the immobilized bait protein are subsequently eluted and subjected to sequencing, either by Edman degradation or by mass spectrometry, thereby revealing the intrinsic binding preferences of the protein7,21. Because phage selections and peptide library screens are both performed in vitro, more control is afforded over the experimental conditions and over the state of the target protein. One drawback of these techniques, however, is that they are competitive in nature; the highest-affinity ligands are preferentially identified and weaker interactors, which may have a biologically relevant role, are often missed. In addition, these methods target all possible sequences within the constraints of the library design and hence often highlight sequences that are nonphysiological (i.e., are not encoded in the genome).
Protein microarrays as a tool for quantitative functional proteomics
In common to all of the methods described above is the fact that they usually produce binary data: proteins are reported either to `interact' or `not interact'. As we seek to extract biological insight from large-scale protein interaction data, it will become increasingly important to obtain reliable, quantitative information that can be incorporated into predictive models of cellular behavior. As a complement to these approaches, protein microarray technology provides a way to study protein–ligand interactions in vitro in a noncompetitive format. Importantly, it can be used to obtain quantitative information on binding affinities, as will be described in detail in the protocols below. Measuring binding affinities serves at least three purposes. First, the additional rigor required to quantify interactions minimizes the amount of incorrect information in the final data set. Most high-throughput methods have alarmingly high rates of false positives and false negatives22–25, limiting their usefulness in generating biological hypotheses. Second, determining binding affinities helps to prioritize which interactions are more likely to be biologically relevant. Finally, quantitative information is particularly useful for modeling studies aimed at predicting protein–protein interactions.
In addition to providing binding affinities, protein microarrays also allow one to assess how well a ligand is recognized by every member of a protein family. As such, they also provide information on binding selectivity. Thus, protein microarrays can be used to determine how cellular systems are insulated from each other and how cross talk is managed within the complex environment of the cell. In addition, they can be used within the context of drug discovery to assess the selectivity of candidate compounds or to identify off-target interactions. For a summary of the advantages and disadvantages of this technique, see Table 1.
TABLE 1.
Advantages and disadvantages of protein domain microarrays.
| Advantages |
| Can be used to obtain quantitative information on binding affinities |
| Affords control over assay conditions and posttranslational modification states |
| High throughput; can be used to determine thousands of binding constants in a single day |
| Can be used to evaluate entire families of domains simultaneously and hence provides information on binding selectivity on a proteome-wide scale |
| Low sample consumption; 1 μg protein is sufficient for > 1,000 assays |
| Low false-negative rate (14% for interactions with KD < 10 μM) |
| Low false-positive rate (14% for interactions with KD < 10 μM) |
| Very low cost per data point once setup is complete |
| Disadvantages |
| Labor-intensive setup; domains must be expressed and purified recombinantly |
| Limited to domains that can be produced as soluble, well-folded protein |
| Limited to domains with moderate to high affinity (KD < 50 μM) |
| Moderate-affinity binders require a secondary assay (e.g., fluorescence polarization) |
| Strictly in vitro assay; does not take expression or colocalization into account |
| Does not take into account avidity afforded by the formation of multiprotein complexes or interactions mediated by other domains in the protein |
In a typical protein microarray experiment, target proteins are spotted in a regular pattern at high spatial density on a solid support, usually a chemically derivatized glass substrate or a glass-supported nitrocellulose membrane (Fig. 1). The spotted proteins become immobilized on the surface and, after a blocking step, are incubated with a labeled probe (e.g., a protein, peptide, nucleic acid or small molecule). After a brief washing step, protein–ligand interactions are identified by detecting and quantifying the label on the probe. If the probe has been labeled with a fluorophore, for example, the array is simply scanned for fluorescence.
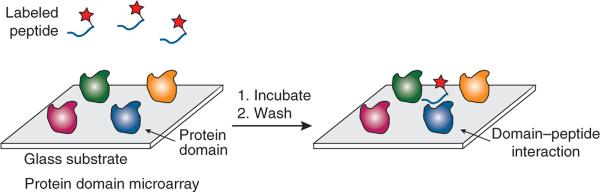
Figure 1.
Protein domain microarrays. Protein domains are immobilized on a solid support (glass substrate) and probed with solution-phase, fluorescently labeled peptides. After an incubation step, the arrays are washed and scanned for fluorescence. Spots comprising domains that recognize the peptide become fluorescent.
Although protein microarrays have been used successfully to conduct genome-wide investigations26–28, one substantial drawback of this technology is that it is extremely labor intensive to clone, express and purify every protein encoded in a genome and many full-length proteins are difficult to produce recombinantly. To avoid these issues, methods have been developed to spot DNA rather than proteins, and then to generate proteins in situ by in vitro transcription/translation29,30. Although these techniques are less labor intensive, they do not provide a way to assess if the proteins are folded correctly and they do not afford control over protein concentrations. In addition, full-length proteins vary widely in their physicochemical properties and so do not behave identically under a common set of conditions; some proteins are highly active on the array surface whereas others are not. To circumvent these limitations, we have chosen to focus our efforts on families of protein interaction domains31–38.
A domain-oriented approach to mapping protein–protein interactions
Most eukaryotic proteins are modular in nature. They comprise both catalytic domains and interaction domains that, to a first approximation, can be abstracted from their host proteins without loss of function39. In general, it is much easier to clone, express and purify isolated domains than it is to work with full-length proteins. It is important to note, however, that many proteins contain several interaction domains and often engage in multivalent interactions or form multiprotein complexes. Uncovering interactions mediated by isolated domains thus provides only part of the information needed to determine how full-length proteins interconnect within the complex environment of the cell. With these caveats in mind, we initially cloned, expressed and purified virtually every Src homology 2 (SH2) and phosphotyrosine binding (PTB) domain encoded in the human genome31. When we spotted these domains on aldehyde-displaying glass substrates and probed them with fluorescently labeled phosphopeptides derived from sites of tyrosine phosphorylation on the ErbB family of receptor tyrosine kinases, we found that at least 102 of the 115 SH2 domain-containing constructs (89%) and at least 27 of the 44 PTB domain-containing constructs (61%) were active on the arrays31. It is likely that many of the `inactive' PTB domains are, in fact, functional, but their role is not to bind to sites of tyrosine phosphorylation40. A domain-oriented strategy thus provides an effective way to circumvent the difficulty associated with producing full-length recombinant proteins and can be used to study the recognition properties of entire families of interaction domains in high throughput.
As mentioned above, one problem with protein microarrays and, indeed, with all other functional proteomics techniques, is that proteins vary considerably in their physicochemical properties. This is true even of closely related family members. For example, we found that human SH2 domains can vary by up to 50-fold in their activity when arrayed on glass surfaces, even if they behave well in solution32. How, then, can we expect to obtain accurate data from any standardized assay performed in high throughput? The best solution, in our opinion, is to generate quantitative data. By measuring binding affinities, we can eliminate the differences in how proteins behave in a standardized assay32.
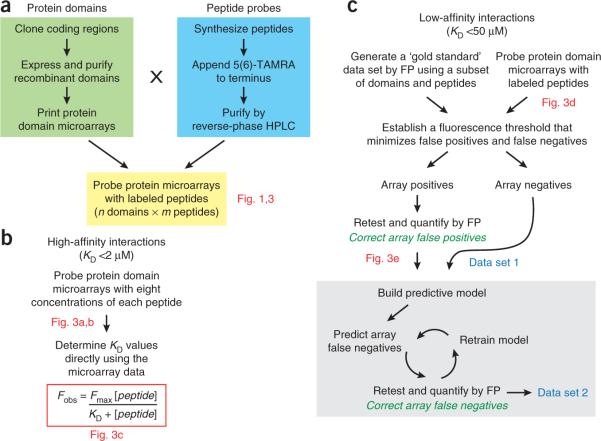
We have developed two alternative strategies for obtaining quantitative information on domain–peptide interactions using protein microarrays (Figs. 2 and 3). First, if the affinities of the interactions are relatively high (KD < 2 μM), as is the case for SH2 and PTB domains, equilibrium dissociation constants can be measured directly on the arrays by probing them with different concentrations of labeled peptide (Figs. 2b and 3a,b). This procedure produces saturation binding curves for every domain–peptide pair, from which binding constants can be derived (Fig. 3c). This strategy has also been used to quantify interactions between proteins and labeled DNA41. Second, if the affinities of the interactions are relatively weak, microarrays of protein domains can be used to rapidly screen many domains against many peptides in an exhaustive manner to highlight putative interactions (Figs. 2c and 3d). Array positives can then be retested and quantified using a lower-throughput assay such as fluorescence polarization (FP) (Fig. 3e). We showed, for example, that microarrays comprising purified, recombinant PDZ domains could be used to screen hundreds of peptides for PDZ domain–peptide interactions36,37. By assaying a subset of the domains and peptides by FP, we established a `gold standard' data set that we then used to calculate false-positive and false-negative rates in the microarray data at different fluorescence intensity thresholds. By choosing an appropriate threshold, we found that the arrays highlighted true biophysical interactions of moderate affinity (KD < 10 μM) with a false-negative rate of 14% and a false-positive rate of 14% (ref. 37). These rates compare very favorably with those observed for other high-throughput methods, such as the yeast two-hybrid system, where false-positive and false-negative rates have been estimated at 50% and 90%, respectively42. We attribute the high fidelity of our protein domain microarrays to the fact that they are prepared under well-controlled in vitro conditions and that they are focused on families of related interaction domains, rather than on full-length proteins with disparate physicochemical properties.
Figure 2.
Quantifying domain–peptide interactions in high throughput using protein domain microarrays. (a) A set of n protein interaction domains are cloned, expressed, purified and arrayed. The microarrays of protein domains are then probed with m fluorescently labeled peptides to reveal the full n × m matrix of domain–peptide interactions. (b) For high-affinity interactions (KD < 2 μM), dissociation constants can be determined directly using protein microarrays. Microarrays of protein domains are probed with eight concentrations of each peptide and the resulting saturation binding curves are used to determine the binding affinity of each domain–peptide interaction. (c) For low-affinity interactions (KD < 50 μM), microarrays of protein domains are probed with fluorescently labeled peptides and a fluorescence threshold is used to divide domain–peptide pairs into putative interactions (array positives) and putative noninteractions (array negatives). A secondary assay (FP) is then used to retest and quantify all array positives. The result is a quantitative interaction data set (data set 1) in which all the false positives in the microarray data set have been eliminated. To remove false negatives, it is necessary to build a model that can predict domain–peptide interactions. The model is then used to highlight suspected false negatives in the microarray data set, which are retested by FP. By performing multiple cycles of prediction, retesting and retraining of the model, many of the microarray false negatives can be corrected. This results in a substantially refined data set (data set 2).
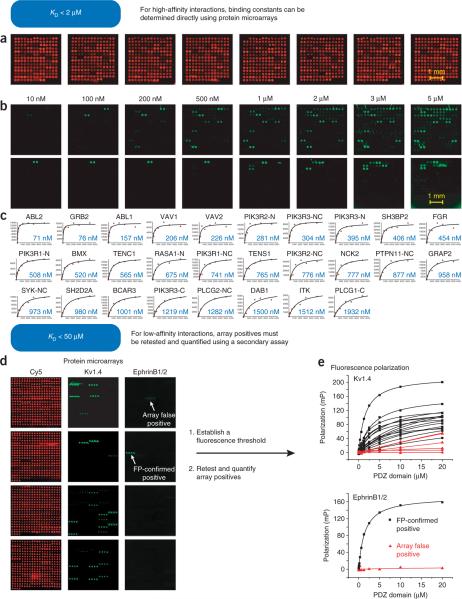
Figure 3.
Using protein domain microarrays to identify and quantify domain–peptide binding interactions. (a) Fluorescent images of eight identical SH2/PTB domain microarrays in separate wells of a 96-well microtiter plate. The fluorescence arises from a trace amount of Cy5-labeled BSA that was added to each protein before arraying. (b) Fluorescent images of SH2/PTB microarrays, probed with eight concentrations of a 5(6)-TAMRA-labeled phosphopeptide derived from ErbB2 pY1139 (5(6)-TAMRA-PLTCSPQPEpYVNQPDVR). (c) Plots showing fluorescence as a function of peptide concentration for 28 high-affinity interactions. The data were fit to equation (1) to determine the KD. (d) PDZ domain microarrays probed with fluorescently labeled peptides. Each PDZ domain was spotted in quadruplicate in wells of 96-well microtiter plates (requiring four wells to include all domains). The Cy5 (red) images are used to identify PDZ domain spots. The green images depict arrays probed with fluorescently labeled peptides. (left, 5(6)-TAMRA-NNGSNAKAVETDV, a promiscuous peptide representing the C terminus of Kv1.4 and right, 5(6)-TAMRANNGQSPANIYYKV, a more selective peptide from Ephrin B1/2).(e) FP titration curves obtained for the array positives identified in (d). Panels a–c31 and d–e36 of this figure are reproduced with permission from the publishers.
As only a small fraction of PDZ domain–peptide combinations result in an interaction, the arrays enabled us to assess a very large number of possible interactions in a noncompetitive way and identify a much smaller number of array positives. We then retested and quantified all array positives by FP (see protocol below). The combined strategy of using protein domain microarrays to highlight potential interactions and FP to quantify binding affinities provides a robust way to obtain high-quality, quantitative data for domains that show moderate-to-weak affinities.
Although this strategy allows one to retest all array positives and thus correct all array false positives, it is much more challenging to identify and correct array false negatives. We addressed this problem with our PDZ domains by constructing mathematical models based on position-specific scoring matrices that could predict domain–peptide interactions36,38. We then used these models to predict which of the array negatives were likely to be false negatives. Predicted false negatives were retested by FP and the corrected data were used to retrain the models. By performing several cycles of prediction, retesting and retraining of the model, we eliminated many of the array false negatives. These computational methods are beyond the scope of this protocol, but are described in detail in our previous reports36,38.
In the protocols that follow, we provide step-by-step instructions on how to prepare, probe and analyze protein domain microarrays to quantify domain–peptide interactions in high throughput. These methods are not restricted to studying interactions with peptides. We have previously shown that protein microarrays can be used to study interactions with labeled small molecules8, and Snyder and co-workers have shown that they can be used to study interactions with phospholipids28. It should be relatively straightforward to adapt our protocols to study interactions between protein domains and fluorescently labeled oligonucleotides, carbohydrates or other biological molecules of interest. To reduce experimental variation and to expedite the processing of thousands of microarrays, we have also developed methods to fabricate protein domain microarrays in individual wells of 96-well microtiter plates31,37 (Fig. 4). The protocols that follow explain how to do this. Alternatively, protein domain microarrays can also be printed on standard 1 inch × 3 inch glass substrates and processed manually. Although the microtiter plate-based methods require some custom-manufactured materials, they are all available by special order from commercial sources at reasonable prices.
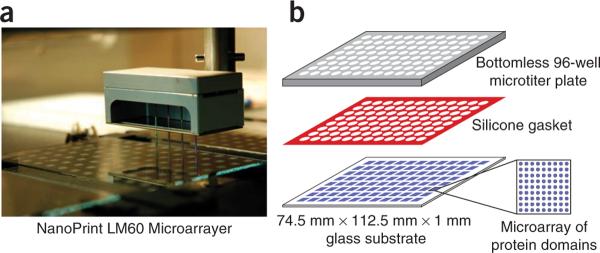
Figure 4.
Microarrays in microtiter plates. (a) NanoPrint Microarrayer spotting recombinant domains onto aldehyde-displaying glass substrates. (b) Attachment of microarrays to the bottom of a bottomless 96-well microtiter plate using an intervening silicone gasket. Panel b of this figure is reproduced, with permission, from the publisher37.
Experimental design
Cloning, expression and purification of protein domains
Domain-based protein microarrays enable the recognition properties of an entire family of protein interaction domains to be assessed simultaneously. The primary advantage of such studies is that they provide insight into binding selectivity on a proteome-wide scale. Our strategy has been to clone, express and purify isolated domains, rather than full-length proteins. The primary requirement for adopting this strategy is that the domains under investigation must fold correctly, be stable and soluble, and retain their function when abstracted from their parent proteins. To date, we have focused entirely on domain families that can be produced recombinantly in Escherichia coli. This greatly simplifies the production of sufficient quantities of protein (a minimum of 40 μl of a 40 μM solution of protein is required). It is also relatively inexpensive. Empirically, we have found that many protein domains are produced efficiently as soluble protein when fused to thioredoxin. We have standardized our cloning and expression efforts by cloning the coding regions of our protein domains into the Gateway-compatible entry vector pENTR-D-TOPO (Invitrogen) by topoisomerase I-mediated directional cloning. We then subclone sequence-verified open reading frames (ORFs) into pET-32-DEST by λ-recombinase-mediated directional subcloning. pET-32-DEST is a Gateway-compatible bacterial expression vector31 based on pET-32c (Novagen). Domains produced using this vector feature N-terminal His6 and thioredoxin tags. To ensure tight regulation of induction and to obtain high yields, it is advisable to produce all domains in E. coli strain BL21(DE3)pLysS (Invitrogen). A brief overview of this process is provided in the protocol below.
Peptide synthesis
We synthesize all our peptides using standard Fmoc chemistry (Box 1). Peptides are synthesized on derivatized Wang resins and therefore feature a free C terminus following deprotection and cleavage. We use a 96-well peptide synthesizer (Apex 396; Aapptec) and the protocol provided below has been optimized for this instrument. We usually synthesize our peptides on a 10-μmol scale, which provides enough material to prepare several aliquots of purified peptide, each of which can be used to probe dozens of protein microarrays. Amino acids and HBTU are predissolved at 62.5 mM in N,N-dimethylformamide (DMF), whereas diisopropylethylamine (DIPEA or Hünig's base) is added neat. Resins that are not Fmoc protected should be pre-swollen in DMF before use. We have found that performing double couplings with five equivalents of each amino acid (instead of single couplings with 10 equivalents) results in peptides of higher purity, although it extends the synthesis time by approximately 1.5 h per residue. We use 5-(and-6)-carboxytetramethylrhodamine (5(6)-TAMRA) as a fluorescent label on our peptides, which we couple to the N terminus before deprotection and cleavage. Purchasing mixed isomers of TAMRA is less expensive than purchasing isomerically pure compound and does not compromise the results of the assay.
Fabrication and processing of protein microarrays
We use aldehyde-displaying glass as a substrate for our protein microarrays and assemble these in 96-well plates as described in Box 2. Proteins attach to the surface through the α-amine at their N terminus or through the ε-amine of accessible lysine residues and hence are shown in many different orientations within each spot. This strategy ensures that their binding sites are accessible to ligand. We typically array our proteins at a concentration of ~40 μM in a buffer that contains 20% glycerol (vol/vol) to prevent evaporation. We print several replicates of each sample and include a small amount (typically 100 nM) of cyanine-5 (Cy5)-labeled bovine serum albumin (BSA) (prepared as described in Box 3) to facilitate image analysis. After a 12-h incubation at room temperature, we quench the unreacted aldehydes on the glass and block the surface by adding a buffer containing BSA. We then probe the arrays with the labeled peptides. After a 1-h incubation at room temperature, the arrays are washed, dried and scanned for both Cy5 and 5(6)-TAMRA fluorescence. The Cy5 image is used to define the location of each spot and the mean fluorescence of replicate spots in the 5(6)-TAMRA image is determined for each domain. We typically subtract the mean intensity of control spots (thioredoxin) and use these background-subtracted values for subsequent analyses.
Determining if a secondary assay is needed
In our experience, fluorescent peptides can be used as probes on protein microarrays up to a concentration of about 5 μM. Many 5(6)-TAMRA-labeled peptides are not soluble in aqueous buffer at higher concentrations and background binding on the arrays becomes prohibitively high. As such, we recommend that a secondary assay (such as FP) be used when studying interactions with KD values greater than 2 μM.
Quantifying domain–peptide interactions by fluorescence polarization
Quantitative, solution phase FP assays can be conducted using the same recombinant domains and 5(6)-TAMRA-labeled peptides as are used for the microarray assays. For FP assays, which are typically conducted in 384-well microtiter plates, we keep the peptide concentration constant (20 nM) and vary the concentration of the protein domain. Protein concentrations can be determined as described in Box 4 and reference 43. Large-scale studies are typically conducted using a maximum domain concentration of 20 μM (to conserve reagents), whereas investigations involving fewer peptides per domain can be conducted with domain concentrations as high as 100 μM. To obtain saturation binding curves, we prepare a twofold serial dilution of the protein domains in the microtiter plate. We typically prepare 12 concentrations of each domain. We then add a constant amount of 5(6)-TAMRA-labeled peptide to each well. For large-scale studies, we have found it useful to automate the dilution of domains and the addition of peptides using standard liquid handling robots. This is not necessary, however, and can readily be accomplished using standard multichannel pipetters.
Quality assurance criteria for determining KD values
For both the protein microarray assay and the FP assay, we fit the resulting data to equations that describe saturation binding (see protocols below). We only report a dissociation constant if the R2 value of the fit is at least 0.9.
MATERIALS
REAGENTS
Bovine serum albumin, fraction VI (Sigma-Aldrich, cat. no. A3294)
pENTR/D-TOPO Cloning Kit (Invitrogen, cat. no. K2400-20)
One Shot BL21(DE3)pLysS Chemically Competent E. coli (Invitrogen, cat. no. C606003)
Subcloning Efficiency DH5α Competent Cells (Invitrogen, cat. no. 18265-017)
Luria Broth EZMix Powder (Sigma-Aldrich, cat. no. L7658)
Luria Broth Agar EZMix Powder (Sigma-Aldrich, cat. no. L7533)
Glycerol (EMD Chemicals, cat. no. GX0185-6)
Ampicillin sodium salt (Sigma-Aldrich, cat. no. A0166)
Kanamycin sulfate (Calbiochem, cat. no. 420411)
Chloramphenicol (Sigma-Aldrich, cat. no. C1919)
Isopropyl β-d-1-thiogalactopyranoside (IPTG; Gold Biotechnology Inc., cat. no. I2481C100)
Ni-NTA His Bind Resin (Novagen, cat. no. 69670-4)
Phenylmethylsulphonyl fluoride (PMSF; Fluka, cat. no. 78830)
Benzamidine hydrochloride (Sigma-Aldrich, cat. no. 43476-0)
BugBuster 10× Protein Extraction Reagent (Novagen, cat. no. 70921-4)
Benzonase nuclease (Novagen, cat. no. 70746-4)
Sodium azide (Sigma-Aldrich, cat. no. S8032)
Illustra NAP-10 columns (GE Healthcare, cat. no. 17-0854-02)
N,N-Dimethylformamide (DMF; EMD Chemicals, cat. no. DX1726P-1) ! CAUTION DMF is an organic solvent and should be handled in a fume hood.
Methanol (Mallinckrodt, cat. no. 3016-16) ! CAUTION Methanol is flammable and toxic.
Ethyl Ether, Anhydrous (EMD Chemicals, cat. no. EX0190-3) ! CAUTION Ether is highly volatile and flammable. It should not be stored for more than 6 months after it is opened as it is prone to peroxide formation; ethylidene peroxide is an extremely brisant and friction-sensitive explosive material.
N,N-Diisopropylethylamine (DIPEA; Sigma-Aldrich, cat. no. 496219) ! CAUTION Toxic and volatile, with a strong odor.
Piperidine (Sigma-Aldrich, cat. no. 571261) ! CAUTION Toxic and volatile, with a strong odor. In addition, this is a regulated substance and paperwork must be filled out before purchase.
HATU (Advanced ChemTech, cat. no. 101398-598) △ CRITICAL Because of the high cost of 5(6)-TAMRA, a highly active coupling agent such as HATU should be used.
HBTU (Advanced ChemTech, cat. no. 101116-588)
Fmoc-Ala-OH (Advanced ChemTech, cat. no. 101114-368)
Fmoc-Cys(Trt)-OH (Advanced ChemTech, cat. no. 101114-452)
Fmoc-Asp(tBu)-OH (Advanced ChemTech, cat. no. 101114-516)
Fmoc-Glu(tBu)-OH (Advanced ChemTech, cat. no. 101114-578)
Fmoc-Phe-OH (Advanced ChemTech, cat. no. 101114-626)
Fmoc-Gly-OH (Advanced ChemTech, cat. no. 101114-780)
Fmoc-His(Trt)-OH (Advanced ChemTech, cat. no. 101114-836)
Fmoc-Ile-OH (Advanced ChemTech, cat. no. 101114-860)
Fmoc-Lys(Boc)-OH (Advanced ChemTech, cat. no. 101114-906)
Fmoc-Leu-OH (Advanced ChemTech, cat. no. 101114-990)
Fmoc-Met-OH (Advanced ChemTech, cat. no. 101115-058)
Fmoc-Asn(Trt)-OH (Advanced ChemTech, cat. no. 101115-086)
Fmoc-Pro-OH (Advanced ChemTech, cat. no. 101115-134)
Fmoc-Gln(Trt)-OH (Advanced ChemTech, cat. no. 101115-190)
Fmoc-Arg(Pbf)-OH (Advanced ChemTech, cat. no. 101115-254)
Fmoc-Ser(tBu)-OH (Advanced ChemTech, cat. no. 101115-304)
Fmoc-Thr(tBu)-OH (Advanced ChemTech, cat. no. 101115-346)
Fmoc-Val-OH (Advanced ChemTech, cat. no. 101115-384)
Fmoc-Trp(Boc)-OH (Advanced ChemTech, cat. no. 101115-446)
Fmoc-Tyr(tBu)-OH (Advanced ChemTech, cat. no. 101115-618)
Fmoc-Asp(tBu)-Wang Resin (Advanced ChemTech, cat. no. 101117-604)
Fmoc-Phe-Wang Resin (Advanced ChemTech, cat. no. 101399-122)
Fmoc-Ile-Wang Resin (Advanced ChemTech, cat. no. 101117-876)
Fmoc-Leu-Wang Resin (Advanced ChemTech, cat. no. 101118-058)
Fmoc-Pro-Wang Resin (Advanced ChemTech, cat. no. 101118-356)
Fmoc-Val-Wang Resin (Advanced ChemTech, cat. no. 101118-688)
5-(and-6)-Carboxytetramethylrhodamine (5(6)-TAMRA; Invitrogen, cat. no. C300)
Trifluoroacetic acid (TFA; EMD Chemicals, cat. no. TX1276-6) ! CAUTION Corrosive acid, restrict use to fume hoods.
Triisopropylsilane (TIPS; Sigma-Aldrich, cat. no. 233781) △ CRITICAL Triisopropylsilane must be included during deprotection and cleavage of peptides to quench carbocations and hence minimize unwanted side reactions.
Ethanedithiol (Fluka, cat. no. 02390) ! CAUTION Volatile stench agent.
Thioanisole (Sigma-Aldrich, cat. no. T-2765) ! CAUTION Volatile stench agent.
Phenol (MP Biomedicals, cat. no. 800672) ! CAUTION Toxic, handle with care and avoid skin contact.
Cy5 mono-Reactive Dye Pack (GE Healthcare, cat. no. PA25001)
Amicon Ultra-4 concentrator, 10 kDa cutoff (Millipore, cat. no. UFC801024)
QIAprep Spin Miniprep Kit (Qiagen, cat. no. 27106)
SafeSeal Microcentrifuge Tubes (Sorenson BioScience Inc., cat. no. 16070)
Fritted poly-prep chromatography columns (Bio-Rad, cat. no. 731-1553)
LR Clonase II Plus (Invitrogen)
EQUIPMENT
Custom-made aldehyde-coated 74.5 mm × 112.5 mm × 1 mm glass substrates (Thermo Fisher Scientific Inc., cat. no. HAR-1101-C60)
ProPlate Gaskets (Grace Bio-labs, cat. no. 204971)
96-Well No-Bottom microtiter plates (Greiner Bio-One, cat. no. 655000)
96-well microtiter plates (Greiner Bio-One, cat. no. 650201)
384-well assay plates, black nonbinding, for FP assays (Corning, cat. no. 3575)
384-well microarray plates, for printing protein microarrays (Genetix, cat. no. X7022)
Storage Mat III (Costar, cat. no. 3080)
Disposable reagent reservoirs, sterile (VWR, cat. no. 82026-350)
Deep-well 96-well microtiter plate (Costar, cat. no. 3960)
14 ml Polypropylene Round-Bottom Tube (Becton Dickinson, cat. no. 352059)
NanoPrint LM60 Microarrayer (Arrayit Corporation) including cooling block for source plate and destination block designed for 16 microtiter-sized glass plates
Silicon Microarray spotting pins (Parallel Synthesis Technologies Inc., cat. no. SMT-S75)
48-pin Silicon printhead assembly (Parallel Synthesis Technologies Inc., cat. no. SMT-H192)
Funnel for silicon 48-pin printhead (Parallel Synthesis Technologies Inc., cat. no. SMT-F48)
Fluorescence microarray scanner—Tecan LS400 Laser Scanner or similar (Tecan, cat. no. LS400)
ArrayPro software or equivalent (Tecan)
Matlab software or equivalent (The MathWorks)
Peptide Synthesizer, Apex 396 or similar (Aapptec, cat. no. Apex 396-DC-FW-M)
Kromasil 100 (5 μm) C18 semi-prep column (Peeke Scientific, cat. no. 100-5-C18 20 × 250)
Kromasil 100 (5 μm) C18 analytical column (Peeke Scientific, cat. no. 100-5-C18 2.1 × 150)
Superdex 200 10/300 GL column (Amersham Biosciences, cat. no 17-5175-01)
Fluorescence polarization plate reader, Analyst AD 96:384 or similar (LJL Biosystems)
Storage Mat Applicator (Corning, cat. no. 3081)
Microtiter plate shaker (Lab Line, cat. no. 4625)
Floor centrifuge, capable of holding both 500 and 30 ml tubes
2-L Baffled flasks (VWR , cat. no. 89083-696)
500-ml Centrifuge tubes (Sorvall, cat. no. 7-9957)
30-ml Oakridge centrifuge tubes (Thermo Fisher Scientific Inc., cat. no. 3119-0010)
P1000 multichannel pipette (Rainin, cat. no. L1000)
P200 multichannel pipette (Rainin, cat. no. L200)
P10 multichannel pipette (Rainin, cat. no. L10)
Mini Bunsen burner
Agilent 1200 series HPLC with fraction collector
REAGENT SETUP
Phosphate-buffered saline (PBS; 100 mM KCl, 20 mM sodium phosphate, pH 7.4)
Blocking Solution (100 mM KCl, 20 mM HEPES, 1% BSA (wt/vol), pH 7.4)
Probing Buffer (100 mM KCl, 20 mM HEPES, 0.1% Tween 20, pH 7.4)
10× Lysis Buffer (500 mM NaH2PO4, 3 M NaCl, 50 mM imidazole, pH 8.0)
Wash Buffer (50 mM NaH2PO4, 300 mM NaCl, 20 mM imidazole, pH 8.0)
Elution Buffer (50 mM NaH2PO4, 300 mM NaCl, 250 mM imidazole, pH 8.0)
Guanidinium Chloride Solution (6 M GdnHCl, 20 mM sodium phosphate, pH 7.4)
Sodium Carbonate Buffer (100 mM sodium carbonate; adjust pH with saturated sodium bicarbonate to pH 9.3)
HPLC Solvent A (ddH2O with 0.1% trifluoroacetic acid)
HPLC Solvent B (Acetonitrile (HPLC grade) with 0.1% trifluoroacetic acid)
EQUIPMENT SETUP
HPLC purification protocol (used for fluorescently labeled synthetic peptides ranging from 1,500 to 2,500 Da)
The following protocol provides general guidelines. Individual users should adjust the protocol depending on the specific properties of the peptides under investigation. Set UV detector to 580 nm to detect 5(6)-TAMRA. In addition, monitoring at 230 and 280 nm allows the user to track unlabeled peptides as well. Using a semi-preparative reverse-phase C18 HPLC column, you should begin by equilibrating the column in 10% Solvent B, 90% Solvent A for 5–10 min at a flow rate of 10 ml per min. Inject crude peptide sample, dissolved in water or DMF, and allow the system to equilibrate for 5–10 min. Separate peptides by running a linear gradient from 10% B to 40% B over 30 min (1% per min). Peptides labeled with 5(6)-TAMRA often elute as a double peak due to the two isomeric forms of the fluorophore. Collect both peaks and pool them. To clean the column, you should run a linear gradient from 40% B to 100% B over 5 min, maintain at 100% B for 10 min and run a linear gradient from 100% B to 10% B over 5 min to restore the column to its initial condition.
Fabricating protein domain microarrays
Over the past 7 years, we have used a variety of methods to fabricate protein domain microarrays. We have found that both noncontact arrayers that use piezoelectric-based delivery and contact arrayers that use split pins are compatible with this technology. The protocols described in Steps 33–50 describe arrays prepared using a contact arrayer and split pins. This is the method we currently favor. As we have not tried every type of pin on the market, we cannot comment on which is best. We currently use SMT-S75 silicon pins manufactured by Parallel Synthesis Technologies Inc. These pins have an uptake volume of ~0.12 μl and a tip size of 75 μm × 75 μm. With these pins, we set the relative humidity in the microarrayer to ~85%. Different pins and different sample compositions may require slightly different settings. For reviews on microarrayers, please see references 44, 45.
PROCEDURE
Cloning, expression and purification of domains ● TIMING 4 d for 30 domains
-
1|
Identify all domain family members of interest and determine consistent truncation parameters. To do this, we use the SMART database46 and typically include 15 residues upstream and 15 residues downstream of the domain sequence.
-
2|
Clone the ORF encoding each domain of interest. We have successfully cloned ORFs from cDNA; subcloned ORFs from previously cloned genes and obtained ORFs through de novo gene synthesis. We typically use Invitrogen's TOPO entry vectors, which are part of Invitrogen's Gateway system. TOPO vectors require a TOPO sequence (CACC) to be included at the 5′ end of the linear DNA being cloned.
-
3|
Follow Invitrogen's Protocol for Directional TOPO Cloning to transfer DNA fragments into the vector pENTR/D-TOPO. Transform DH5α competent cells with 2 μl of the reaction mixture and plate onto LB agar plates containing 34 μg ml−1 kanamycin.
-
4|
From single colonies, amplify bacterial clones by inoculating 5 ml of LB broth containing 34 μg ml−1 kanamycin in a 14 ml round-bottom tube. Shake tubes at 200 r.p.m., 37 °C for 16 h. Purify plasmids using Qiagen's QIAprep Spin Miniprep kits and sequence verify clones using M13 forward and reverse primers (M13 forward: TGTAAAACGACGGCCAGT; M13 reverse: CAGGAAACAGCTATGAC).
-
5|
Transfer each ORF to a Gateway-compatible destination vector using LR Clonase II Plus according to the manufacturer's specifications. Verify that the resulting construct is correct either by restriction enzyme analysis or by DNA sequencing. For protein production in bacteria, we use the Gateway-compatible E. coli expression vector pET-32-DEST31. This vector was constructed by inserting the Gateway Reading Frame Cassette C into pET-32c (Novagen) that had been digested with EcoRV. Transform competent DH5α E. coli cells with 2 μl of the reaction mixture and plate cells onto LB agar plates containing 100 μg ml−1 ampicillin. Incubate at 37 °C overnight. Inoculate 5 ml of LB broth containing 100 μg ml−1 ampicillin in a 14 ml round-bottom tube with a single colony and shake at 200 r.p.m., 37 °C for 16 h. Purify plasmids using a QIAprep Spin Miniprep Kit (Qiagen).
■ PAUSE POINT Vectors can be stored indefinitely at −20 °C.
-
6|
Transform BL21(DE3)pLysS E. coli cells with 1 μl of the purified plasmid. Plate cells onto LB agar plates containing 100 μg ml−1 ampicillin and 34 μg ml−1 chloramphenicol.
-
7|
Inoculate 2 ml of LB broth containing 100 μg ml−1 ampicillin and 34 μg ml−1 chloramphenicol in a 14 ml round-bottom tube with a single colony and shake at 200 r.p.m., 37 °C for 16 h.
-
8|
Add 1 ml culture to 500 ml LB broth containing 100 μg ml−1 ampicillin and 34 μg ml−1 chloramphenicol in a 2 L baffled flask. Shake at 200 r.p.m., 37 °C until the absorbance of the cultures at 600 nm is 0.8 (A600 = 0.8; 1 cm path length).
This step will take approximately 4 h.
-
9|
Cool the cultures to room temperature in a sink filled with cool water. Once at room temperature, move the flasks to a shaker at room temperature and add IPTG to a final concentration of 1.0 mM. Continue to shake at approximately 25 °C for 12–16 h (overnight).
-
10|
Transfer media to 500 ml centrifuge bottles and pellet bacteria at 11,150g for 15 min.
-
11|
Discard the supernatant and add 5 ml of 0.5× lysis buffer containing protease inhibitors (1 mM PMSF and 1 mM benzamidine hydrochloride). Supernatant should be disinfected with one part bleach for every 10 parts supernatant before disposal.
-
12|
Break apart cell pellet using a 2 ml pipette and short periods of agitation with a bench-top vortex. Transfer suspension to a 30 ml Oakridge tube.
-
13|
Add 500 μl of 10× BugBuster and 5 μl benzonase nuclease. Incubate for 1.5 h with rocking at 4 °C. If pellet lysates are particularly aggregated, incubation can be increased to 2.5 h.
-
14|
Add 5 ml of 1.5× lysis buffer containing protease inhibitors (1 mM PMSF and 1 mM benzamidine hydrochloride) to each sample.
-
15|
Centrifuge each sample at 35,800g for 30 min, 4 °C.
-
16|
Although the samples are centrifuging, prepare Ni-NTA resin. For each sample to be purified, transfer 600 μl of resin to a 50 ml Falcon tube and mark the level of the resin on the tube. Centrifuge at 3,000g for 1 min and remove supernatant. Fill the Falcon tube with 1× lysis buffer and centrifuge at 3,000g for 1 min. Discard supernatant. Add 1× lysis buffer to the original marked volume and resuspend the resin.
-
17|
Decant the supernatant from the cell lysates (Step 15) into 15 ml Falcon tubes and store on ice. Add 600 μl of the resuspended resin to each sample. Allow proteins to bind to the resin by rotating samples at 4 °C for 30 min (batch binding).
-
18|
Set disposable fritted columns on a rack suspended above a waste collection container. Precondition each column by allowing 1 ml of ddH2O to pass through, followed by 1 ml of 1× lysis buffer.
-
19|
Transfer cell lysate/resin slurry to column and allow to drain.
-
20|
Add 10 ml of wash buffer to each column and allow to drain.
-
21|
Plug each column and add 0.5 ml of elution buffer to resin. Incubate for 5 min and then unplug the column and collect the flow-through in a 1.7 ml microcentrifuge tube. Repeat with an additional 0.5 ml of elution buffer.
-
22|
Perform a buffer exchange to transfer the purified proteins into PBS. We use NAP-10 columns for this step, rather than dialysis membranes, as this procedure is quick and can be performed on many samples simultaneously. Equilibrate the NAP-10 columns with PBS by flushing three times with 5 ml of PBS. Add exactly 1 ml of purified protein and drain to waste. If volume is less than 1 ml, add PBS to make up the difference. Add 1.5 ml of PBS to the column and collect the flow-through in a 1.7 ml microcentrifuge tube. This fraction contains the purified protein in PBS buffer.
? TROUBLESHOOTING
-
23|
Centrifuge samples on a tabletop centrifuge at 14,000g for 2 min to remove any precipitate.
-
24|
Determine protein concentration (Box 4). Adjust concentration to 75 μM. If the protein concentration is less than 75 μM, use the sample for subsequent experiments, but note its actual concentration. This information is useful if unexpected negatives are observed on the arrays.
-
25|
Assess the purity of the protein by SDS-polyacrylamide gel electrophoresis. In our laboratory, we have chosen to restrict our studies to proteins with > 80% purity.
-
26|
Assess the aggregation state of each domain by analytical size exclusion column chromatography. We use a Superdex 200 10/300 GL column run in PBS at 0.75 ml min−1 and typically observe monomeric domains of 30–40 kDa eluting at ~18 min after injection. We restrict our subsequent studies to domains that are predominantly monomeric or dimeric.
? TROUBLESHOOTING
-
27|
Prepare protein domains for long-term storage at − 80 °C. If the domains will be used to prepare protein microarrays, mix three parts protein with one part 80% glycerol and divide into 100 μl aliquots. If proteins will be used for fluorescent polarization assays, do not add glycerol and divide into 100–1,000 μl aliquots. Flash freeze the aliquots in liquid nitrogen and store at − 80 °C.
■ PAUSE POINT Protein domains can typically be stored at − 80 °C for up to 6 months in buffer. Proteins can be stored for longer periods at − 80 °C when stabilized in 20% glycerol.
Synthesis and purification of labeled peptides ● TIMING 1 month for 100 peptides
-
28|
Design peptides for protein domain microarray experiments. For peptides that are recognized by SH2 and PTB domains, we include nine residues upstream and seven residues downstream of established sites of tyrosine phosphorylation. We include a C-terminal Asp residue to promote solubility and cap the N terminus with 5(6)-TAMRA. This results in an 18-residue peptide. The 5(6)-TAMRA serves both as a chromophore for quantification purposes and as a fluorophore for visualization of the protein microarrays. For peptides that are recognized by PDZ domains, we include the last 10 residues of the protein of interest. Longer peptides are not compatible with fluorescent polarization assays. We also include a short Asn-Asn-Gly linker at the N terminus to promote solubility and then cap the peptide at its N terminus with 5(6)-TAMRA. This results in a 13-residue peptide.
-
29|
Synthesize peptides on the solid phase using standard Fmoc chemistry. Box 1 outlines our preferred methods that we have optimized for the Apex 396 Peptide Synthesizer (Aapptec).
-
30|
After the peptides have been synthesized, labeled, deprotected, cleaved and precipitated, dissolve them in 1 ml DMF.
-
31|
Purify the peptides by reverse-phase HPLC using a C18 semi-preparative column. In our experience, 5(6)-TAMRA-labeled peptides, including phosphopeptides, typically elute between 20% and 40% HPLC Solvent B. For guidelines on HPLC methods, please see EQUIPMENT SETUP.
-
32|
Identify fractions containing the desired peptide product by MALDI-TOF mass spectrometry. Pool these fractions and evaluate the purity of the sample by analytical reverse-phase HPLC. We typically use a Kromasil 100 (5 μm) C18 column (150 mm length; 2.1 mm ID). The best results are achieved in the microarray assay if the peptide is at least 95% pure. To assess yield, you can dilute the purified peptide in a 1:1 mixture of HPLC Solvents A and B and measure absorbance at 548 nm. The molar extinction coefficient of 5(6)-TAMRA is 85,500 M−1 cm−1 in ddH2O. Divide the sample into smaller aliquots, freeze in liquid nitrogen and lyophilize to dryness.
■ PAUSE POINT Lyophilized peptides can be stored at − 20 °C. Storing peptides in solution for extended periods of time results in unwanted oxidation.
Fabricating protein domain microarrays ● TIMING 2 d preparation, then 12 h per four plates of arrays
-
33|
Before printing purified domains, perform a test print run using BSA. Prepare a solution of 40 μM BSA and 0.4 μM Cy5-BSA (Box 3) in PBS with 20% glycerol (see REAGENT SETUP). Prepare 180 μl of solution for each domain to be printed. For example, if you are planning to print 50 domains, prepare 9.0 ml of BSA solution for the test run.
-
34|
Using a multichannel pipette, you can add 40 μl BSA solution to all the wells of a 384-well microarray source plate that will be used during the actual print run. Note: in the actual print run, each sample will be present in four separate and alternating wells of the source plate (see Step 47).
-
35|
Assemble the NanoPrint arrayer such that a cooling block is placed under the source plate (plate containing the BSA). Destination plates (aldehyde-displaying glass substrates) on which the arrays will be printed should not be cooled.
-
36|
Attach the funnel to the printhead, and load one pin. This pin will be used to determine the coordinates for each glass plate. This can be performed using a pin that is no longer functional assuming the tip of the pin is intact. Use fine-point tweezers when loading the pins, according to the manufacturer's instructions.
-
37|
Attach the printhead to the printing arm of the arrayer. Be careful not to damage the tip of the pin.
-
38|
Using the manual control option of Arrayit software, you can slowly lower the pin to the bottom left corner of each plate to be used in the print run. Set x and y coordinates using this value. Next, test all four corners of every plate. Use the lowest of these four z-values as the z-value for each plate to ensure that each pin makes contact throughout the print run.
▲ CRITICAL STEP Although tedious, setting z-heights on the arrayer is critical for the production of consistent arrays.
-
39|
Detach the printhead from the arrayer arm and remove the pin. Attach the funnel to the printhead and load four good pins in a square pattern with 9 mm spacing between each pin.
-
40|
Detach the funnel from the printhead and attach the foam top. Replace the foam in the top of the printhead every couple of months for best results. Foam of the correct size and density can be obtained from Parallel Synthesis Technologies, Inc.
-
41|
With the aid of a mini Bunsen burner, hold the tip of each pin under the flame for 2 s. Repeat so that each pin is flamed twice. Attach the printhead to the printing arm of the arrayer. Pins should be flamed after every 12 h of printing to eliminate organic sediment that builds up in the tip of the pin.
-
42|
Use the Arrayit software package to set up a method for the print run. In our hands, print runs are most consistent when silicon pins are flamed every 12 h. For this reason, we attempt to limit all print runs to 12 h or pause longer print runs in 12 h increments. Twelve-hour print runs allow for the printing of four destination plates, with 96 microarrays per plate (16 × 16 spots per array). One blot plate is also used for each print run.
-
43|
Set up the print run with the following parameters: (1) set the cooling block under the source plate to 2.5 °C; (2) set the relative humidity in the arrayer to 85%; (3) `ink' the pins with a dwell time of 4 s in the source plate; (4) perform 20 touches on the blot plate (reusable glass substrate) after each inking; (5) spot the samples on the destination plates (aldehyde-displaying glass substrates) in quadruplicate; (6) re-ink the pins every two destination plates (192 touches); (7) perform two cycles of washing and drying the pins between successive protein samples. Use ddH2O for the washes and do not sonicate the pins.
-
44|
Attach the destination plates to bottomless 96-well microtiter plates using an intervening silicone gasket (Box 2). It is a good idea to practice this using BSA microarrays.
-
45|
Scan the arrays using a Tecan LS400 Laser Scanner. To visualize Cy5-BSA, we use a 633 nm laser with a Cy5 filter. To visualize 5(6)-TAMRA-labeled peptides, we use a 543 nm laser with a Cy3 filter.
? TROUBLESHOOTING
-
46|
Once satisfied with the results of the BSA print run, prepare a master plate of purified protein domains. Master plates should be prepared in 96-well microtiter plates. For each well, combine 178.2 μl of purified protein domain with 1.8 μl of 5 mg ml−1 Cy5-labeled BSA and mix well. To prepare Cy3-BSA, see Box 3. Include purified thioredoxin as a negative control.
-
47|
Prepare a source plate for arraying from the master plate. Source plates are prepared using low-volume 384-well microarray plates (Genetix). Each well of the source plate should contain 40 μl of the sample to be printed (purified domain mixed with Cy5-BSA). Each sample (protein domain) should be introduced into four separate wells of the source plate such that the four pins in the microarrayer will have access to these four aliquots simultaneously. As the arrays are printed with 9 mm spacing on the destination plate (to match 96-well spacing), the protein samples should be introduced into alternating wells of the 384-well source plate. For example, if the four pins are set in a square pattern in the print head of the arrayer, the first protein sample should be placed into wells A1, A3, C1 and C3 of the source plate.
-
48|
Store source plate(s) at 4 °C until ready to begin arraying. The domains we have worked with to date are all stable at 4 °C in 20% glycerol for several weeks.
-
49|
Perform Steps 35–44 using the purified protein domains and fresh aldehyde-displaying glass substrates (destination plates).
-
50|
After attaching the arrays to the bottomless 96-well microtiter plates, allow the arrays to incubate in the dark for 4 h and then freeze them at − 80 °C.
■ PAUSE POINT Arrays can be stored at − 80 °C for up to 1 year.
Probing protein domain microarrays ● TIMING 3 h
-
51|
Working with one array plate at a time, remove from − 80 °C, remove seal, and immediately add 195 μl of blocking solution to each well (see REAGENT SETUP). This step should be performed reasonably quickly to ensure that the array spots do not thaw before the blocking solution is added. The blocking solution typically freezes when it is added to the plate.
-
52|
Incubate the plate at room temperature for 20 min to allow the BSA solution to thaw.
-
53|
Repeat Steps 51 and 52 up to three additional times. It is reasonable to probe and process four plates at a time.
-
54|
Shake the plates on a secured microtiter plate shaker at 600 r.p.m. for 1 h.
-
55|
Although the arrays are blocking, fill row A of a deep-well 96-well microtiter plate with 1 ml of a 5 μM solution of each labeled peptide. Each peptide should be dissolved in probing buffer (see REAGENT SETUP).
? TROUBLESHOOTING
-
56|
Prepare dilutions of each peptide in a serial manner down the columns of the plate, using the buffer and transfer volumes provided in table 2.
-
57|
Remove blocking solution from the array plate by rapidly inverting the plate. Replace with 195 μl of probing buffer and shake for 1 min. Invert the plate to remove probing buffer.
-
58|
Transfer 100 μl of labeled peptide from each well of the peptide source plate to each well of the array plate. Each well in the peptide source plate contains enough volume to probe four wells in the array plate. We typically probe our arrays in duplicate.
-
59|
Shake the array plates on a microtiter plate shaker at 600 r.p.m. for 1 h at room temperature.
-
60|
Empty the array plates by inverting them and add 200 μl of probing buffer. Shake for 1 min as before. Repeat with 200 μl of ddH2O. Empirically, we have found that a final wash with ddH2O does not disrupt domain–peptide interactions on the arrays.
-
61|
Remove ddH2O by inverting the plates and place array plates upside down over a deep-well 96-well microtiter plate. To dry arrays, we can centrifuge at 1,000g for 5 min.
■ PAUSE POINT Probed, dried arrays can be stored at room temperature, in the dark, for several weeks.
-
62|
Scan the arrays with a 633-nm laser using a Tecan LS400 Laser Scanner (or equivalent). Set the resolution to 10 μm per pixel and the PMT to 200. If the resulting image contains saturated pixels, lower the PMT setting and rescan. Scan the arrays with a 543-nm laser, starting with a PMT setting of 255 and decreasing as necessary. Although repeated scanning results may provide some photobleaching of the fluorophores, we have not found this to be a large problem at these settings.
▲ CRITICAL STEP Ensure that the final scans do not contain saturated pixels.
? TROUBLESHOOTING
-
63|
Save all files as 16-bit TIFF images.
▲ CRITICAL STEP Do not compress files to 8-bit format. This compromises subsequent analyses.
TABLE 2.
Dilution of peptides.
| Row | [Peptide] (nM) | Probing buffer (μl) | Transfer volume from preceding row (μl) |
|---|---|---|---|
| A | 5,000 | Stock | |
| B | 3,000 | 381 | 572 |
| C | 2,000 | 266 | 533 |
| D | 1,000 | 380 | 380 |
| E | 500 | 340 | 340 |
| F | 200 | 391 | 260 |
| G | 100 | 231 | 231 |
| H | 10 | 378 | 42 |
Alignment of spots ● TIMING 1 d for 4 arrays
-
64|
Import image files into a software package that enables the facile identification and quantification of microarray spots. We use ArrayPro, which was packaged with the Tecan LS400 Laser Scanner.
-
65|
Use the 633 nm (Cy5) scan to define the locations of all spots on the microarrays and quantify all intensities. We typically use a circle of constant diameter (250 μm). Most software packages feature automated spot finding, but we strongly recommend inspecting and adjusting these automated alignments manually.
-
66|
Steps 66–69 are specific to the ArrayPro software. Save the location of each of these circles in a file independent of the 633 nm image. In ArrayPro, this file is called the `grid' file.
-
67|
If the intensity of any spot for a given peptide probe is saturated, use a scan at a lower PMT setting for that peptide. We generally scan each plate at three PMT settings when acquiring the 543 nm image: 255, 200 and 150.
-
68|
Import the grids that were determined for the 633 nm image to the 543 nm image. Occasionally, a minor positional offset will be encountered. If detected by eye (it is generally obvious), move all of the grids together to align with the spots.
▲ CRITICAL STEP Do not move individual circles in the grid file at this stage.
-
69|
Quantify all spots in the 543 nm image using the circles in the grid file.
-
70|
If using ArrayPro, export the net intensities and maximum densities for all of the spots. In ArrayPro, maximum densities are used to determine if any of the spots has saturated pixels. If using a different software package, ensure that there is a way to identify if a spot contains saturated pixels.
Analyzing intensities ● TIMING 10 min using a Matlab script
-
71|
Compute the ratio of the 543 nm fluorescence intensity to the 633 nm fluorescence intensity for each spot. We will refer to this as the `signal'. Calculate the mean signal of replicate spots in each well.
-
72|
To remove outliers among replicate spots, we exclude the signal furthest from the mean if its value is < 50% of the mean or > 150% of the mean. Recalculate the mean signal using the remaining measurements and repeat this process to exclude a second point if it also deviates too far from the mean. This procedure should be performed for every domain on every array and for every scan (every PMT setting).
-
73|
For every protein–peptide pair, compare the mean signal obtained at the highest peptide concentration (typically 5 μM) with the mean signal of the control spots (thioredoxin). If the mean signal of the domain spots is less than twice that of thioredoxin, do not count this as an interaction. For high-affinity interactions (KD < 2 μM), follow Steps 74–80. These steps detail the procedure that we currently use to derive equilibrium dissociation constants from protein microarray data. The procedure assumes that the arrays have been probed with eight concentrations of each peptide, ranging from 5 μM down to 10 nM. For low-affinity interactions (KD < 50 μM), follow Steps 81–86. These steps provide a broad overview on how we currently use protein microarrays to highlight putative interactions, which we then retest and quantify by FP (Steps 87–96). These steps are based on methods we established to study PDZ domains37.
For high-affinity interactions ● TIMING 10 min using a Matlab script
-
74|
Perform the following analysis for each domain–peptide pair that passes the criterion of Step 73. For each domain–peptide pair, there are data on eight arrays (eight peptide concentrations). For each array, choose the scan with the highest PMT setting that does not include any saturated pixels for that domain.
-
75|
Subtract the mean signal of thioredoxin control spots from the mean signal of domain spots on each array. You should now have eight values, one for each peptide concentration.
-
76|
Normalize these eight values to a single PMT voltage setting (the highest setting) as follows. First, plot the intensity of all of the spots on a microarray plate at a given PMT voltage setting (x axis) against the intensity of all of the spots at the highest PMT voltage setting (y axis). Exclude all spots that contain saturated pixels. The points on this plot should lie on a straight line. Use the slope of this line as the conversion factor. For example, to convert signals obtained at a PMT voltage of 200 to their equivalent values at a PMT voltage of 255, multiply their values by the slope of the corresponding line.
-
77|Fit the resulting data, Fobs, to equation (1) using Matlab's nonlinear fit function:
where Fmax is the maximum fluorescence at saturation, [peptide] is the total peptide concentration, and KD is the equilibrium dissociation constant.(1) -
78|
Record the domain–peptide pair as an interaction if KD < 2 μM and R2 > 0.9.
-
79|
If the criteria of Step 78 are not met, delete each of the eight data points one at a time and repeat the fit. If the criteria of Step 78 are met for one of these deletions, record the interaction. If more that one deletion causes the criteria to be met, use the KD associated with the lower R2 value.
-
80|
If the criteria of Step 78 are still not met, delete the point that results in the highest R2 value and repeat Step 78 (i.e., remove a second point). If after the removal of a second point the criteria are still not met, do not record the domain-peptide pair as an interaction. This is the last step of the procedure for studying high-affinity interactions.
▲ CRITICAL STEP As a precaution, all saturation binding curves and fits should be inspected by eye.
For low-affinity interactions ● TIMING 4 h for a set of 64 interactions
-
81|
Randomly select a subset of domains and peptides (e.g., ~20 of each). Quantify all possible domain–peptide interactions by FP (see Steps 87–96).
-
82|
Generate several `gold standard' data sets by applying different affinity thresholds to the quantitative FP data. For example, to generate a gold standard data set that includes all interactions with KD ≤10 μM, label domain–peptide interactions with KD ≤10 μM as `true positives' and noninteractions or interactions with KD > 10 μM as `true negatives'.
-
83|
For each peptide concentration used in the microarray studies (Steps 51–73), divide the mean signal for each domain by the mean signal for thioredoxin. This is the `fold over background' or FOB value. Apply different FOB thresholds to the microarray data, focusing on the domains and peptides used to generate the gold standard data sets. Label as `array positives' any domain–peptide pairs that exceed the FOB threshold; label all other pairs as `array negatives'.
-
84|
On the basis of these assignments, we calculate the false-positive and false-negative rates of the microarrays at each FOB threshold relative to each gold standard data set. At a given FOB threshold, false positives are domain–peptide interactions that exceed the FOB threshold but are not present in the gold standard data set; false negatives are domain–peptide interactions that do not exceed the threshold but are nevertheless found in the gold standard data set. Plot the false-positive and false-negative rates of the microarrays relative to each gold standard data set as a function of FOB threshold.
-
85|
Using these plots, we choose an appropriate FOB threshold to apply to the full microarray data set. The choice of threshold depends on the goals of the experiment. If the goal is to miss as few true interactions as possible, a low FOB threshold should be chosen as this minimizes the false-negative rate. This comes, however, at the expense of incurring a high false-positive rate. A high false-positive rate requires retesting a lot of array positives to eliminate false positives. If, however, it is desirable to minimize the amount of retesting, a high FOB threshold should be chosen. This minimizes the false-positive rate, but causes many true interactions to be missed (incurs a high false-negative rate).
? TROUBLESHOOTING
-
86|
Retest and quantify all array positives by FP (see below). The result is a quantitative interaction data set that has been corrected for all false positives. Correcting false negatives is a much harder problem and requires the construction of a predictive model (Fig. 2c). Standardized procedures on how to do this are beyond the scope of these protocols, but the reader is referred to our reports on predictive modeling of PDZ domain–peptide interactions36,38.
Retesting and quantifying domain–peptide interactions by fluoresce polarization ● TIMING 4 h for a set of 64 interactions
-
87|
Preparation of peptide stocks for FP (Steps 87–90; 2 h for a set of 10). Dissolve 5(6)-TAMRA-labeled peptides in ddH2O at a concentration of at least 5 μM. Centrifuge peptide solutions at 20,800g for 5 min at room temperature to remove any insoluble material.
-
88|
Determine the concentration of the solution by measuring absorbance at 548 nm. The extinction coefficient of 5(6)-TAMRA is 85,500 M−1 cm−1 in ddH2O.
? TROUBLESHOOTING
-
89|
We have found that the best signal-to-noise ratio is achieved when the 5(6)-TAMRA-labeled peptide is used at a final concentration of 20 nM in the FP assay. With this as the target concentration, prepare a 100 nM peptide stock solution by mixing 1 ml of the 5 μM peptide solution with 5 ml of 10× PBS, 500 μl of 2% (wt/vol) NaN3, 5 ml of 1% (wt/vol) BSA and 38.5 ml ddH2O.
-
90|
100 nM peptide stocks should be stored at 4 °C. Any remaining solution of 5 μM peptide should be divided into 1 ml aliquots, lyophilized and stored at −20 °C.
∎ PAUSE POINT Dissolved peptide stocks can be stored at 4 °C for up to 2 months. Lyophilized peptide aliquots can be stored at −20 °C for several years.
-
91|
Fluorescent polarization assays (Steps 91–98; 4 h for a set of 10): thaw aliquots of the purified domains (Step 27) on ice and centrifuge at 20,800g for 5 min, 4 °C to remove an insoluble material. Use a 2 M stock solution of DTT to bring the DTT concentration to 1.25 mM. Determine the concentration of the protein domains by absorbance spectroscopy (Box 4). Do not rely on previous determinations, as material is often lost on freezing and thawing. Dilute the domains with PBS containing 1.25 mM DTT to a concentration 25% higher than the highest concentration to be assayed.
▴ CRITICAL STEP Titrations are most accurate when multiple concentration points are available both above and below the KD. Additional experiments using different concentrations of protein domains may be required to meet this criterion.
-
92|
Perform twofold serial dilutions of the proteins domains in a deep-well 96-well microtiter plate. Use FP Assay Buffer as the diluent. The volume will depend on the number of different peptides under investigation. Each well of the final assay will require 40 μl protein solution. Generating 10% excess volume is recommended.
▴ CRITICAL STEP Errors can easily be propagated during serial dilutions and care should be taken during each mixing step.
-
93|
Using a 12-channel pipette, you can transfer 40 μl of protein solution from each well of the deep-well 96-well microtiter plate to a black 384-well microtiter plate. Note: protein solution is added to every other well of the 384-well plate due to the spacing of the multichannel pipette. Keep this in mind when analyzing the final data.
-
94|
Use a multichannel pipette to transfer 10 μl of the 100 nM peptide stock from a disposable reagent reservoir to each well of the assay plate and mix thoroughly by pipetting up and down several times. Control wells should also be included that contain only FP assay buffer (i.e., no protein domain).
-
95|
Incubate plates at room temperature for 1 h. Centrifuge at 1,000g for 5 min at room temperature.
-
96|
Measure FP using a suitable fluorescence plate reader. Use a 561 nm filter set for 5(6)-TAMRA-labeled probes and obtain 10 readings per well with an integration time of 0.1 s per reading.
-
97|Calculate KD by fitting the observed FP, FPobs, to equation (2) using Matlab's nonlinear fit function:
where FPmax is the maximum polarization at saturation, FPo is the polarization in the absence of protein domain, [domain] is the total concentration of protein domain and [peptide] is the total concentration of peptide (20 nM).(2) -
98|
We generally require R2 to be greater than 0.9 to ensure data quality. Each binding curve and its associated fit, however, should be inspected by eye.
● TIMING
Steps 1–27, Cloning, expression and purification of domains: 4 d for 30 domains
Steps 28–32, Synthesis and purification of labeled peptides: 1 month for 100 peptides
Steps 33–50, Fabricating protein domain microarrays: 2 d preparation, then 12 h per four plates of arrays
Steps 51–63, Probing protein domain microarrays: 3 h
Steps 64–70, Alignment of spots: 1 d for 4 arrays
Steps 71–73, Analyzing intensities: 10 min using a Matlab script
Steps 74–80, For high-affinity interactions: 10 min using a Matlab script
Steps 81–86, For low-affinity interactions: 4 h for a set of 64 interactions
Steps 87–98, Interactions by fluoresce polarization: 4 h for a set of 64 interactions
? TROUBLESHOOTING
Troubleshooting advice can be found in table 3.
TABLE 3.
Troubleshooting table.
| Step | Problem | Possible cause | Solution |
|---|---|---|---|
| 22, 26 | Protein domain cannot be expressed and purified | Domain does not fold properly when abstracted from its full-length parent protein; domain does not fold correctly in E. coli (is expressed as an inclusion body) | Clone domain with longer flanking regions; express domain in yeast or insect cells. The structural biology literature is a useful source for alternative strategies to abstract domains from their parent proteins and to express and purify different families of protein domains |
| Box 1 | Peptide cannot be synthesized | Steric interference from protected side chains can reduce coupling efficiency | Double or triple coupling may help to promote the reaction. Alternatively, using PEG-linked resins may help |
| 45 | Spots on arrays are not uniform | Contact printing is highly dependent on glycerol concentration, depends on time of the pins on the glass substrate, humidity and buffer composition | Optimize each of these parameters for your arrayer and protein domains. We have found that the most important parameters to optimize are humidity and glycerol concentration |
| 55, 88 | Peptides are not soluble enough to quantify by absorbance spectroscopy | Peptides are insoluble in water | Dissolve peptides in methanol and measure absorbance at 540 nm. Use a molar extinction coefficient of 95,000 M−1 cm−1. Remove methanol by evaporation. Dissolve in DMSO, then dilute in probing buffer. The final concentration of DMSO should not exceed 2% (vol/vol) |
| 55 | Peptide is not soluble in probing buffer | Problem may be either kinetic or thermodynamic in nature | Dissolve the peptide in DMSO, then dilute in probing buffer as above |
| 62, 85 | High fluorescence background on the arrays | Peptide is binding nonspecifically to the glass substrate | Shorten peptide or add hydrophilic residues to the peptide. Eliminate peptide from data set |
| Bright speckles appear all over the arrays | Peptide is precipitating | Dissolve the peptide in DMSO, then dilute in probing buffer as above. Shorten peptide or add hydrophilic residues |
ANTICIPATED RESULTS
Domain-based protein microarrays provide a powerful way to obtain quantitative information on protein–protein interactions in high throughput. By focusing on families of peptide-binding domains, we have been able to optimize array-based assays so that the resulting data sets are unbiased, reasonably large and are focused on physiologically relevant sequences. Although this approach is most efficient when used to study domains that mediate high-affinity interactions (e.g., SH2 and PTB domains), we have also used protein domain microarrays to obtain quantitative interaction data sets for weaker-binding protein domains (e.g., PDZ domains). In Figure 3, we provide examples of the type of data obtained using protein domain microarrays. Equilibrium binding constants can be calculated directly from the array data when studying high-affinity interactions (Fig. 3a–c). Alternative, quantitative data can be obtained in two steps when studying weaker interactions (Fig. 3d,e). Overall, we have used our SH2/PTB domain microarrays to gain insight into receptor tyrosine kinases signaling31–35 and we have used our PDZ domain microarrays to gain insight into how binding selectivity is distributed across the mammalian proteome36 as well as to construct predictive models of PDZ domain-mediated peptide recognition36,38.
BOX 1 | PEPTIDE SYNTHESIS ● TIMING 3 D.
Fmoc deprotection protocol
-
1.
Add 1.4 ml of DMF to the resin, agitate for 3 min and drain. Repeat twice.
-
2.
Add 1.4 ml of 25% (vol/vol) piperidine in DMF, agitate for 7 min and drain. Repeat once.
-
3.
Add 1.4 ml of DMF to the resin, agitate for 3 min and drain. Repeat twice.
Coupling protocol
-
4.
Add 800 μl of amino-acid solution (5 equivalents), 760 μl of HBTU coupling agent (4.75 equivalents) and 100 μl of DIPEA, and agitate for 1 h. Repeat once.
5-(and-6)-Carboxytetramethylrhodamine (5(6)-TAMRA) coupling
-
5.
In a glass scintillation vial, add two equivalents of 5(6)-TAMRA, 1.9 equivalents of HATU (more expensive than HBTU but provides better yields) and 100 μl of DIPEA. Dissolve in 900 μl of DMF and mix well.
-
6.
Add the solution to the N-terminally deprotected resin and agitate overnight or up to 24 h. Because 5(6)-TAMRA does not contain chiral centers, longer incubation times are permitted.
-
7.
Add 1.4 ml of DMF to the resin, agitate for 3 min and drain. Repeat twice.
Trifluoroacetic acid (TFA) cleavage reaction
-
8.
Prepare a cleavage mixture of 94% TFA (vol/vol), 2% ddH2O (vol/vol), 2% phenol (wt/vol) and 2% triisopropylsilane (vol/vol) at a volume of 1 ml per reaction. If methionine is present in the peptide, include 2% each of the reducing agents thioanisole and ethan-edithiol to minimize oxidation.
-
9.
Wash resin with 1.4 ml of methanol three times, followed by three washes with 1.4 ml of diethyl ether to remove residual DMF and to facilitate drying.
-
10.
Add 1 ml of cleavage solution to the resin and incubate at room temperature for 1 h.
-
11.
Add 50 ml of diethyl ether to a Falcon tube and set it below a disposable fritted column. Transfer the cleavage mixture into the fritted column and drain into the ether. The peptides should visibly precipitate in the ether. An additional 0.5 ml of TFA can be used to rinse the resin to recover more peptide.
-
12.
Chill the ether in an ice bath for 20 min. Once cooled, invert Falcon tube several times and decant the precipitated peptide slurry into a fresh fritted column. Dispose of the ether and allow the precipitated peptide to dry in a fume hood (residual ether evaporates).
? TROUBLESHOOTING
BOX 2 | HOW TO ASSEMBLE ARRAYS ● TIMING 5 M.
Using a Storage Mat Applicator, we attach a silicone gasket to the bottom of a bottomless 96-well microtiter plate, being careful to attach the correct side of the gasket to the 96-well plate, as indicated by the manufacturer (Fig. 4). This can be performed on a bench-top and does not require a clean room. The use of powder-free gloves, however, is critical as fingerprints can produce artifacts on the arrays.
Detach protective covering from the back of the gasket.
With microtiter plate bottom-side up (i.e., gasket-side up), carefully place the glass plate with the printed proteins over the gasket, ensuring that the arrays are centered in each well. The printed proteins should be on the bottom side of the glass and hence accessible inside the wells of the microtiter plate. Note that once the glass plate makes contact with the gasket, it cannot be removed or adjusted. Be careful to note the orientation of the proteins for later data processing.
Using a Storage Mat Applicator, we press the glass plate against the microtiter plate to form a tight seal. There should be no visible air bubbles at any of the points of contact.
Apply a plastic seal over the top of the microtiter plate to protect the arrays.
BOX 3 | LABELING BSA WITH CY5 ● TIMING 2 H.
Prepare a 1 mg ml−1 solution of BSA in sodium carbonate buffer (see REAGENT SETUP).
Add 1 ml of BSA solution to one Cy5 mono-Reactive Dye Pack.
Incubate at room temperature, in the dark, for 30 min.
Equilibrate four NAP10 columns with PBS (see REAGENT SETUP).
Mix 1 ml of BSA/Cy5 solution with 3 ml of PBS.
Add 1 ml of diluted BSA/Cy5 solution to each NAP-10 column.
Elute Cy5-BSA from each column with 1.5 ml of PBS.
Combine all samples of Cy5-BSA and concentrate using Ultra-4 concentrator to ~160 μl.
BOX 4 | DETERMINING PROTEIN CONCENTRATION ● TIMING 10 M.
Mix 20 μl of protein solution with 80 μl of 6 M guanidinium chloride solution. This step is necessary to ensure that the protein is denatured and hence its chromophoric residues are uniformly exposed to solvent.
Calculate the molar extinction coefficient for each domain of interest. Count 5,690 M−1 cm−1 for each Trp, 1,280 M−1 cm−1 for each Tyr and 120 M−1 cm−1 for each Cys43.
Record absorbance at 280 nm and subtract absorbance at 600 nm (baseline correction).
ACKNOWLEDGMENTS
We thank J.-R. Chen, G. Koytiger and M. Sevecka for helpful contributions to these protocols, particularly in regards to data processing and analysis. This work was supported by awards from the W.M. Keck Foundation, the Arnold and Mabel Beckman Foundation, and the Camille and Henry Dreyfus Foundation, and by grants from the National Institutes of Health (1 RO1 GM072872 and 1 R33 CA128726). A.K. was supported in part by the National Institutes of Health Molecular, Cellular and Chemical Biology Training grant (5 T32 GM07598) and A.G. was the recipient of an NSF Graduate Research Fellowship.
Footnotes
COMPETING FINANCIAL INTERESTS The authors declare competing financial interests (see the HTML version of this article for details).
Reprints and permissions information is available online at http://npg.nature.com/reprintsandpermissions/.
References
- 1.Fields S, Song O. A novel genetic system to detect protein-protein interactions. Nature. 1989;340:245–246. doi: 10.1038/340245a0. [DOI] [PubMed] [Google Scholar]
- 2.Chien CT, Bartel PL, Sternglanz R, Fields S. The 2-hybrid system—a method to identify and clone genes for proteins that interact with a protein of interest. Proc. Natl. Acad. Sci. USA. 1991;88:9578–9582. doi: 10.1073/pnas.88.21.9578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Johnsson N, Varshavsky A. Split ubiquitin as a sensor of protein interactions in vivo. Proc. Natl. Acad. Sci. USA. 1994;91:10340–10344. doi: 10.1073/pnas.91.22.10340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gavin AC, et al. Functional organization of the yeast proteome by systematic analysis of protein complexes. Nature. 2002;415:141–147. doi: 10.1038/415141a. [DOI] [PubMed] [Google Scholar]
- 5.Ho Y, et al. Systematic identification of protein complexes in Saccharomyces cerevisiae by mass spectrometry. Nature. 2002;415:180–183. doi: 10.1038/415180a. [DOI] [PubMed] [Google Scholar]
- 6.Smith GP. Filamentous fusion phage—novel expression vectors that display cloned antigens on the virion surface. Science. 1985;228:1315–1317. doi: 10.1126/science.4001944. [DOI] [PubMed] [Google Scholar]
- 7.Songyang Z, et al. SH2 domains recognize specific phosphopeptide sequences. Cell. 1993;72:767–778. doi: 10.1016/0092-8674(93)90404-e. [DOI] [PubMed] [Google Scholar]
- 8.MacBeath G, Schreiber SL. Printing proteins as microarrays for high-throughput function determination. Science. 2000;289:1760–1763. doi: 10.1126/science.289.5485.1760. [DOI] [PubMed] [Google Scholar]
- 9.Ito T, et al. A comprehensive two-hybrid analysis to explore the yeast protein interactome. Proc. Natl. Acad. Sci. USA. 2001;98:4569–4574. doi: 10.1073/pnas.061034498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Uetz P, et al. A comprehensive analysis of protein-protein interactions in Saccharomyces cerevisiae. Nature. 2000;403:623–627. doi: 10.1038/35001009. [DOI] [PubMed] [Google Scholar]
- 11.Giot L, et al. A protein interaction map of Drosophila melanogaster. Science. 2003;302:1727–1736. doi: 10.1126/science.1090289. [DOI] [PubMed] [Google Scholar]
- 12.Li S, et al. A map of the interactome network of the metazoan C. elegans. Science. 2004;303:540–543. doi: 10.1126/science.1091403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rual JF, et al. Towards a proteome-scale map of the human protein–protein interaction network. Nature. 2005;437:1173–1178. doi: 10.1038/nature04209. [DOI] [PubMed] [Google Scholar]
- 14.Stelzl U, et al. A human protein–protein interaction network: a resource for annotating the proteome. Cell. 2005;122:957–968. doi: 10.1016/j.cell.2005.08.029. [DOI] [PubMed] [Google Scholar]
- 15.Tarassov K, et al. An in vivo map of the yeast protein interactome. Science. 2008;320:1465–1470. doi: 10.1126/science.1153878. [DOI] [PubMed] [Google Scholar]
- 16.Gavin AC, et al. Proteome survey reveals modularity of the yeast cell machinery. Nature. 2006;440:631–636. doi: 10.1038/nature04532. [DOI] [PubMed] [Google Scholar]
- 17.Fuh G, et al. Analysis of PDZ domain–ligand interactions using carboxyl-terminal phage display. J. Biol. Chem. 2000;275:21486–21491. doi: 10.1074/jbc.275.28.21486. [DOI] [PubMed] [Google Scholar]
- 18.Tonikian R, Zhang Y, Boone C, Sidhu SS. Identifying specificity profiles for peptide recognition modules from phage-displayed peptide libraries. Nat. Protoc. 2007;2:1368–1386. doi: 10.1038/nprot.2007.151. [DOI] [PubMed] [Google Scholar]
- 19.Tonikian R, et al. A specificity map for the PDZ domain family. PLoS Biol. 2008;6:e239. doi: 10.1371/journal.pbio.0060239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang Y, et al. Convergent and divergent ligand specificity among PDZ domains of the LAP and zonula occludens (ZO) families. J. Biol. Chem. 2006;281:22299–22311. doi: 10.1074/jbc.M602902200. [DOI] [PubMed] [Google Scholar]
- 21.Songyang Z, et al. Recognition of unique carboxyl-terminal motifs by distinct PDZ domains. Science. 1997;275:73–77. doi: 10.1126/science.275.5296.73. [DOI] [PubMed] [Google Scholar]
- 22.Aloy P, Russell RB. Potential artefacts in protein-interaction networks. FEBS Lett. 2002;530:253–254. doi: 10.1016/s0014-5793(02)03427-0. [DOI] [PubMed] [Google Scholar]
- 23.Bader JS, Chaudhuri A, Rothberg JM, Chant J. Gaining confidence in high-throughput protein interaction networks. Nat. Biotechnol. 2004;22:78–85. doi: 10.1038/nbt924. [DOI] [PubMed] [Google Scholar]
- 24.Deane CM, Salwinski L, Xenarios I, Eisenberg D. Protein interactions: two methods for assessment of the reliability of high throughput observations. Mol. Cell. Proteomics. 2002;1:349–356. doi: 10.1074/mcp.m100037-mcp200. [DOI] [PubMed] [Google Scholar]
- 25.Phizicky E, Bastiaens PI, Zhu H, Snyder M, Fields S. Protein analysis on a proteomic scale. Nature. 2003;422:208–215. doi: 10.1038/nature01512. [DOI] [PubMed] [Google Scholar]
- 26.Popescu SC, et al. MAPK target networks in Arabidopsis thaliana revealed using functional protein microarrays. Genes Dev. 2009;23:80–92. doi: 10.1101/gad.1740009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ptacek J, et al. Global analysis of protein phosphorylation in yeast. Nature. 2005;438:679–684. doi: 10.1038/nature04187. [DOI] [PubMed] [Google Scholar]
- 28.Zhu H, et al. Global analysis of protein activities using proteome chips. Science. 2001;293:2101–2105. doi: 10.1126/science.1062191. [DOI] [PubMed] [Google Scholar]
- 29.He M, Taussig MJ. Single step generation of protein arrays from DNA by cell-free expression and in situ immobilisation (PISA method) Nucleic. Acids Res. 2001;29:e73. doi: 10.1093/nar/29.15.e73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ramachandran N, et al. Self-assembling protein microarrays. Science. 2004;305:86–90. doi: 10.1126/science.1097639. [DOI] [PubMed] [Google Scholar]
- 31.Jones RB, Gordus A, Krall JA, MacBeath G. A quantitative protein interaction network for the ErbB receptors using protein microarrays. Nature. 2006;439:168–174. doi: 10.1038/nature04177. [DOI] [PubMed] [Google Scholar]
- 32.Gordus A, MacBeath G. Circumventing the problems caused by protein diversity in microarrays: implications for protein interaction networks. J. Am. Chem. Soc. 2006;128:13668–13669. doi: 10.1021/ja065381g. [DOI] [PubMed] [Google Scholar]
- 33.Kaushansky A, et al. System-wide investigation of ErbB4 reveals 19 sites of Tyr phosphorylation that are unusually selective in their recruitment properties. Chem. Biol. 2008;15:808–817. doi: 10.1016/j.chembiol.2008.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kaushansky A, Gordus A, Chang B, Rush J, MacBeath G. A quantitative study of the recruitment potential of all intracellular tyrosine residues on EGFR, FGFR1 and IGF1R. Mol. Biosyst. 2008;4:643–653. doi: 10.1039/b801018h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gordus A, et al. Linear combinations of docking affinities explain quantitative differences in RTK signaling. Mol. Syst. Biol. 2009;5:235. doi: 10.1038/msb.2008.72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Stiffler MA, et al. PDZ domain binding selectivity is optimized across the mouse proteome. Science. 2007;317:364–369. doi: 10.1126/science.1144592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Stiffler MA, Grantcharova VP, Sevecka M, MacBeath G. Uncovering quantitative protein interaction networks for mouse PDZ domains using protein microarrays. J. Am. Chem. Soc. 2006;128:5913–5922. doi: 10.1021/ja060943h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chen JR, Chang BH, Allen JE, Stiffler MA, MacBeath G. Predicting PDZ domain–peptide interactions from primary sequences. Nat. Biotechnol. 2008;26:1041–1045. doi: 10.1038/nbt.1489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pawson T, Nash P. Assembly of cell regulatory systems through protein interaction domains. Science. 2003;300:445–452. doi: 10.1126/science.1083653. [DOI] [PubMed] [Google Scholar]
- 40.Yan KS, Kuti M, Zhou MM. PTB or not PTB—that is the question. FEBS Lett. 2002;513:67–70. doi: 10.1016/s0014-5793(01)03305-1. [DOI] [PubMed] [Google Scholar]
- 41.Boutell JM, Hart DJ, Godber BL, Kozlowski RZ, Blackburn JM. Functional protein microarrays for parallel characterisation of p53 mutants. Proteomics. 2004;4:1950–1958. doi: 10.1002/pmic.200300722. [DOI] [PubMed] [Google Scholar]
- 42.von Mering C, et al. Comparative assessment of large-scale data sets of protein–protein interactions. Nature. 2002;417:399–403. doi: 10.1038/nature750. [DOI] [PubMed] [Google Scholar]
- 43.Edelhock H. Spectroscopic determination of tryptophan and tyrosine in proteins. Biochemistry. 1967;6:1948–1954. doi: 10.1021/bi00859a010. [DOI] [PubMed] [Google Scholar]
- 44.Barbulovic-Nad I, et al. Bio-microarray fabrication techniques—a review. Crit. Rev. Biotechnol. 2006;26:237–259. doi: 10.1080/07388550600978358. [DOI] [PubMed] [Google Scholar]
- 45.George RA. The printing process: tips on tips. Methods Enzymol. 2006;410:121–135. doi: 10.1016/S0076-6879(06)10006-3. [DOI] [PubMed] [Google Scholar]
- 46.Schultz J, Milpetz F, Bork P, Ponting CP. SMART, a simple modular architecture research tool: identification of signaling domains. Proc. Natl. Acad. Sci. USA. 1998;95:5857–5864. doi: 10.1073/pnas.95.11.5857. [DOI] [PMC free article] [PubMed] [Google Scholar]