Abstract
Leprosy continues to be endemic in parts of China. To track the occurrence of leprosy and determine at risk communities, molecular strain typing based on variable number of tandem repeats (VNTRs) was applied in Qiubei County, Wenshan Prefecture, Yunnan Province of the People’s Republic of China, a multiethnic region that is home to four predominant ethnic minorities. A previous study, conducted between 2002 and 2005, provided the first descriptions of Mycobacterium leprae strains in the region. M. leprae strains in Qiubei are highly conserved, so only sufficiently polymorphic loci can distinguish strains. A balance between mutation rate and loci stability is needed, so that secondary transmissions can be identified as genotypic matches. The long incubation period of leprosy necessitated an extension of the study to assess the validity of VNTR typing and observe allelic shifts in the same multiethnic population. From 2006 to early 2010 the extension was performed to yield a cumulative total of 164 enrolled patients and 130 skin samples suitable for VNTR typing. Patient demographic information revealed that the case detection rate among certain minority populations in the county is considerably higher than the national rate. Cluster analysis of allele frequencies showed similar strain types within family groups and neighboring townships. Allele frequencies were not found to significantly differ between genders or clinical presentations. The percentage of cases showing near-matching genotypes varied with geography; showing a considerably higher rate in the northern townships. The northern townships continue to show strain types falling into the groups previously defined. Southern genotypes were distinct from those in the north, but clonal genetic relationships were indiscernible in the south. Social interactions and the physical, residential and occupational environments may be more conducive to transmission of community strains in the north.
Keywords: Leprosy, Molecular Epidemiology, Strain typing, VNTR
1. INTRODUCTION
Leprosy control strategies have been designed to stop transmission of the pathogen Mycobacterium leprae through early case detection and antimicrobial multi-drug therapy (MDT), as recommended by the World Health Organization (WHO, 1982, 2009). In 1998, at the 15th International Leprosy Conference in Beijing, the Chinese government announced that leprosy had reached the nationwide elimination target. Despite this, approximately 1,500–1,800 new patients have been detected annually in China since 1998 - mainly in the southwestern provinces (NCLC, 2007), with socioeconomic disparities contributing to the detection rate and prevalence of leprosy in these geographically confined areas (Chen et al., 2000; Shen et al., 2005; Meima et al., 1997; Saikawa, 1981).
Qiubei County, Wenshan Prefecture, Yunnan Province, has one of the highest rates of leprosy in China. The annual detection rate of leprosy in all of China is 0.115 per 100,000 persons (NCLC, 2007), while our research reports an annual detection rate of 4.5 per 100,000 persons in Qiubei County. Leprosy has long been endemic in Qiubei, where population surveys have demonstrated seroprevalence of M. leprae antigens as high as 30% in the general population (Weng et al., 2000, 2005); making Qiubei an excellent site for molecular studies of M. leprae. A population-based investigation of leprosy complemented with molecular strain typing of the pathogen M. leprae is a powerful technique for tracking the transmission of leprosy. In 2002 we undertook such a program and mapped variable number-tandem repeat (VNTR) genomic loci in skin samples of leprosy patients in Qiubei County.
Molecular techniques have been used extensively to study the transmission of human tuberculosis –another disease primarily attributed to person-to-person transmission of a slow growing Mycobacterium. Several families of genetic markers have been tested in M. tuberculosis clinical isolates. These markers have included: insertion sequences (IS6110) (van Embden et al., 1993), a single direct repeat (DR) region interspersed with unique spacers (Goguet de la Salmonière et al., 1997), multiple interspersed repeat units (MIRUs) scattered across the genome (Allix-Béguec et al., 2008), single nucleotide polymorphisms (SNPs) (Filliol e al., 2006), and insertion-deletions (Gordon et al., 1999). With each marker system providing a different level of strain resolution, the combined diversity of available molecular techniques enables the study of questions ranging in scale from outbreak tracking (Tabet et al., 1994) to evolution of the M. tuberculosis complex (Brosch et al., 2002).
In clinical investigations of tuberculosis, strain typing techniques have been used to distinguish recent transmission from spatially and temporally remote sources of infection, such as reactivated or immigrant cases (van Soolingen et al., 1999). Strain typing methods, and their interpretations, continue to evolve, but such techniques have nevertheless played an important public health role by guiding contact tracing efforts (de Vries et al., 2009), and illustrating associations between genotype and phenotype (Parwati et al., 2010). The success of molecular techniques in the study of tuberculosis transmission serves as an illustration of how leprosy research can benefit from strain typing methods. Strain typing can shed light on how M. leprae circulates through an affected population, giving clues as to the best way in which to interrupt the stable pattern of incidence that has burdened some regions (Weng et al., 2007). However, there are clinical and practical differences between tuberculosis and leprosy. Active case searching and contact tracing have become limited due to the decentralization of leprosy care and the redirection of funds to other health priorities (Feasey et al., 2010). Contact tracing investigations of leprosy are impeded by the long incubation period of leprosy (estimated at five to seven years) (WHO, 2006), the wide range of clinical manifestations (Ridley and Jopling, 1966), and the powerful historical social stigma associated with the disease (Nicholls et al., 2003). Isolation of the pathogen for in vitro propagation is infeasible. Though tuberculosis serves as an important analogy, these distinctions mandate an approach specifically tailored to the biological and cultural uniqueness of leprosy.
Genome sequencing of several stains (Monot et al., 2009) has shown the M. leprae genome to be virtually invariant since a historical bottleneck event that resulted in massive genome decay (Cole et al., 2001). The identification of four major lineages of M. leprae, SNP types 1 through 4, has provided a robust illustration of the global dissemination of leprosy (Monot et al., 2005). To provide a finer scale of strain typing, 16 additional SNP subtypes were developed (Monot et al., 2009). The SNP typing system has provided a valuable framework at the global level, but is often insufficient to distinguish strains at the regional and community level (Monot et al., 2008; Sakamuri et al., 2009a, 2009b; Weng et al. 2007). For such situations, more rapidly evolving genetic markers are required. To this end, VNTR loci at microsatellite and minisatellite regions of the M. leprae genome were explored (Groathouse et al., 2004; Matsuoka et al., 2004; Sakamuri et al., 2009a).
Concerns have been raised regarding VNTR typing, including the possibility of multiple infections within a single patient (Monot et al., 2008; Young et al., 2008), and variations in the rate of M. leprae replication across anatomical niches (Young et al., 2004; Young et al., 2008). In order to objectively assess the suitability of VNTRs for disease tracking, empirical data must be drawn from endemic situations; ideally, with consecutive case ascertainment over time periods at least the length of the average leprosy incubation period. To date, only two multiyear molecular epidemiology investigations have been reported; our initial study in Qiubei (Weng et al., 2007; Xing et al., 2009) and another in Cebu, Philippines (Sakamuri et al., 2009a, 2009b). These investigations demonstrate that VNTR typing methods are able to distinguish M. leprae subgroups within unresolved SNP lineages (type 3 in Qiubei and type 1 in Cebu) and illuminate recent transmission through the shared VNTR profiles found in multicase families (MCFs) and community infections.
Before our first Qiubei study, the methods for strain typing of clinical leprosy samples were just beginning to emerge. Our initial molecular investigation ran for over three years, and provided the first descriptions of M. leprae VNTR genotypes in the region. We identified several clusters of patients whose M. leprae specimens shared similar VNTR profiles and uncovered characteristic features of leprosy transmission in Qiubei (Weng et al., 2007). In collaboration with Colorado State University, a program in the molecular epidemiology of leprosy has been established at the Beijing Tropical Medicine Research Institute (BTMRI). Field support has been provided by the Qiubei County Leprosy Control Station, which is also responsible for maintaining patient medical records and distributing MDT provided by the National Centre for Leprosy Control. Although Qiubei is nearly 3,000 km from Beijing, researchers from BTMRI periodically visit the field site to meet with clinicians and staff, retrieve skin biopsy samples, and collect relevant patient data. This research program represents a best effort at complete case inclusion, so as to study the transmission patterns and population genetics of M. leprae in a genuinely endemic setting. The support of these organizations has made a continuation of our previous study possible; affording us the opportunity to research a complete incubation period, and thus capture secondary infections and genotypic shifts.
We present the cumulative findings from a total of 164 enrolled cases spanning 8 years, from 2002 to 2010, with the aim of furthering understanding of the leprosy genome and aiding intervention measures by the prevailing disease control programs.
2. MATERIALS AND METHODS
2.1. Field site, study population and the source of M. leprae
Qiubei County, composed of 14 townships, is located in Wenshan Prefecture of Yunnan Province and is inhabited by a diversity of ethnic peoples (Zhang and Chen, 1999). Leprosy case ascertainment and diagnosis of leprosy occurred at the Skin Diseases Control Stations (SDCS). Clinical presentation was classified according to the Ridley-Jopling scheme (Ridley and Jopling, 1966). In addition to date of diagnosis, basic demographic information such as gender, year of birth, ethnicity, and township of residence were collected. Population data for townships and ethnicities were acquired from the National Bureau of Statistics of China (NBSC, 2010). The molecular epidemiology investigations were approved by ethical committees of SDCS, Beijing Tropical Medicine Research Institute (BTMRI), and Colorado State University (CSU). Patient skin biopsies were collected at SDCS in Qiubei after diagnosis and formal consent from patients. DNA of M. leprae was extracted from skin biopsies for strain typing as described previously (Weng et al., 2007).
2.2. M. leprae DNA extraction, VNTR amplification and DNA sequencing of PCR products
The methods for sample processing have been described previously (Weng et al., 2007). A brief summary follows. After collection and fixation of skin biopsies in ethanol, DNA extraction was performed using the DNeasy® tissue kit (QIAGEN, Valencia, CA) and subjected to VNTR amplification of the loci (AC)9, (GTA)9, (AT)17, (AC)8a, (AT)15, (TA)10 and 6–7. The primer sequences and amplicon sizes were as previously reported (Weng et al., 2007). The presence of PCR product was detected by agarose gel electrophoresis. For copy number (allele) determination, the amplicons were then either submitted for DNA sequencing or for fragment length analysis (FLA) according to published procedures (Kimura et al., 2009; Weng et al., 2007). Applied Biosystems™ software Sequence Scanner© and GeneMapper© were used to manually view and analyze the chromatograms. Representative chromatograms and gels images may be found in previously published works (Groathouse et al., 2004; Kimura et al., 2009; Sakamuri et al., 2009b).
2.3. Statistical analysis of VNTR data and genotype clustering
The patient cohort was classified according to township of residence [BDS, GH, GZ, JP, NJ, PZ, SD, SLY, SP, TX, WL, XD, YJ, YZ] (see Weng et al., 2007 for full township names), ethnicity [Zhuang, Miao, Han, Yi, Other], sex [male, female], and clinical presentation [TT, BT, BB, BL, LL]. Detection rate of cases for each subpopulation was calculated as (number of cases)•(105)/(population • years).
The allele frequencies for each of the subpopulations were calculated using the Microsatellite Toolkit 3.1.1© (Park, 2001) in Excel 2007©. Minitab 16© was used for statistical analysis and preparation of the subpopulation clustering dendrogram, principal component graphs and allele distribution histograms. Clustering of subpopulations was performed using a complete linkage method and a squared Euclidean distance measure (Michalakis and Excoffier, 1996). The relationships among township and ethnicity populations were analyzed using PCA performed upon the covariance matrix of allele frequencies within subpopulations.
The software Arlequin 3.5© (Excoffier and Lischer, 2010) was used to initially screen for identical genotypes using the loci (AC)9, 6–7, (AC)8a, (TA)10 and (GTA)9. Manual modifications to the clustering assignments were made to allow for a variation in one or two repeats for (TA)10 and (GTA)9 alleles greater than 12 repeats due to increased variability in these ranges. Percent clustering was determined by taking the number of clustered cases divided by the total cases suitable for analysis, with the 95% confidence intervals adjusted using a finite population correction factor of 0.56 (130 genotyped cases taken from 189 total cases).
3. RESULTS AND DISCUSSION
3.1. Field site, population characteristics and case detection rate
Total case ascertainment in Qiubei County was 189 cases from 2002 to 2010, but not all cases were enrolled in the study. Patients under the age of fourteen and 21 cases from 2002 to 2004 were not enrolled. Thus, the combined studies in Qiubei County yielded a total of 164 enrolled cases since 2002. As demographic data were unavailable for cases that were not enrolled, they have not been included in the analysis, and case detection rates should be interpreted as minimum bounds. The first case genotyped was reported in April 2002, and the last in March 2010, reflecting a total duration of eight years across the previous and follow-up study, with individual durations of 45 and 51 months, sequentially. Between 2006 and 2010, 95 new cases were detected in 13 of the 14 townships. YJ was the only township where no cases were reported in either study period.
Population growth for each ethnicity was linear (minimum R2=0.95) and did not exceed 1.2% per year for any ethnicity (data not shown). Administrative regions were redefined after 2005, which merged the townships of GH with SLY, and WL with YJ. Given the minimal impact of growth on case detection and the unavailability of individual data for some townships, the 2005 population data was used to calculate the detection rate for the entire study period.
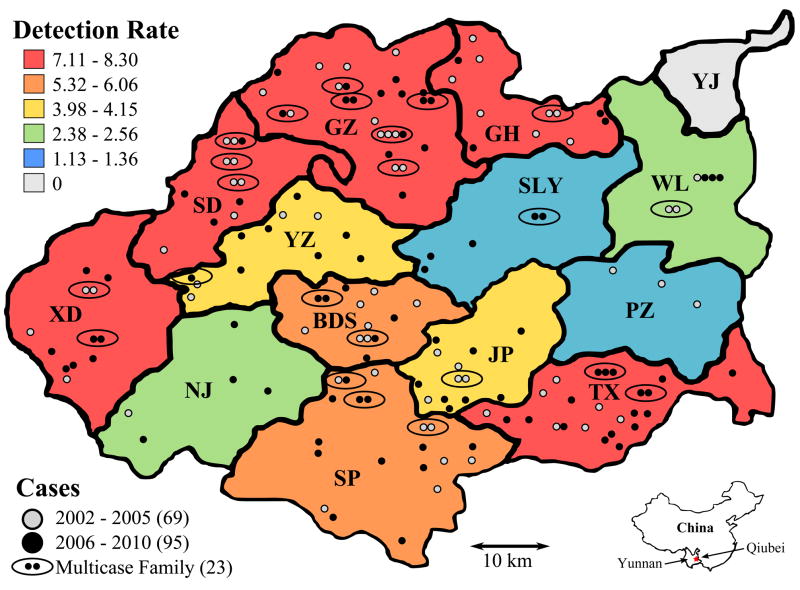
The distribution of the cases according to township of residence, ethnic group and type of leprosy is summarized in Table 1. A choropleth map showing variations in the detection rate across the county, as well as the approximate geographic location of each case, is present in Figure 1.
Table 1. Summary of the leprosy cases in Qiubei County over an 8 year period (2002–2010).
Population statistics, case detection rate, and the distribution of cases according to township, ethnicity and clinical presentation. The township of YJ does not appear in the table due to zero reported cases.
| Townshipa | Township Populationb | Number of Patients | Detection Rated | Ethnicity
|
Type of Leprosye |
||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Zhuang | Miao | Han | Yi | Other | TT | BT | BB | BL | LL | ||||
| GH | 4.1% | 11 | 7.44 | 7 | 4 | 0 | 0 | 0 | 0 | 4 | 0 | 6 | 1 |
| GZ | 10.0% | 29 | 7.98 | 24 | 3 | 1 | 1 | 0 | 3 | 8 | 1 | 16 | 1 |
| SD | 4.0% | 11 | 7.55 | 0 | 10 | 0 | 1 | 0 | 0 | 4 | 1 | 6 | 2 |
| YZ | 7.3% | 11 | 4.15 | 0 | 9 | 1 | 1 | 0 | 0 | 8 | 0 | 5 | 0 |
| BDS | 6.7% | 13 | 5.32 | 0 | 8 | 5 | 0 | 0 | 0 | 2 | 0 | 3 | 0 |
| JP | 9.0% | 13 | 3.98 | 3 | 7 | 1 | 0 | 2 | 0 | 0 | 0 | 3 | 0 |
| NJ | 5.4% | 5 | 2.56 | 0 | 2 | 0 | 3 | 0 | 0 | 3 | 1 | 7 | 0 |
| SLY | 12.1% | 6 | 1.36 | 3 | 1 | 0 | 1 | 1 | 1 | 10 | 3 | 6 | 0 |
| SP | 9.1% | 20 | 6.06 | 3 | 11 | 5 | 1 | 0 | 2 | 9 | 1 | 12 | 0 |
| TX | 9.3% | 24 | 7.11 | 16 | 6 | 2 | 0 | 0 | 0 | 0 | 0 | 4 | 2 |
| XD | 4.0% | 12 | 8.30 | 1 | 6 | 2 | 3 | 0 | 0 | 2 | 0 | 7 | 3 |
| PZ | 7.3% | 3 | 1.13 | 2 | 0 | 1 | 0 | 0 | 1 | 5 | 0 | 5 | 0 |
| WL | 6.9% | 6 | 2.38 | 1 | 5 | 0 | 0 | 0 | 0 | 1 | 0 | 5 | 0 |
|
| |||||||||||||
| Ethnic Populationc | 27.8% | 14.8% | 37.7% | 16.3% | 3.5% | ||||||||
| Total | 94.9%f | 164 | 4.50 | 60 | 72 | 18 | 11 | 3 | 7 | 56 | 7 | 85 | 9 |
| Detection Rated | 5.93 | 13.35 | 1.31 | 1.85 | 2.37 | 0.19 | 1.54 | 0.19 | 2.33 | 0.25 | |||
Two or three letter coding system has been used to represent the names of the 14 townships in Qiubei as previously defined (Weng et al., 2007).
Percentage of the total Qiubei population within a township in 2005.
Percentage of the total Qiubei population belonging to a specific ethnicity in 2005.
Case detection rate is reported as the number of cases detected per 100,000 persons per year.
Ridley-Jopling classification of leprosy based on histopathology of skin biopsy [tuberculoid, TT; borderline tuberculoid, BT; borderline borderline, BB; borderline lepromatous, BL; and lepromatous, LL] (Ridley and Jopling, 1966).
The total population within townships is less than 100% due to the fact that the YJ township and non-township populations are not reported.
Figure 1. Leprosy case detection map of Qiubei County.
A choropleth map showing the variation in case detection rate (per 100,000 persons per year) and the approximate geographic location of each case and multicase family.
Of particular note is that the detection rate among both the Zhuang and Miao peoples is significantly higher than among the Han majority within Qiubei. The Miao in particular show a 10-fold higher rate than the majority ethnicity. These trends are indicative of a health disparity in the region (Krieger et al., 2005).
3.2. Trends in leprosy transmission in Qiubei based on M. leprae VNTR typing
Multilocus VNTR typing methods were applied in this study, as local strains cannot be distinguished using known SNP markers (Monot et al., 2005; Monot et al., 2009; Weng et al., 2007). In the previous study, eight VNTR loci (AC)9, 6–7, (GTA)9, (AT)17, (AC)8a, (AT)15, (TA)18 and (GAA)21 were mapped and analyzed. For the post-2005 DNA samples, loci (TA)18 and (GAA)21 were excluded due to the lack of concordance or reproducibility in reading the number of repeat units from sequence. The locus (TA)10 was added to the current panel of markers. It was previously determined that the loci (AC)8b, (GGT)5, 12–5, 18–8, 21–3, 23–3, 27–5 and rpoT lacked sufficient polymorphism within Qiubei County for analysis, having predominate values of 8, 4, 3, 7, 2, 2, 5 and 3, respectively (Weng et al., 2006; Xing et al., 2009). Out of the 164 study cases, 130 were suitable for genotyping. The VNTR patterns of the 130 typed cases are shown in Table 2.
Table 2. Demographic, clinical and M. leprae VNTR data for a subset of 130 leprosy patients.
The table is divided into two sections. The first is comprised of cases clustered according to VNTR halotypes. The data are not arranged in chronological order of diagnosis or patient code. In the second section, the unclustered cases are sorted according to patient code. The codes for cases and multicase families through 2005 are as before (Weng et al., 2007). All cases identified after 2005 are shown in bold font. An * indicate cases studied previously (Xing et al., 2009). The Group column refers to the groupings assigned previously (Weng et al., 2007), with those entries in parenthesis being the closest shared parent branch for those that were not assigned into one of the five groupings [A, B, C, D, E]. Missing or indeterminate data are identified by a ‘?’.
| Code | Multicase Family | Ethnicity | Gender | Township | Date Diagnosed | Leprosy Type | Alleles
|
Group | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| AC9 | 6—7 | AC8a | TA10 | GTA9 | AT17 | AT15 | ||||||||
|
Clustered
| ||||||||||||||
| 241 | Zhuang | M | GH | Apr-05 | LL | 8 | 8 | 11 | 10 | 9 | 13 | 17 | A | |
| 244 | Zhuang | M | GZ | May-05 | BT | 8 | 8 | 11 | 10 | 9 | 13 | 14 | A | |
| 271 | Zhuang | M | GH | Sep-05 | BL | 8 | 8 | 11 | 10 | 9 | 13 | 15 | A | |
| 273 | Miao | M | YZ | Sep-05 | BL | 8 | 8a | 11 | 10 | 9 | 13 | 15 | A | |
| 277 | F6 | Zhuang | M | GH | Sep-05 | BT | 8a | 8 | 11 | 10 | 9 | 13 | 14 | A |
| 296* | F12 | Zhuang | M | GZ | May-06 | BL | 8 | 8 | 11 | 10 | 9 | 13 | 16 | |
| 302* | F13 | Zhuang | M | GZ | Aug-06 | BL | 9 | 8 | 10 | 10 | 9 | 13 | 18 | |
| 403 | F17 | Zhuang | F | GZ | Oct-07 | BL | 9 | 8 | 10 | 10 | 9 | 13 | 15 | |
| 135 | F1 | Zhuang | M | GZ | Apr-04 | BL | 9 | 8 | 11 | 10 | 9 | 13 | 16 | B |
| 239 | F13 | Zhuang | M | GZ | Apr-05 | BL | 9 | 8 | 11 | 10 | 9 | 13 | 16 | B |
| 240 | Miao | M | YZ | Apr-05 | BT | 9 | 8 | 11 | 10 | 9 | 13 | 16 | B | |
| 245 | Zhuang | M | GZ | May-05 | LL | 9 | 8 | 11 | 10 | 9 | 13 | 16 | B | |
| 297 | F12 | Zhuang | M | GZ | Jun-06 | BT | 9 | 8 | 11 | 10 | 9 | 13 | 16 | |
| 298* | Zhuang | M | GZ | Jun-06 | BL | 9 | 8 | 11 | 10 | 9 | 13 | 16 | ||
| 299* | Zhuang | M | GZ | Jun-06 | BL | 9 | 8 | 11 | 10 | 9 | 14 | 15 | ||
| 312* | F | YZ | Oct-06 | BL | 9 | 8 | 11 | 10 | 9 | 14 | 15 | |||
| 317 | F1 | Zhuang | F | GZ | Oct-06 | TT | 9 | 8 | 11 | 10 | 9 | 13 | 18 | |
| 359 | F17 | Zhuang | F | GZ | Sep-07 | BL | 9 | 8 | 11 | 10 | 9 | 12 | 15 | |
| 462 | Zhuang | M | GZ | Feb-08 | BL | 9 | 8 | 11 | 10 | 9 | 13 | 16 | ||
| 554 | Zhuang | M | GZ | Jan-10 | BL | 9 | 8 | 11 | 10 | 9 | 13 | 16 | ||
| 556 | Han | M | XD | Mar-10 | BT | 9 | 8 | 11 | 10 | 9 | 13 | 14 | ||
| 237 | F1 | Zhuang | M | GZ | Apr-05 | BL | 9 | 8 | 12 | 10 | 9 | 13 | 16 | B |
| 287* | F1 | Zhuang | M | GZ | Mar-06 | BL | 9 | 8 | 12 | 10 | 9 | 13 | 16 | |
| 242 | Han | M | BDS | Apr-05 | BL | 10 | 8 | 11 | 10 | 9 | 13 | 15 | (B) | |
| 415 | Zhuang | M | SLY | May-08 | BL | 10 | 8 | 11 | 10 | 9 | 13 | 15 | ||
| 439 | Miao | F | GH | Aug-08 | BT | 10 | 8 | 11 | 10 | 9 | 13 | 15 | ||
| 289* | Han | M | TX | Mar-06 | BL | 9 | 7 | 8 | 12 | 10 | 18 | 14 | ||
| 291 | Miao | M | YZ | Apr-06 | BT | 9 | 7 | 8 | 12 | 10 | 18 | 14 | ||
| 136 | F3 | Miao | M | SD | Mar-04 | BL | 9 | 7 | 10 | 11 | 11 | 14 | 16 | C |
| 268 | F14 | Miao | M | GZ | Jul-05 | BL | 9 | 7 | 10 | 11 | 11 | 13 | 14 | C |
| 147 | F2 | Miao | F | GZ | Sep-03 | BL | 9 | 7 | 9 | 14 | 25 | 17 | 17 | (E) |
| 150 | F2 | Zhuang | M | GZ | Sep-03 | BT | 9 | 7 | 9 | 13 | 25 | ? | 16 | (E) |
| 247 | F5 | Miao | M | Jun-05 | BL | 9 | 8 | 9 | ? | 44 | 15 | 16 | E | |
| 249 | F5 | Miao | F | SD | Jul-05 | BL | 9 | 8 | 9 | 15 | 44 | 17 | 17 | E |
| 274 | F9 | Miao | M | XD | Oct-05 | BL | 8 | 9 | 13 | 12 | 26a | 16 | 13 | E |
| 360 | Miao | M | XD | Sep-07 | BL | 8 | 9 | 13 | 12 | 26 | 16 | 13 | ||
| 318 | F15 | Miao | M | BDS | Dec-06 | BL | 8 | 9 | 12 | 12 | 13 | 10 | ? | |
| 319 | F15 | Miao | M | BDS | Oct-07 | BT | 8 | 9 | 12 | 12 | 13 | 10 | ? | |
| 398 | F16 | Yi | F | XD | Oct-07 | BL | 8 | 9 | 10 | 15 | 13 | 14 | 16 | |
| 399 | F16 | Yi | F | XD | Oct-07 | LL | 8 | 9 | 10 | 15 | 13 | 14 | 14 | |
| 138 | Zhuang | M | TX | Feb-04 | BL | 8 | 9 | 8 | 20 | 15 | 12 | 19 | (A/B/C) | |
| 459 | Zhuang | M | TX | Dec-08 | BL | 8 | 9 | 8 | 21 | 15 | 12 | 18 | ||
| 405 | F22 | Zhuang | M | SLY | Jan-08 | BL | 8 | 8 | 9 | 9 | 45 | 15 | 15 | |
| 523 | F22 | Zhuang | M | SLY | Jun-09 | BL | 8 | 8 | 9 | 9 | 46 | 15 | 15 | |
| 139 | Miao | M | SP | Dec-03 | BT | 8 | 8 | 9 | 9 | 36 | 15 | 15 | E | |
| 149 | Miao | F | SP | Mar-04 | BL | 8 | 8 | 9 | 9 | 35 | 15 | 15 | E | |
| 362 | Miao | M | SP | Aug-07 | BB | 8 | 8 | 9 | 9 | 22 | 16 | 15 | ||
| 363 | Miao | F | JP | Aug-07 | BL | 8 | 8 | 9 | 9 | 23 | 15 | 15 | ||
| 300* | F21 | Zhuang | M | TX | Jul-06 | BL | 8 | 8 | 10 | 12 | 25 | 15 | 15 | |
| 441 | Zhuang | M | GZ | Aug-08 | BT | 8 | 8 | 10 | 12 | 26 | 20 | 19 | ||
| 460 | Zhuang | M | TX | Feb-09 | BT | 8 | 8 | 10 | 12 | 26 | 20 | 18 | ||
| 461 | F21 | Zhuang | M | TX | Mar-09 | BT | 8 | 8 | 10 | 12 | 26 | 14 | 18 | |
| 463 | F21 | Zhuang | F | TX | Mar-09 | BT | 8 | 8 | 10 | 12 | 26 | 20 | 14 | |
| 341 | Yi | M | NJ | May-07 | BL | 8 | 8 | 9 | 12 | 21 | 16 | 15 | ||
| 408 | Miao | M | SD | Mar-08 | BL | 8 | 8 | 9 | 12 | 21 | 16 | 15 | ||
| 144 | Miao | F | BDS | Apr-04 | BT | 8 | 7 | 8 | 11 | 25 | 16 | 15 | D | |
| 198 | F8 | Miao | F | JP | Nov-04 | BT | 8 | 7 | 8 | 11 | 25 | 16 | 15 | D |
| 199 | Miao | F | SD | Oct-04 | BT | 8 | 7 | 8 | 11 | 25a | 16 | 16 | D | |
| 339 | Miao | M | GH | Mar-07 | BL | 8 | 7 | 9 | 12 | 23 | 16 | 15 | ||
| 553 | Miao | M | SLY | Dec-09 | BL | 8 | 7 | 9 | 12 | 22 | 15 | 15 | ||
| 233 | F10 | Miao | F | WL | Jan-05 | BL | 8 | 7 | 10 | 17 | 30 | 11 | 26a | E |
| 234 | F10 | Miao | F | WL | Jan-05 | BL | 8 | 7 | 10 | 17 | 28 | 11 | 28 | E |
| 417 | Miao | M | WL | Jun-08 | LL | 8 | 7 | 10 | 17 | 30 | 11 | 22 | ||
| 152 | Miao | M | TX | Dec-03 | TT | 7 | ?a | 8 | 12 | 13 | 14 | ?a | (C) | |
| 450 | Miao | M | TX | Oct-08 | BL | 7 | 6 | 8 | 12 | 13 | 14 | 19 | ||
|
Unclustered
| ||||||||||||||
| 53 | Miao | M | JP | Aug-03 | BT | 8 | 7 | 9 | 10 | 37 | ? | ? | ? | |
| 71 | Zhuang | F | TX | Sep-02 | BT | 8 | 7 | 9 | 15 | 31 | ? | ? | ? | |
| 77 | Miao | M | BDS | Jul-02 | LL | 8 | 7 | 9 | 12 | 13 | 14 | 14 | (A/B/C) | |
| 86 | Han | M | GZ | Apr-02 | BB | 8 | 8 | 10 | 14 | 21 | 12 | 12 | (A/B/C) | |
| 132 | Zhuang | M | GH | Feb-04 | BT | 8 | 7 | 10 | 10 | 38a | 16 | 19 | E | |
| 133 | F7 | Han | M | BDS | Mar-04 | BL | 8 | 8 | 11 | 15 | 11 | 16 | 13 | ? |
| 134 | Zhuang | F | GH | Dec-03 | BL | 10 | 7 | 11 | 9 | 9 | 13 | 15 | (B) | |
| 137 | Miao | F | XD | Mar-04 | BL | 8 | 8 | 8 | 10 | 9 | 13 | 17 | A | |
| 140 | Han | F | PZ | Feb-04 | BL | 8 | 8 | 10 | 16 | 19 | 13 | 12 | (A/B/C) | |
| 141 | Han | M | PZ | Mar-04 | BL | 8 | 7 | 10 | 19 | 20 | 11 | 16 | (A/B/C) | |
| 143 | Miao | M | GH | Apr-04 | BL | 9 | 8 | 12 | 11 | 19 | 15 | 12 | D | |
| 146 | Yi | M | XD | Dec-03 | LL | 8 | 7 | 9 | 13 | 24 | 16 | 15 | (E) | |
| 151 | Han | M | TX | Dec-03 | BL | 8 | 9 | 11 | 10 | 18 | 13 | 13 | (A/B/C) | |
| 153 | Han | M | BDS | Feb-04 | LL | 8 | 8 | 8 | 11 | 26a | 16 | 15 | D | |
| 154 | F4 | Miao | M | SD | Dec-02 | BL | 9 | 7 | 11 | 12 | 13 | 14 | 14 | C |
| 196 | F11 | Han | M | SP | Oct-04 | BT | 8 | 7 | 7 | 11 | 16 | 12 | 14 | (C) |
| 235 | Miao | M | TX | Feb-05 | BL | 8 | 7 | 8 | 9 | 11 | 12 | 14 | (A/B) | |
| 236 | Zhuang | M | TX | Mar-05 | ? | 8 | 10 | 8 | 11 | 11 | 11 | 18 | (C) | |
| 238 | F7 | Han | M | BDS | Mar-05 | BT | 8 | 9a | 10 | 14 | 9 | 11 | 14 | (A/B) |
| 243 | Miao | M | SP | Apr-05 | BT | 8 | 7 | 8 | 9 | 30 | 11 | 14 | E | |
| 246 | F3 | Miao | M | SD | Jun-05 | ? | 9 | 7 | 10 | 12 | 11 | 14 | 16 | C |
| 248 | Miao | M | YZ | Jun-05 | TT | 9 | 8 | 11 | 13 | 13 | 10 | 22 | (C) | |
| 269 | Miao | F | TX | Sep-05 | BL | 7 | 7 | 10 | 17 | 36 | 11 | 16 | E | |
| 270 | F11 | Han | M | SP | Sep-05 | TT | 10 | 7 | 10 | 10 | 9 | 11 | ? | ? |
| 272 | Zhuang | M | GZ | Sep-05 | TT | 8 | 8 | 9 | 12 | 29 | 13 | 15 | E | |
| 275 | F6 | Zhuang | M | GH | Oct-05 | BL | 8 | 8 | 9 | 12 | 22 | 15 | 15 | D |
| 276 | Zhuang | M | WL | Oct-05 | BL | 8 | 8 | 10 | 9 | 9 | 14 | 15 | A | |
| 285 | Han | M | SP | Dec-05 | BT | 8 | 9 | 11 | 19 | 17 | 16 | 15 | ? | |
| 286 | Miao | M | YZ | Mar-06 | BT | 8 | 8 | 12 | 13 | 23 | 18 | 13 | ||
| 288 | Zhuang | F | TX | Mar-06 | TT | 8 | 7 | 8 | 12 | 9 | ? | 15 | ||
| 290 | F14 | Miao | M | GZ | Apr-06 | BT | 9 | 6 | 10 | 12 | 12 | 14 | 15 | |
| 301* | Miao | F | NJ | Jul-06 | BL | 8 | 9 | 9 | 13 | 15 | 13 | 14 | ||
| 316 | Zhuang | M | GZ | Oct-06 | BT | 7 | 7 | 8 | 22 | 14 | 13 | 13 | ||
| 320 | F19 | Zhuang | F | TX | Feb-07 | BL | 8 | 9 | 8 | 19 | 13 | 13 | 21 | |
| 321 | F7 | Han | M | BDS | Jan-07 | BB | 8 | 8 | 10 | 14 | 11 | 15 | 12 | |
| 338 | F23 | Miao | M | SP | Mar-07 | BL | 8 | 8 | 9 | 12 | 17 | 17 | 16 | |
| 340 | Tu | M | JP | Apr-07 | BL | 8 | 9 | 9 | 11 | 26 | 15 | 17 | ||
| 342 | Miao | M | BDS | May-07 | BT | 7 | 8 | 9 | 13 | 18 | 16 | 19 | ||
| 345 | Miao | M | BDS | Jul-07 | BL | 8 | 9 | 12 | 12 | 12 | 10 | 21 | ||
| 346 | Miao | M | XD | Jul-07 | LL | 8 | 8 | 9 | 13 | 26 | 16 | 19 | ||
| 361 | Miao | M | YZ | Aug-07 | BL | 8 | 8 | 12 | 17 | ? | 14 | 13 | ||
| 365 | Yi | M | YZ | Aug-07 | BT | 9 | 8 | 8 | 9 | 11 | 15 | 14 | ||
| 400 | Zhuang | M | SP | May-06 | BL | 8 | 9 | 10 | 16 | 22 | 8 | ? | ||
| 401 | Miao | M | WL | Aug-06 | BL | 8 | 7 | 10 | 10 | 13 | 16 | 14 | ||
| 402 | Yi | M | NJ | Feb-07 | BT | 8 | 9 | 10 | 24 | 13 | 13 | 15 | ||
| 406 | Miao | M | JP | Jan-08 | BL | 9 | 8 | 10 | 14 | 16 | 16 | 13 | ||
| 414 | Bai | M | SLY | May-08 | BL | 7 | 8 | 9 | 13 | 19 | 10 | 18 | ||
| 416 | Miao | M | SP | May-08 | BL | 9 | 8 | 12 | 15 | 21 | 13 | 13 | ||
| 438 | Han | F | SP | Jul-08 | BT | 8 | 8 | 9 | 9 | 24 | 10 | 14 | ||
| 448 | Miao | M | BDS | Aug-08 | BL | 8 | 7 | 8 | 11 | 13 | 8 | 14 | ||
| 449 | F20 | Miao | F | YZ | Sep-08 | BL | 9 | 9 | 9 | 12 | 12 | 17 | 13 | |
| 471 | Han | M | XD | May-09 | BL | 8 | 8 | 11 | 13 | 20 | 14 | 14 | ||
| 472 | Zhuang | M | GZ | Mar-09 | BT | 8 | 8 | 9 | 12 | 25 | 18 | 23 | ||
| 473 | F23 | Miao | M | SP | Mar-09 | BT | 8 | 8 | 9 | 12 | 19 | 13 | 25 | |
| 474 | Zhuang | M | JP | Mar-09 | BL | 8 | 7 | 9 | 16 | 27 | 11 | 21 | ||
| 475 | Miao | M | SP | Apr-09 | BT | ? | 7 | 9 | 11 | 27 | 11 | 15 | ||
| 476 | Miao | M | SP | Apr-09 | BL | 8 | 9 | 12 | 12 | 14 | 10 | 18 | ||
| 520 | Miao | F | TX | Oct-09 | BL | 8 | 7 | 11 | 14 | 16 | 15 | 14 | ||
| 521 | Miao | M | YZ | Jul-09 | BL | 9 | 8 | 8 | 12 | 22 | 17 | ? | ||
| 522 | Miao | M | XD | Jul-09 | BL | 8 | 9 | 12 | 12 | 22 | 14 | 13 | ||
| 524 | Yi | M | SD | Aug-09 | BL | 9 | 10 | 9 | 10 | 27 | 16 | 19 | ||
| 527 | Zhuang | M | TX | Oct-09 | BL | 9 | 8 | 9 | 15 | 39 | 17 | 16 | ||
| 550 | Zhuang | F | JP | Dec-09 | BL | 8 | 8 | 8 | 11 | 22 | 15 | 18 | ||
| 551 | Miao | M | NJ | Nov-09 | BT | 8 | 7 | 8 | 12 | 13 | 8 | 14 | ||
| 555 | Miao | M | WL | Feb-10 | LL | 8 | 7 | 11 | 12 | 12 | 14 | 13 | ||
Alleles differing from those reported in Weng et al., 2007.
3.3. Informative subpopulations and regional groupings of VNTR allele frequencies
To illuminate the patterns of leprosy transmission in Qiubei and the factors that contribute to these, a variety of demographic and clinical subpopulation definitions were tested. The allele frequencies of the total population and subpopulations were analyzed with a panel of statistical methods.
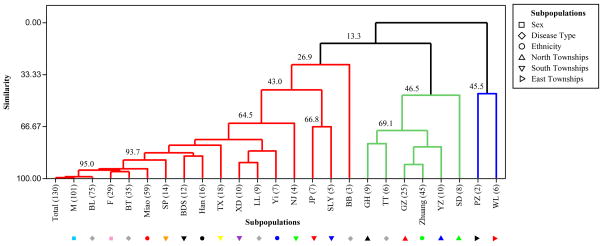
In Figure 2 the level of similarity of allele frequency distributions with respect to township of origin, ethnicity, gender, and clinical presentation is shown. Three primary branches emerged, shown in red, green and blue.
Figure 2. Similarity dendrogram for M. leprae genotypes in Qiubei.
The dendrogram is based on the distribution of allele frequencies for various subpopulation definitions: gender [male, female], clinical type [TT, BT, BB, BL and LL], township of residence [BDS, GH, GZ, JP, NJ, PZ, SD, SP, SLY, TX, WL, XD, YZ] and ethnicity [Zhuang, Miao, Han, Yi]. The number of cases for each subpopulation is shown in parenthesis.
To address the question of whether genetic differences were present in the M. leprae samples of different patient genders and clinical types, the distribution of alleles in these subpopulations were compared relative to the total population (Total in Figure 2). No significant distinction was found when considering the allele frequencies of either gender groups (95.0% similarity) or those presenting with either BT or BL type disease (95.0% similarity). This implies a lack of distinction in terms of strain type for these subpopulations and that segregating the sample space into gender or clinical presentation would not be informative. The TT type shows clustering away from the overall population distribution, but this is assumed to be sampling error, as only a small number of TT cases (n=6) could be reliably typed due to low bacterial index (BI). Further investigation may be warranted.
Qiubei is a multiethnic region whose member minorities are distributed unevenly across the 14 townships. To evaluate the strain types within this demographic context, the samples were grouped according to the township of residence and self-reported ethnicities of the patients. The allele frequencies of strains in the townships of BDS, TX, XD, SP, JP, NJ and SLY which are found in the south, west and center of Qiubei County (43.0% minimum similarity) are more similar to those of the total population, rather than to those in the townships of GH, GZ, SD and YZ (46.5% minimum similarity) that are situated in the north of the county and are separated from the former in the dendrogram. The townships of WL and PZ (45.5% similarity) are found on the eastern edge of the county.
At the level of ethnicities, a high degree of similarity is seen between the Miao subpopulation and the overall allele frequencies (95.0% similarity). The alleles of the Han subgroup also branch together with that of the total population. The Zhuang subgroup is separated, and clusters with the North townships of GH and GZ, which is generally in concordance with the ethnic compositions of these two townships (see Table 3) and therefore exhibits the expected sampling bias. However, the SD and YZ townships, where Miao people form a higher composition than Zhuang people (see Table 3), are also clustered with the Zhuang allele frequencies, yet the majority of patients in SD and YZ are Miao. Thus, transmission of leprosy, as judged by allele frequencies, is related to geographical proximity of townships beyond the ethnic distributions found locally. Based on the branching on the dendrogram and the townships that cluster within each branch, it has been possible to classify the population into three regions: North, South and East.
Table 3. Distribution of ethnicities in each township of Qiubei County.
| Townshipa | Populationb |
||||
|---|---|---|---|---|---|
| Zhuang | Miao | Han | Yi | Other | |
| GH | 47.24% | 7.88% | 43.46% | 1.39% | 0.04% |
| GZ | 61.86% | 8.58% | 19.94% | 9.62% | 0.00% |
| SD | 4.04% | 41.04% | 13.41% | 41.51% | 0.00% |
| YZ | 11.80% | 21.97% | 36.23% | 19.06% | 10.94% |
| BDS | 14.48% | 23.89% | 30.12% | 28.14% | 3.37% |
| JP | 43.38% | 8.55% | 33.62% | 6.25% | 8.19% |
| NJ | 3.35% | 15.80% | 41.57% | 39.28% | 0.00% |
| SLY | 11.42% | 5.99% | 58.34% | 15.99% | 8.26% |
| SP | 8.52% | 19.36% | 42.28% | 29.83% | 0.00% |
| TX | 32.58% | 20.29% | 33.39% | 13.17% | 0.57% |
| XD | 13.55% | 46.36% | 7.81% | 32.21% | 0.06% |
| PZ | 56.27% | 3.37% | 35.25% | 2.57% | 2.55% |
| WL | 42.21% | 9.11% | 47.90% | 0.09% | 0.69% |
| YJ | 28.76% | 0.06% | 54.67% | 0.07% | 16.45% |
Two or three letter coding system has been used to represent the names of the 14 townships in Qiubei (Weng et al., 2007).
Percentage of the total township population for a specific ethnicity in 2005.
3.4. Principal component analysis (PCA) of M. leprae VNTR genotype data
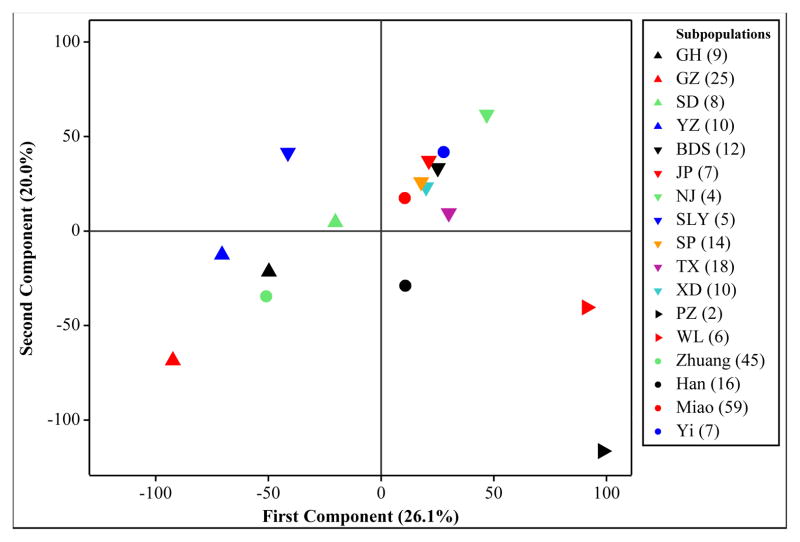
PCA has been exploited in population genetics to reduce large amounts of measurements into a few principal components. PCA can be used as a tool for detecting population substructure, geographical associations, and migration patterns (Novembre and Stephens, 2008; Rendine et al., 1986). A PCA was performed on the allele frequencies for each of the ethnicity and township subpopulations using all seven loci reported (Figure 3A, B).
Figure 3.
Figure 3A. The score plot for the first two principal components of M. leprae VNTR allele frequencies classified according to patient ethnicity and township
Colored arrowhead symbols have been used for each of the townships of the North, South and East regions with the direction of the arrowheads indicating region membership. The first two principal components account for 46.1% of the variation in the data.
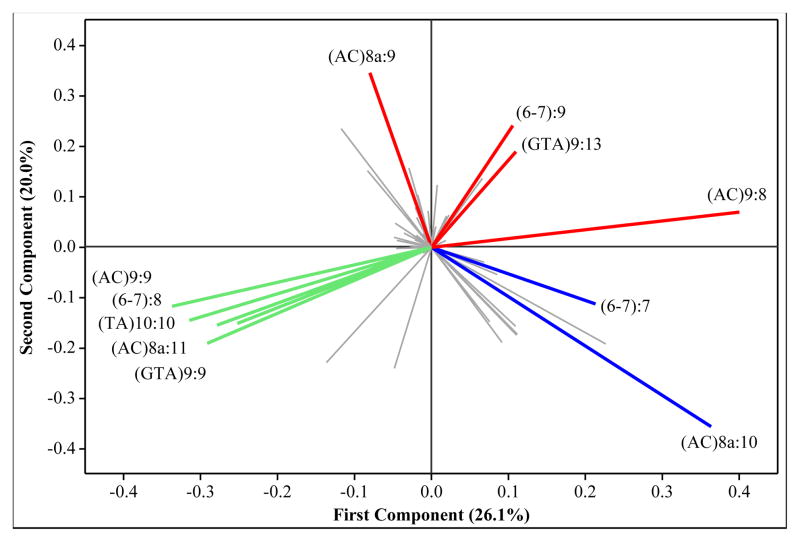
Figure 3B. The loading plot for the first two principal components of M. leprae VNTR allele frequencies classified according to patient ethnicity and township
The position of alleles on the loading plot corresponds to the location of townships and ethnicities on the score plot. Alleles with similar directionality are positively correlated, and the length of the projection line represents the magnitude of an allele’s contribution. Thus, those alleles that are most casual to the distribution seen on the score plot will show similar directionality from the origin when compared to subpopulations on the score plot. Here there are a large number of alleles clustered in the center. These alleles are not informative at the township or ethnicity subpopulation level, owing to lack of diversity or low overall frequency. The colors of the projections highlight alleles having higher frequencies in particular regions; East, North and South are shown in blue, green and red, respectively.
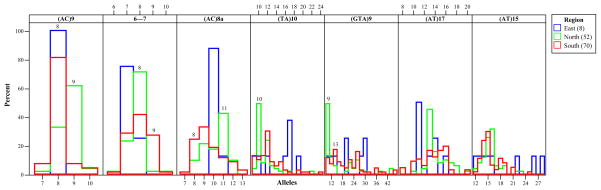
The PCA plot of the townships recapitulates the associations detected by the dendrogram; i.e., differentiation of the North, South and East townships. In addition, examination of the corresponding loading plot for PC1 vs PC2 (Figure 3B) reveals the alleles that contribute to the observed distribution of subpopulations. The North is distinguished by the frequency of the following alleles: 9, 8, 11, 10, 9 for the loci (AC)9, 6–7, (AC)8a, (TA)10 and (GTA9), respectively (Figure 4). The genotype matching this set of alleles is found almost exclusively in the North (12 North vs 1 South), though each individual allele can be found in moderate proportions in other locations. In contrast, the distinguishing alleles in the South are: 8, 9, 8, and 13 for the loci (AC)9, 6–7, (AC)8a, and (GTA)9, respectively (Figure 4). These alleles predominate in the South, but not together as a dominant genotype. Therefore, recent clonal genetic relationships are not obvious in the South.
Figure 4. Histogram of allele frequencies by region.
The variation among alleles across the East, North and South regions is shown in blue, green and red, respectively. Specific distinguishing alleles are indicated on the graph as a number above the bar outline.
3.5. Clustering of leprosy according to individual M. leprae VNTR genotypes
Clustering of pathogen genotypes has been used in the study of infectious diseases as a means of estimating recent transmission against a background of the total strain types which represents relapses, reactivation, immigrant, and emigrant cases. In general, as clustering increases (pairs or larger groups of shared genotypes) transmission is assumed to be active. Percent clustering is influenced by a number of sampling and test factors, such as the method of DNA typing, the length of the study, and the age range of the individuals within the study population. Nevertheless, it is a useful method when supported by epidemiological investigations. (Houben and Glynn, 2009).
Samples were assigned into clusters on the basis of having identical, or highly similar, genotypes. Percent clustering is reported as a lower bound due to the tendency of clustering by the method of shared genotypes to underestimate actual percent clustering (Murray and Alland, 2002). The potential impact of homoplasy when assigning clusters was not considered, as it would not affect the total percent clustered. Classification of unclustered cases as either relapse or immigrant cases was not investigated.
Under these assumptions, the lower bounds for percent clustering, based on a five locus VNTR genotype, for both study periods are comparable (45.4 ± 8.5% vs 50.7 ± 5.8%) (Table 4). The data indicate that nearly half of the patients have M. leprae strains that are found in at least one other patient, which implies active regional transmission. Though this is an expected result, we find a more in-depth picture of transmission patterns than from case detection alone.
Table 4. Percent clustering of genotyped cases.
Percent clustering is shown for township, ethnicity and clinical presentation subpopulations. The case detection rate is also presented for comparison. The range of the 95% confidence interval is indicated as a ± value. A CI is not listed for entries in parenthesis.
| Subpopulation | Percent Clustered | Detection Rate |
|---|---|---|
| Total | 48.5 ± 4.8% | 4.50 |
| 2002–2005 | 45.4 ± 8.5% | 4.04 |
| 2006–2010 | 50.7 ± 5.8% | 4.91 |
| North | 65.4 ± 7.2% | 6.72 |
| GH | (55.6%) | 7.44 |
| GZ | (80.0%) | 7.98 |
| SD | (62.5%) | 7.55 |
| YZ | (40.0%) | 4.15 |
| South | 37.1 ± 6.3% | 4.60 |
| BDS | (33.3%) | 5.32 |
| JP | (28.6%) | 3.98 |
| NJ | (25.0%) | 2.56 |
| SLY | (80.0%) | 1.36 |
| SP | (21.4%) | 6.06 |
| TX | (38.9%) | 7.11 |
| XD | (50.0%) | 8.30 |
| East | 37.5% ± 18.8% | 1.74 |
| PZ | (0.0%) | 1.13 |
| WL | (50.0%) | 2.38 |
| Han | 18.8% ± 10.7% | 1.31 |
| Miao | 44.1 ± 7.1% | 13.35 |
| Yi | 42.9 ± 20.5% | 1.85 |
| Zhuang | 66.7 ± 7.7% | 5.93 |
| TT | 16.7 ± 16.7% | 0.19 |
| BT | 48.6 ± 9.3% | 1.54 |
| BB | 33.3 ± 29.9% | 0.19 |
| BL | 53.3 ± 6.3% | 2.33 |
| LL | 44.4 ± 18.2% | 0.25 |
In many subpopulations we see similar patterns of percent clustering and detection rate, in that those with higher clustering percentage tend toward higher case detection rates. However, this is not a universal correlation. The Miao detection rate is more than double that of the Zhuang rate, yet the Miao percent clustering is approximately 23% lower than that of the Zhuang. Assuming there is no nonhuman reservoir of M. leprae in the region (Truman and Fine, 2010), this implies an underlying difference in the transmission patterns across the two ethnic minorities exhibiting the most prominent health disparity in the county.
3.6. Relationships to previously defined groups
In the previous study a set of genotypes indentified as A/B was shown to occur predominately in the North (Weng et al., 2007). A minimum spanning network of genotypes based on an infinite allele mutation model (not shown), again identified this transmission supercluster (Groups A/B, Table 2). Members of the A group have not been reported since 2006, and may have died out, while the B group has expanded from 6 cases in 2002–2005 to 12 new cases during the 2006 to 2010 time frame. This further implies ongoing transmission of community strains within the North part of the county. Interestingly, the allele at locus (AC)9 of these strains is predominantly 9 repeats in GZ, and 8 or 10 in GH. A variant with a 10 at (AC)9 is seen in the adjacent BDS and SLY townships. Future monitoring may provide valuable insight into the evolution of these community strains.
3.7. Multicase families
Transmission of leprosy in family units remains a serious concern. From the total 164 enrolled cases, 53 belong to 23 multicase families. Eleven of these families were indentified in the 2002–2005 study, while the remaining twelve where found in the 2006–2010 time period.
Within the genotyped cases, 42% (19/45) of the Zhuang and 29% (17/59) of the Miao detected cases are from multicase families. From the total genotype clusters 67% (14/21) of them contain at least one multicase family patient (Table 2). Patients from families F1, F6, F12, F13 and F17 exhibit a change of only one repeat in a single allele from other multicase families in another cluster. This strongly implies an active transmission chain in, and developing from, multicase families. However, not all multicase families appear to share the same infection sources, as seen in multicase families F6 and F11 which show significant genotypic differences. We hypothesize that the source of infection was outside the home, for at least one patient, in each of these two family cases.
3.8. Relapse versus re-infection
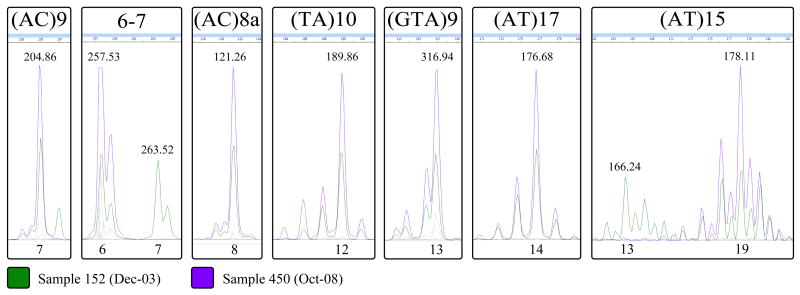
During this study one case relapsed. However, this was not known at the time of sample collection or VNTR analysis in the laboratory. Samples 152 and 450 are from the same patient in the TX township, taken at different clinical episodes of leprosy (originally TT, later BL form). The length of time between the first and second episode was approximately 5 years (Dec-03 to Oct-08). These two samples were not considered in the analysis of genotypically clustered cases, though they appear in the Clustered section of Table 2. The VNTR profiles indicate that relapse rather than re-infection was the cause of the second episode of leprosy. The originally reported allele at locus 6–7 for sample 152 was 7 (Weng et al., 2007), which differs from the value of 6 reported for sample 450 (Table 2). The alleles for sample 152 were previously determined by sequencing, which can obscure mixed alleles. FLA was performed to compare alleles in 152 and 450, which revealed mixed alleles at loci 6–7 and (AT)15 in sample 152 (Figure 5). However, concordance was seen among all other loci tested and each allele in sample 450 was also present in sample 152. The patient originally presented with TT type leprosy, which provides little genetic material for analysis. The mixed allelic signatures in sample 152 may be due to poor quality of source DNA, but may also reflect active evolution or co-infection wherein only one variant survived initial treatment.
Figure 5. Fragment length analysis chromatograms for the relapse patient.
The two samples shown are from a single patient, the first (152) from initial diagnosis of leprosy and the second (450) from a second presentation approximately 5 years later. The relevant peaks are shown for each locus used in this study. Peak intensities have been adjusted for the sake of clarity, and non-informative dye colors have been grayed out. The size of each fragment, in base pairs, is listed above the dominant peaks. Copy number (allele) is listed below each peak. For loci 6–7 and (AT)15 multiple alleles are seen for sample 152. Inset within each allelic peak in sample 450 (purple, full-height) is a perfectly matched allele from sample 152 (green, half-height).
4. CONCLUSIONS
Pathogen genotyping and cluster analysis are means of identifying both recent and unsuspected transmission, which can then be used to inform control programs on disease trends and aid in the implementation of corrective measures (Scott et al., 2004). Longitudinal VNTR strain typing of M. leprae in an endemic population, at first during a 43 month period (69 patients, though only 68 were previously reported) and extended by an additional 51 months (95 patients), has been informative and instructive at multiple levels. Though overall genetic diversity of M. leprae in Qiubei is minimal, by using an appropriate selection of seven polymorphic VNTR loci we were able to identify clustered genotypes; track the temporal, spatial, and ethnogeographic distribution of leprosy; observe VNTR genotype evolution; and identify at-risk populations.
Within the sample size available for population level analysis, genotypes were not found to significantly differ based on clinical form or patient gender. Geographical clustering was evident, indicating that transmission continued to occur among people living within short distances of each other (in the same or neighboring township). Certain regional strains of M. leprae continued to present, suggesting a lack of adequate control in the infected populations. No major regional shifts in dominant alleles were seen between the two study periods. The majority of genotype clusters (19/21) had samples from both study periods, with multiple clusters (6/21) showing histories of longer than 2 years (Table 2). In particular, the B group in the North continued to expand from 2004 through 2010. In one patient, it was demonstrated that relapse, not reinfection, was the cause of disease recurrence after 5 years.
Strain typing was performed using a single skin biopsy from each patient as the source of M. leprae. As the majority of MCFs demonstrated shared genotypes, this implies that these skin derived specimens represent the dominantly transmitted strain type, and perhaps the source of infection. Job et al. have shown that untreated multibacillary patients shed M. leprae via their skin and nasal secretions, and that close contacts are at risk for exposure to the pathogen (Job et al., 2008). Allelic differences have been seen across bacterial samples taken from dermal and nervous tissue (Young et al., 2004; Young et al., 2008), however the transmissibility of M. leprae residing in nervous tissue remains to be clarified. Subtle genotypic shifts were seen in some related strains, such as the (AC)8a allele in members of MCFs F13 and F17 (Table 2). Of the seven loci mapped, the stutter prone locus (AT)15 (Table 2, Figure 5) had the greatest variability within MCFs and clusters.
The combination of epidemiological and molecular data was able to provide depth beyond what either data set could provide alone. The epidemiological data revealed a disparity in the leprosy burden across ethnic populations within Qiubei, with case detection rates of the Zhuang and Miao significantly higher than that of the Han majority (Table 4). We were able to capture a similar story with the molecular data; where, as an estimate of active transmission, the percent of clustered genotypes for both the Zhuang and Miao also surpassed the value for the Han majority (Table 4). Further, there were marked quantitative distinctions between the case detection rates and percent of clustered genotypes that extended the story. We found that although the detection rate among the Miao was higher than that of the Zhuang, the percent of clustered genotypes among the Miao was lower; thus implying a more fragmented transmission pattern among the Miao. Analogous to the molecular clusters identified by VNTR strain typing, MCFs represent contact linked clusters of infection. We found that the percent of MCFs among the Zhuang was similarly higher than for the Miao, reinforcing the conclusion of a more fragmented transmission pattern among the Miao. The underlying reason for the difference in transmission patterns is unclear, but it may be rooted in geographic, socioeconomic and cultural factors.
Genotypic clustering serves as a method to measure the temporal concomitance of transmission events. When overall clustering is high and individual clusters are large, a recent pathogen invasion may be implied (Tanaka and Francis, 2006). In the townships of the North, where the Zhuang form the majority, genotypic clustering was found to be highest, and the largest observed supercluster, the A/B group, was found almost exclusively in the North. In the South, clusters were smaller and genotypes were more diverse, implying a more historical endemicity. Thus, the difference between the Zhuang and Miao transmission patterns may be a product of geography, with the underlying cause being a more recent introduction of leprosy in the Zhuang communities of the North. “Recent” must be interpreted cautiously, as it may refer to a time long before the beginning of this study.
Chen et al. found that within endemic regions of China the median delay between self-reported onset of symptoms and diagnosis is 23 months (Chen et al., 2000). The long delay before treatment of symptomatic leprosy presents substantial opportunity for secondary transmission of M. leprae. When socioeconomic development is slower, the delay in treatment is significantly longer, likely due to less accessible health care systems (Chen et al., 2000). In the South, the pace of economic development is faster. As a consequence, communication and transportation infrastructure is superior to that of the North, and more employment opportunities outside farming exist, such as in the construction, service and administrative fields (personal communication). Therefore, a difference in treatment delay may also explain the difference in transmission patterns across ethnicities and regions (Shen et al., 2010). Where health care accessibility may affect entire villages, as in the North, community clusters may dominate. Shen et al. compared the case finding methods in China and found that passive methods, such as skin clinics and referrals, contribute to fewer cases in Southwestern endemic provinces than in the less endemic Eastern provinces (52.2% vs 83.7%) (Shen et al., 2010). In contrast, methods such as contact, clue and group surveys resulted in more detected cases. Just as with patient access to care, active case detection is dependent on adequate communication and transportation infrastructure. As such, regional differences in the efficiency of case finding methods may also exist in Qiubei. In the North, where cluster sizes are large, active case finding methods may need to be strengthened.
Another contributing factor may be local customs. Among the Zhuang in the North it is common for extended families to live in adjoined community housing structures, while for the Miao it is common for people to leave one place and move to another in search of suitable farm land (personal communication). Families that live in closely connected communities may share community strains, leading to higher percent clustering. Those families that choose to relocate may carry local strains with them, and promote a higher number of unclustered cases. This study has been based only in Qiubei and leprosy in adjacent counties has not been explored. In a few family histories there is knowledge of recent migration of patients or family members from Guangnan and Luxi counties. Women traditionally relocate to the spouse’s family after marriage, which may also contribute to the migration of people from one county or township to another, and promote the dispersal of M. leprae genotypes. Detailed family histories may have been illuminating, but were not undertaken, or feasible, for all patients. Further contributing culture factors may include language barriers between ethnicities and amenability to BCG vaccination (personal communication).
As seen by the overall concordance between molecular and epidemiologically derived conclusions, VNTR strain typing can estimate transmission patterns when patient history and demographic data are unavailable, though the greatest degree of resolution is found by combining molecular and patient history data. The findings in this study provide adequate validation for continued application of VNTRs, in contrast to the conclusions by Monot et al. that VNTRs may be too dynamic for use as epidemiological markers in leprosy (Monot et al., 2008). Recognizing that homoplasy and convergent evolution of VNTR are certainly possible, this study shows that valuable information can be gained from longitudinal VNTR strain typing. When closely and continuously monitored, VNTR strain typing may be used to estimate the half-life of both strain types and individual alleles, and provide a means for surveillance of transmission. Other candidate M. leprae microsatellite loci exist and exploration of these additional loci may identify more distant relationships among the currently unclustered strains (Zhang et al, 2005). Furthermore, universally adopting FLA as the typing system would yield more rapid and economic typing of multiple samples, provide a consistent framework for comparing the results of different studies, permit multiplexing of loci, and allow for the detection of mixed allelic signatures (Kimura et al., 2009). We conclude that VNTRs, when appropriately used, are sensitive tools for stain typing that have sufficient resolution at the regional and community level where SNP types may be invariant.
Acknowledgments
We gratefully acknowledge the technical assistance of Dr. Rama Murthy Sakamuri, the support of Dr. Li Huan-Ying and staff at BTMRI, and the cooperation of clinicians and field staff at the Qiubei Skin Disease Control Stations.
This work was supported by the National Natural Science Foundation of China grant 30670111; and NIH/NIAID grant RO1-AI-63457, ARRA grant supplement RO1-AI-63457 S1 and contract NO1 AI-25469.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Allix-Béguec C, Fauville-Dufaux M, Supply P. Three-year population-based evaluation of standardized mycobacterial interspersed repetitive-unit-variable-number tandem-repeat typing of Mycobacterium tuberculosis. J Clin Microbiol. 2008;46(4):1398–1406. doi: 10.1128/JCM.02089-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brosch R, Gordon SV, Marmiesse M, Brodin P, Buchrieser C, Eiglmeier K, Garnier T, Gutierrez C, Hewinson G, Kremer K, Parsons LM, Pym AS, Samper S, van Soolingen D, Cole ST. A new evolutionary scenario for the Mycobacterium tuberculosis complex. Proc Natl Acad Sci USA. 2002;99(6):3684–3689. doi: 10.1073/pnas.052548299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen XS, Li WZ, Jiang C, Ye GY. Leprosy in China: delay in the detection of cases. Ann Trop Med Parasitol. 2000;94(2):181–188. doi: 10.1080/00034980057527. [DOI] [PubMed] [Google Scholar]
- Cole ST, Eiglmeier K, Parkhill J, James KD, Thomson NR, Wheeler PR, Honoré N, Garnier T, Churcher C, Harris D, Mungall K, Basham D, Brown D, Chillingworth T, Connor R, Davies RM, Devlin K, Duthoy S, Feltwell T, Fraser A, Hamlin N, Holroyd S, Hornsby T, Jagels K, Lacroix C, Maclean J, Moule S, Murphy L, Oliver K, Quail MA, Rajandream MA, Rutherford KM, Rutter S, Seeger K, Simon S, Simmonds M, Skelton J, Squares R, Squares S, Stevens K, Taylor K, Whitehead S, Woodward JR, Barrell BG. Massive gene decay in the leprosy bacillus. Nature. 2001;409(6823):1007–1011. doi: 10.1038/35059006. [DOI] [PubMed] [Google Scholar]
- de Vries G, van Hest RA, Burdo CC, van Soolingen D, Richardus JH. A Mycobacterium tuberculosis cluster demonstrating the use of genotyping in urban tuberculosis control. BMC Infect Dis. 2009;9:151. doi: 10.1186/1471-2334-9-151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Excoffier L, Lischer HEL. Arlequin suite ver 3.5: A new series of programs to perform population genetics analyses under Linux and Windows. Mol Ecol Resour. 2010;10:564–567. doi: 10.1111/j.1755-0998.2010.02847.x. [DOI] [PubMed] [Google Scholar]
- Feasey N, Wansbrough-Jones M, Mabey DC, Solomon AW. Neglected tropical diseases. Br Med Bull. 2010;93:179–200. doi: 10.1093/bmb/ldp046. [DOI] [PubMed] [Google Scholar]
- Filliol I, Motiwala AS, Cavatore M, Qi W, Hazbón MH, Bobadilla del Valle M, Fyfe J, García-García L, Rastogi N, Sola C, Zozio T, Guerrero MI, León CI, Crabtree J, Angiuoli S, Eisenach KD, Durmaz R, Joloba ML, Rendón A, Sifuentes-Osornio J, Ponce de León A, Cave MD, Fleischmann R, Whittam TS, Alland D. Global phylogeny of Mycobacterium tuberculosis based on single nucleotide polymorphism (SNP) analysis: insights into tuberculosis evolution, phylogenetic accuracy of other DNA fingerprinting systems, and recommendations for a minimal standard SNP set. J Bacteriol. 2006;188(2):759–772. doi: 10.1128/JB.188.2.759-772.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goguet de la Salmonière YO, Li HM, Torrea G, Bunschoten A, van Embden J, Gicquel B. Evaluation of spoligotyping in a study of the transmission of Mycobacterium tuberculosis. J Clin Microbiol. 1997;35(9):2210–2214. doi: 10.1128/jcm.35.9.2210-2214.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon SV, Brosch R, Billault A, Garnier T, Eiglmeier K, Cole ST. Identification of variable regions in the genomes of tubercle bacilli using bacterial artificial chromosome arrays. Mol Microbiol. 1999;32(3):643–655. doi: 10.1046/j.1365-2958.1999.01383.x. [DOI] [PubMed] [Google Scholar]
- Groathouse NA, Rivoire B, Kim H, Lee H, Cho SN, Brennan PJ, Vissa VD. Multiple polymorphic loci for molecular typing of Mycobacterium leprae. J Clin Microbiol. 2004;42:1666–1672. doi: 10.1128/JCM.42.4.1666-1672.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houben R, Glynn J. A systematic review and meta-analysis of molecular epidemiological studies of tuberculosis: development of a new tool to aid interpretation. Trop Med Int Health. 2009;14:892–909. doi: 10.1111/j.1365-3156.2009.02316.x. [DOI] [PubMed] [Google Scholar]
- Job CK, Jayakumar J, Kearney M, Gillis TP. Transmission of leprosy: a study of skin and nasal secretions of household contacts of leprosy patients using PCR. Am J Trop Med Hyg. 2008;78:518–521. [PubMed] [Google Scholar]
- Kimura M, Sakamuri RM, Groathouse NA, Rivoire BL, Gingrich D, Krueger-Koplin S, Cho SN, Brennan PJ, Vissa VD. Rapid variable-number tandem-repeat genotyping for Mycobacterium leprae clinical specimens. J Clin Microbiol. 2009;47:1757–1766. doi: 10.1128/JCM.02019-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krieger N, Chen JT, Waterman PD, Rehkopf DH, Subramanian SV. Painting a truer picture of US socioeconomic and racial/ethnic health inequalities: the Public Health Disparities Geocoding Project. Am J Public Health. 2005;95:312–323. doi: 10.2105/AJPH.2003.032482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuoka M, Zhang L, Budiawan T, Saeki K, Izumi S. Genotyping of Mycobacterium leprae on the basis of the polymorphism of TTC repeats for analysis of leprosy transmission. J Clin Microbiol. 2004;42:741–745. doi: 10.1128/JCM.42.2.741-745.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meima A, Gupte MD, van Oortmarssen GJ, Habbema JD. Trends in leprosy case detection rates. Int J Lepr Other Mycobact Dis. 1997;65(3):305–319. [PubMed] [Google Scholar]
- Michalakis Y, Excoffier LA. Generic estimation of population subdivision using distances between alleles with special reference for microsatellite loci. Genetics. 1996;142:1061–1064. doi: 10.1093/genetics/142.3.1061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monot M, Honoré N, Balière C, Ji B, Sow S, Brennan PJ, Cole ST. Are variable-number tandem repeats appropriate for genotyping Mycobacterium leprae? J Clin Microbiol. 2008;46(7):2291–2297. doi: 10.1128/JCM.00239-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monot M, Honoré N, Garnier T, Araoz R, Coppee JY, Lacroix C, Sow S, Spencer JS, Truman RW, Williams DL, Gelber R, Virmond M, Flageul B, Cho SN, Ji B, Paniz-Mondolfi A, Convit J, Young S, Fine PE, Rasolofo V, Brennan PJ, Cole ST. On the origin of leprosy. Science. 2005;308:1040–1042. doi: 10.1126/science/1109759. [DOI] [PubMed] [Google Scholar]
- Monot M, Honoré N, Garnier T, Zidane N, Sherafi D, Paniz-Mondolfi A, Matsuoka M, Taylor GM, Donoghue HD, Bouwman A, Mays S, Watson C, Lockwood D, Khamesipour A, Dowlati Y, Jianping S, Rea TH, Vera-Cabrera L, Stefani MM, Banu S, Macdonald M, Sapkota BR, Spencer JS, Thomas J, Harshman K, Singh P, Busso P, Gattiker A, Rougemont J, Brennan PJ, Cole ST. Comparative genomic and phylogeographic analysis of Mycobacterium leprae. Nat Genet. 2009;41:1282–1289. doi: 10.1038/ng.477. [DOI] [PubMed] [Google Scholar]
- Murray M, Alland D. Methodological problems in the molecular epidemiology of tuberculosis. Am J Epidemiol. 2002;155:565–571. doi: 10.1093/aje/155.6.565. [DOI] [PubMed] [Google Scholar]
- Nation Bureau of Statistics of China. Personal communication. 2010.
- Nation Center for Leprosy Control, China CDC (NCLC) [accessed August, 2010.];Leprosy Situation in China in 2006. 2007 http://www.lepinfo.org/English/statistics/stext2007/sta040501.htm.
- Nicholls PG, Wiens C, Smith WC. Delay in presentation in the context of local knowledge and attitude towards leprosy - the results of qualitative fieldwork in Paraguay. Int J Lepr Other Mycobact Dis. 2003;71:198–209. doi: 10.1489/1544-581X(2003)71<198:DIPITC>2.0.CO;2. [DOI] [PubMed] [Google Scholar]
- Novembre J, Stephens M. Interpreting principal component analyses of spatial population genetic variation. Nat Genet. 2008;40:646–649. doi: 10.1038/ng.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park SDE. PhD thesis. University of Dublin; 2001. Trypanotolerance in West African Cattle and the Population Genetic Effects of Selection. [Google Scholar]
- Parwati I, van Crevel R, van Soolingen D. Possible underlying mechanisms for successful emergence of the Mycobacterium tuberculosis Beijing genotype strains. Lancet Infect Dis. 2010;10(2):103–111. doi: 10.1016/S1473-3099(09)70330-5. [DOI] [PubMed] [Google Scholar]
- Rendine S, Piazza A, Cavalli-Sforza LL. Simulation and Separation by Principal Components of Multiple Demic Expansions in Europe. Am Nat. 1986;128:681–706. [Google Scholar]
- Ridley RS, Jopling WH. Classification of leprosy according to immunity—a five group system. Int J Lepr. 1966;34:225–273. [PubMed] [Google Scholar]
- Saikawa K. The effect of rapid socio-economic development on the frequency of leprosy in a population. Lepr Rev. 1981;52(1):167–175. doi: 10.5935/0305-7518.19810067. [DOI] [PubMed] [Google Scholar]
- Sakamuri RM, Harrison J, Gelber R, Saunderson P, Brennan PJ, Balagon M, Vissa V. A continuation: study and characterisation of Mycobacterium leprae short tandem repeat genotypes and transmission of leprosy in Cebu, Philippines. Lepr Rev. 2009a;80(3):272–279. [PubMed] [Google Scholar]
- Sakamuri RM, Kimura M, Li W, Kim HC, Lee H, Kiran MD, Black WC, 4th, Balagon M, Gelber R, Cho SN, Brennan PJ, Vissa V. Population-based molecular epidemiology of leprosy in Cebu, Philippines. J Clin Microbiol. 2009b;47(9):2844–2854. doi: 10.1128/JCM.02021-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott JN, McNabb J, Kammerer S, Hickey AC, Braden CR, Nong S, Rosenblum LS, Navin TR. Added epidemiologic value to tuberculosis prevention and control of the investigation of clustered genotypes of Mycobacterium tuberculosis isolates. Am J Epidemiol. 2004;160:589–597. doi: 10.1093/aje/kwh253. [DOI] [PubMed] [Google Scholar]
- Shen JP, Gupte MD, Jiang C, Manickam P, Yu MW, Li WZ. Trends of case detection and other indicators of leprosy in China during 1985–2002. Chin Med Sci J. 2005;20(2):77–82. [PubMed] [Google Scholar]
- Shen J, Zhou M, Xu X, Ray A, Zhang G, Yan L. A big challenge in case finding at low endemic situation: analysis on 1462 new leprosy patients detected in China in 2007. Lepr Rev. 2010;81:176–183. [PubMed] [Google Scholar]
- Tabet SR, Goldbaum GM, Hooton TM, Eisenach KD, Cave MD, Nolan CM. Restriction fragment length polymorphism analysis detecting a community-based tuberculosis outbreak among persons infected with human immuno-deficiency virus. J Infect Dis. 1994;169:189–192. doi: 10.1093/infdis/169.1.189. [DOI] [PubMed] [Google Scholar]
- Tanaka MM, Francis AR. Detecting emerging strains of tuberculosis by using spoligotypes. Proc Natl Acad Sci USA. 2006;103(41):15266–15271. doi: 10.1073/pnas.0603130103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Truman R, Fine PE. ‘Environmental’ sources of Mycobacterium leprae: issues and evidence. Lepr Rev. 2010;81(2):89–95. [PubMed] [Google Scholar]
- van Embden JD, Cave MD, Crawford JT, Dale JW, Eisenach KD, Gicquel B, Hermans P, Martin C, McAdam R, Shinnick TM. Strain identification of Mycobacterium tuberculosis by DNA fingerprinting: recommendations for a standardized methodology. J Clin Microbiol. 1993;31(2):406–409. doi: 10.1128/jcm.31.2.406-409.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Soolingen D, Borgdorff MW, de Haas PE, Sebek MM, Veen J, Dessens M, Kremer K, van Embden JD. Molecular epidemiology of tuberculosis in the Netherlands: a nationwide study from 1993 through 1997. J Infect Dis. 1999;180(3):726–736. doi: 10.1086/314930. [DOI] [PubMed] [Google Scholar]
- Weng XM, Chen SY, Ran SP, Zhang CH, Li HY. Immuno-histopathology in the diagnosis of early leprosy. Int J Lepr Other Mycobact Dis. 2000;68(4):426–433. [PubMed] [Google Scholar]
- Weng XM, Wang Z, Liu J, Kimura M, Black WC, 4th, Brennan PJ, Li H, Vissa VD. Identification and distribution of Mycobacterium leprae genotypes in a region of high leprosy prevalence in China: a 3-year molecular epidemiological study. J Clin Microbiol. 2007;45:1728–1734. doi: 10.1128/JCM.00018-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weng XM, Wen Y, Tian XJ, Wang HB, Tan XJ, Li HY. Preliminary study on the genotyping of Mycobacterium leprae on 50 isolates from China. Zhonghua Liu Xing Bing Xue Za Zhi. 2006;27(5):402–405. [PubMed] [Google Scholar]
- Weng XM, Wen Y, Yuan L, Yang R, Long H, Shaofeng L, Zhang W, Hong B, Wang Y, Zao X, Guo Y, Yang G, Li H. The detection of PGL- IgM and M. leprae in nasal secretion in the application of leprosy epidemiology. China J Lepr Skin Dis. 2005;21(6):425–430. [Google Scholar]
- World Health Organization. Chemotherapy of leprosy for control programmes. WHO Tech Rep Ser. 1982;675:18–21. [PubMed] [Google Scholar]
- World Health Organization. Global strategy for further reducing the leprosy burden and sustaining leprosy control activities 2006–2010. Lepr Rev. 2006;77(3):IX–X. 1–50. [PubMed] [Google Scholar]
- World Health Organization. Global leprosy situation, 2009. Wkly Epidemiol Rec. 2009;84:333–340. [PubMed] [Google Scholar]
- Xing Y, Liu J, Sakamuri RM, Wang Z, Wen Y, Vissa VD, Weng X. VNTR typing studies of Mycobacterium leprae in China: assessment of methods and stability of markers during treatment. Lepr Rev. 2009;80:261–271. [PubMed] [Google Scholar]
- Young SK, Ponnighaus JM, Jain S, Lucas S, Suneetha S, Lockwood DN, Young DB, Fine PE. Use of short tandem repeat sequences to study Mycobacterium leprae in leprosy patients in Malawi and India. PLoS Negl Trop Dis. 2008;2(4):e214. doi: 10.1371/journal.pntd.0000214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young SK, Taylor GM, Jain S, Suneetha LM, Suneetha S, Lockwood DN, Young DB. Microsatellite mapping of Mycobacterium leprae populations in infected humans. J Clin Microbiol. 2004;42(11):4931–4936. doi: 10.1128/JCM.42.11.4931-4936.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Budiawan T, Matsuoka M. Diversity of potential short tandem repeats in Mycobacterium leprae and application for molecular typing. J Clin Microbiol. 2005;43(10):5221–5229. doi: 10.1128/JCM.43.10.5221-5229.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Chen X. Local Chronicles of Qiubei County. Zhong Hua Publishing House; Beijing: 1999. [Google Scholar]