Abstract
Plackett-Burman design was employed for screening 18 nutrient components for the production of inulinase using Garlic as substrate by Streptomyces sp. in solid-state fermentation (SSF). From the experiments, 4 nutrients, namely, NH4NO3, MnSO4·7H2O, Soya bean cake, and K2HPO4 were found to be most significant nutrient components. Hence, these 4 components are selected. The selected components were optimized using response surface methodology (RSM). The optimum conditions are NH4NO3—6.63 mg/gds, MnSO4·7H2O—26.16 mg/gds, Soya bean cake—60.6 mg/gds, and K2HPO4—5.24 mg/gds. Under these conditions, the production of inulinase was found to be 76 U/gds.
1. Introduction
The current tendency in the development of new food products is that they present functional properties and that their consumption provides some health benefit. One of the aspects directly related to human health is the question of the amount of sugar and fat present. One alternative that would reduce the amount of sugar without losing flavor is the addition of fructooligosaccharides (FOSs) as a sweetener [1]. Inulin (a polyfructan consisting of linear β-2,l-linked fructose) is a linear biopolymer made up of fructose residues linked by β-2,1 bonds [2] that constitute the storage carbohydrate in the roots and tubers of different plants [3, 4]. Its hydrolysis by microbial inulinases (β-2,1-D-fructan fructanohydrolase, EC 3.2.1.7) appears to be an interesting alternative for the production of high fructose, calorie-reduced sweeteners, which are presently produced mainly by enzymatic isomerization of prehydrolyzed corn starch [2, 5–8]. This polymer is a recognized source for the production of ultra-high fructose syrups, with D-fructose content over 95% by exoenzymatic hydrolysis, the production of oligofructose syrups by endoenzymatic hydrolysis [8], or for ethanol production by fermentation. Fructose is a GRAS (generally regarded as safe) sweetener that is widely used in many foods and beverages instead of sucrose because it is up to 1.5 times sweeter than sucrose, has lower production costs, and has functional properties that enhance flavor, color, and product stability [9]. Fructose production by inulin hydrolysis is more advantageous than conventional processes based on starch, which includes the action of α-amylase, amyloglucosidase, and glucose isomerase, yielding only 45% of fructose in the final product due to the thermodynamic equilibrium of the reaction. Inulinase-based hydrolysis of inulin can yield products with 95% fructose [10, 11]. Microbial inulinases are extensively produced through submerged fermentation [12, 13]. Few studies on the production of inulinase by solid state fermentation have been recently reported [14–16]. Many hydrolytic enzymes such as lipase, protease, and phytase have been commercially produced by yeasts. Among the enzymes, inulinase has received much attention as it can be widely applied to the production of fuel ethanol and ultra-high fructose syrup from inulin. Inulin is a linear β-(2,1)-linked fructose polymer that occurs as a reserve carbohydrate in Jerusalem artichoke, dahlia tubers, or chicory root [7, 8]. Fructose is widely used in many foods and beverages instead of sucrose. Inulin can be converted into fructose by chemical approach. However, the chemical approach is currently associated with some drawbacks [7, 17]. Inulinase which catalyzes hydrolysis of inulin is produced by many microorganisms, such as Kluyveromyces, Aspergillus, Staphylococcus, Xanthomonas, and Pseudomonas. Yeasts such as Kluyveromyces fragilis, K. marxianus, Candida kefyr, Debaryomyces cantarelli, and fungi, Penicillium, and Aspergillus species are the common inulinase producers [7]. Solid state fermentation (SSF) offers numerous advantages for the production of bulk chemicals and enzymes due to low-cost substrates, simplified downstream and environmental-friendly process, reduced energy requirement, reduced wastewater produced, high yield of fermentation products, high volumetric productivity, increased product recovery, and simplicity of bioreactor design compared to submerged fermentation (SmF) [18]. The use of the sequential strategy of the experimental design is a useful tool for process optimization. In the present work, the optimization of inulinase production by Streptomyces sp. in batch process, using garlic as substrate, was carried out using a sequential strategy of the experimental design.
2. Materials and Methods
2.1. Actinomycete Strain
Microorganism used in this work is well preserved in the laboratory. Streptomyces sp. MTCC-3119 is a stock of the Microbial Type Culture collection Centre (MTCC), Chandigarh, India. The strain was maintained on solid medium at 5°C. The medium composition was comprised off the following: Yeast extract 4.0 g; Malt extract 10.0 g; Glucose 4.0 g; Agar 20.0 g; Distilled water 1.0 L; pH 7.2–7.4. Cells were harvested from slants and used to inoculate liquid medium.
2.2. Pretreatment of Substrate
Garlic (bulbs) were washed thoroughly with cold water, sliced, and then dried at 100°C for 72 h. The dried slices were then milled to a fine powder with a hammer mill. After milling, the resultant powder was used directly as a carbon source [19].
2.3. Solid State Fermentation
Pretreated garlic powder was used as substrate for inulinase production. Fermentation was carried out in Erlenmeyer flasks (250 mL) with 10 g of pretreated garlic powder, supplemented with nutrients concentrations defined by the experimental design. Moisture was adjusted to 65%. Each flask was covered with hydrophobic cotton and autoclaved at 121°C for 20 min. After cooling, each flask was inoculated with 2 mL of the suspension previously prepared and incubated for 96 hrs in a chamber with temperature and humidity control. The whole contents were incubated at 37°C [19]. During the preliminary screening process, the experiments were carried out for 5 days, and it was found that at the 24 hr, the maximum production occurs. Hence, experiments are carried out for 24 hrs. All the experiments were carried out in triplicate and the average values are reported.
2.4. Extraction of Inulinase
After fermentation, 5 volumes of distilled water were added to the fermented matter, and the contents were agitated for 30 minutes at 200 rpm on a rotary shaker (at 28°C). Then the sample was centrifuged at 15000 rpm for 20 minutes, and the supernatants were analyzed by DNS method [20].
2.5. Optimization of Inulinase Production
Response surface methodology (RSM) consists of a group of empirical techniques used for evaluation of relationship between cluster of controlled experimental factors and measured response. A prior knowledge with understanding of the related bioprocesses is necessary for a realistic modeling approach. To determine which variables significantly affect inulinase production by Streptomyces sp., Plackett-Burman design was used. Eighteen variables (Table 1) were screened in 20 experimental runs (Table 2), and insignificant ones were eliminated in order to obtain a smaller, manageable set of factors. The low level (−1) and high level (+1) of each factor are listed in (Table 1). The statistical software package “Design Expert 7.1.5” was used for analyzing the experimental data. Once the critical factors were identified through the screening, the central composite design (CCD) was used to obtain a quadratic model, consisting of factorial trials and star points to estimate quadratic effects and central points to estimate the pure process variability with inulinase production as response. RSM was employed to optimize the selected four significant nutrient components, namely, NH4NO3, MnSO4·7H2O, Soya bean cake, and K2HPO4 which enhances the inulinase production. The four independent variables were studied at five different levels (Table 3), and sets of 30 experiments were carried out (Table 4). The statistical software package “Design Expert 7.1.5” was used to analyze the experimental data. All variables were taken at a central coded value of zero. The minimum and maximum ranges of variables investigated are listed in (Table 3). Upon the completion of experiments, the average maximum inulinase was taken as the response (Y). A multiple regression analysis of the data was carried out for obtaining an empirical model that relates the response measured to the independent variables. A second-order polynomial equation is
Table 1.
Nutrient screening using a Plackett-Burman design.
| Variable | Levels (mg/g dry substrate) | ||
|---|---|---|---|
| Nutrient code | Nutrient | Low (−1) | High (+1) |
| A | Yeast extract | 1.0 | 5.0 |
| B | Beef extract | 5.0 | 15.0 |
| C | MnSO4·7H2O | 10.0 | 50.0 |
| D | K2HPO4 | 2.0 | 7.0 |
| E | Soya bean cake | 40.0 | 80.0 |
| F | MgSO4·7H2O | 0.2 | 1.2 |
| G | NH4Cl | 1.0 | 3.0 |
| H | KCl | 0.5 | 1.5 |
| J | (NH4)2HPO4 | 5.0 | 30.0 |
| K | NH4NO3 | 5.0 | 10.0 |
| L | ZnSO4·7H2O | 10.0 | 50.0 |
| M | (NH4)2SO4 | 6.0 | 10.0 |
| N | Corn steep liquor | 40.0 | 80.0 |
| O | Peptone | 5.0 | 15.0 |
| P | Dextrose | 10.0 | 30.0 |
| Q | FeSO4·7H2O | 0.05 | 2.0 |
| R | KH2PO4 | 10.0 | 60.0 |
| S | Urea | 10.0 | 30.0 |
Table 2.
Plackett-Burman experimental design matrix for screening of important variables for inulinase production.
| Run no. | A | B | C | D | E | F | G | H | J | K | L | M | N | O | P | Q | R | S | Inulinase activity (U/g dry substrate) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 1 | −1 | 1 | −1 | 1 | 1 | 1 | 1 | −1 | −1 | 1 | 1 | −1 | 1 | 1 | −1 | −1 | −1 | 27.21 |
| 2 | 1 | −1 | 1 | 1 | 1 | 1 | −1 | −1 | 1 | 1 | −1 | 1 | 1 | −1 | −1 | −1 | −1 | 1 | 49.34 |
| 3 | −1 | −1 | −1 | −1 | 1 | −1 | 1 | −1 | 1 | 1 | 1 | 1 | −1 | −1 | 1 | 1 | −1 | 1 | 52.97 |
| 4 | −1 | −1 | −1 | −1 | −1 | −1 | −1 | −1 | −1 | −1 | −1 | −1 | −1 | −1 | −1 | −1 | −1 | −1 | 15.02 |
| 5 | −1 | −1 | −1 | 1 | −1 | 1 | −1 | 1 | 1 | 1 | 1 | −1 | −1 | 1 | 1 | −1 | 1 | 1 | 22.83 |
| 6 | 1 | 1 | −1 | 1 | 1 | −1 | −1 | −1 | −1 | 1 | −1 | 1 | −1 | 1 | 1 | 1 | 1 | −1 | 27.66 |
| 7 | −1 | −1 | 1 | 1 | −1 | 1 | 1 | −1 | −1 | −1 | −1 | 1 | −1 | 1 | −1 | 1 | 1 | 1 | 15.20 |
| 8 | −1 | 1 | −1 | 1 | 1 | 1 | 1 | −1 | −1 | 1 | 1 | −1 | 1 | 1 | −1 | −1 | −1 | −1 | 28.78 |
| 9 | 1 | −1 | −1 | −1 | −1 | 1 | −1 | 1 | −1 | 1 | 1 | 1 | 1 | −1 | −1 | 1 | 1 | −1 | 20.24 |
| 10 | −1 | 1 | −1 | 1 | −1 | 1 | 1 | 1 | 1 | −1 | −1 | 1 | 1 | −1 | 1 | 1 | −1 | −1 | 14.88 |
| 11 | 1 | −1 | 1 | 1 | −1 | −1 | −1 | −1 | 1 | −1 | 1 | −1 | 1 | 1 | 1 | 1 | −1 | −1 | 21.16 |
| 12 | −1 | 1 | 1 | 1 | 1 | −1 | −1 | 1 | 1 | −1 | 1 | 1 | −1 | −1 | −1 | −1 | 1 | −1 | 30.90 |
| 13 | 1 | 1 | 1 | −1 | −1 | 1 | 1 | −1 | 1 | 1 | −1 | −1 | −1 | −1 | 1 | −1 | 1 | −1 | 50.19 |
| 14 | 1 | 1 | −1 | −1 | 1 | 1 | −1 | 1 | 1 | −1 | −1 | −1 | −1 | 1 | −1 | 1 | −1 | 1 | 10.33 |
| 15 | 1 | −1 | −1 | 1 | 1 | −1 | 1 | 1 | −1 | −1 | −1 | −1 | 1 | −1 | 1 | −1 | 1 | 1 | 11.93 |
| 16 | 1 | 1 | 1 | 1 | −1 | −1 | 1 | 1 | −1 | 1 | 1 | −1 | −1 | −1 | −1 | 1 | −1 | 1 | 11.23 |
| 17 | −1 | −1 | 1 | −1 | 1 | −1 | 1 | 1 | 1 | 1 | −1 | −1 | 1 | 1 | −1 | 1 | 1 | −1 | 45.40 |
| 18 | 1 | 1 | −1 | −1 | −1 | −1 | 1 | −1 | 1 | −1 | 1 | 1 | 1 | 1 | −1 | −1 | 1 | 1 | 16.32 |
| 19 | −1 | 1 | 1 | −1 | −1 | −1 | −1 | 1 | −1 | 1 | −1 | 1 | 1 | 1 | 1 | −1 | −1 | 1 | 41.36 |
| 20 | −1 | 1 | 1 | −1 | 1 | 1 | −1 | −1 | −1 | −1 | 1 | −1 | 1 | −1 | 1 | 1 | 1 | 1 | 45.99 |
Table 3.
Ranges of the independent variables used in RSM.
| Variable | Levels (mg/g dry substrate) | |||||
|---|---|---|---|---|---|---|
| Code | −2 | −1 | 0 | +1 | +2 | |
| NH4NO3 | X1 | 5.0 | 6.2 | 7.5 | 8.7 | 10.0 |
| MnSO4·7H2O | X2 | 10.0 | 20.0 | 30.0 | 40.0 | 50.0 |
| Soya bean cake | X3 | 40.0 | 50.0 | 60.0 | 70.0 | 80.0 |
| K2HPO4 | X4 | 2.0 | 3.2 | 4.5 | 5.7 | 7.0 |
Table 4.
Central composite design (CCD) of factors in coded levels with enzyme activity as response.
| Run no. | X1 | X2 | X3 | X4 | Inulinase activity (U/g dry substrate) | |
|---|---|---|---|---|---|---|
| Experimental | Predicted | |||||
| 1 | −1 | 1 | −1 | −1 | 32.25 | 29.92 |
| 2 | 1 | 1 | 1 | 1 | 19.38 | 24.17 |
| 3 | 1 | 1 | −1 | −1 | 25.78 | 25.83 |
| 4 | 0 | 0 | −2 | 0 | 41.94 | 35.42 |
| 5 | −1 | 1 | −1 | 1 | 40.83 | 47.50 |
| 6 | 0 | 0 | 0 | 0 | 58.43 | 59.43 |
| 7 | 0 | 0 | 0 | 0 | 60.75 | 59.43 |
| 8 | −1 | −1 | −1 | 1 | 54.69 | 55.25 |
| 9 | 0 | 0 | 0 | 0 | 59.23 | 59.43 |
| 10 | 1 | 1 | −1 | 1 | 15.32 | 12.92 |
| 11 | 2 | 0 | 0 | 0 | 23.85 | 26.08 |
| 12 | 1 | −1 | 1 | −1 | 44.89 | 47.83 |
| 13 | −1 | −1 | −1 | −1 | 23.92 | 28.17 |
| 14 | 1 | −1 | −1 | 1 | 14.37 | 24.67 |
| 15 | 1 | −1 | 1 | 1 | 50.03 | 42.92 |
| 16 | −2 | 0 | 0 | 0 | 45.18 | 42.75 |
| 17 | −1 | 1 | 1 | −1 | 25.30 | 24.67 |
| 18 | 0 | 0 | 0 | 0 | 60.34 | 59.43 |
| 19 | −1 | −1 | 1 | 1 | 45.59 | 55.50 |
| 20 | 0 | 2 | 0 | 0 | 19.42 | 23.42 |
| 21 | −1 | 1 | 1 | 1 | 45.69 | 40.75 |
| 22 | −1 | −1 | 1 | −1 | 36.80 | 29.92 |
| 23 | 1 | −1 | −1 | −1 | 31.86 | 28.08 |
| 24 | 0 | 0 | 0 | 2 | 46.05 | 37.25 |
| 25 | 1 | 1 | 1 | −1 | 48.29 | 38.58 |
| 26 | 0 | 0 | 0 | 0 | 57.44 | 59.43 |
| 27 | 0 | −2 | 0 | 0 | 45.30 | 40.42 |
| 28 | 0 | 0 | 0 | 0 | 60.23 | 59.43 |
| 29 | 0 | 0 | 2 | 0 | 42.40 | 48.42 |
| 30 | 0 | 0 | 0 | −2 | 16.42 | 24.58 |
| (1) |
where Y is the measured response, β0 is the intercept term, βi are linear coefficients, βii are quadratic coefficient, βij are interaction coefficient, and Xi and Xj are coded independent variables. The optimal concentrations of the critical variables were obtained by analyzing 3D plots. The statistical analysis of the model was represented in the form of analysis of variance (ANOVA).
2.6. Assay of Enzyme Activity
Enzymes were assayed by measuring the concentration of reducing sugars released from inulin or sucrose. The reaction mixture containing 1 mL of diluted crude enzyme and 4 mL of 2% inulin or 2% sucrose (dissolved in 0.1 M acetate buffer, pH 5.0) was incubated at 50°C. After incubating for 30 min, aliquots of 0.5 mL were withdrawn and increase in reducing sugar was estimated by a 3,5-dinitrosalicylic acid method [20] using calibration curve obtained with a standard solution of fructose [21]. Absorbance was read at 575 nm. A higher absorbance indicated a high level of reducing sugar produced and consequently a high enzyme activity. One unit of inulinase activity (U) was defined as the amount of enzyme, which forms 1 μmol fructose per min. Results of the determination of inulinase activity were presented in units of activity/gram of dry substrate (U/gds.).
3. Results and Discussion
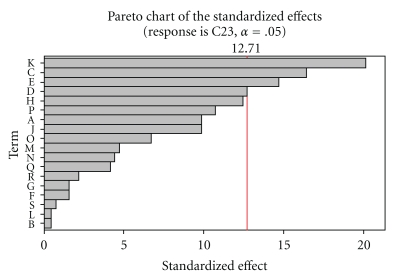
Plackett-Burman experiments (Table 2) showed a wide variation in inulinase activity. This variation reflected the importance of optimization to attain higher productivity. From the Pareto chart (Figure 1), the variables, namely, NH4NO3, MnSO4·7H2O, Soya bean cake, and K2HPO4 were selected for further optimization to attain a maximum production of inulinase.
Figure 1.
Pareto chart showing the effect of media components on inulinase activity.
The levels of factors (NH4NO3, MnSO4·7H2O, Soya bean cake, and K2HPO4) and the effect of their interactions on inulinase production were determined by central composite design of RSM. Thirty experiments were performed at different combinations of the factors shown in Table 3. The predicted and observed responses along with design matrix are presented in Table 4, and the results were analyzed by ANOVA. The second-order regression equation provided the levels of inulinase activity as the function of NH4NO3, MnSO4·7H2O, Soya bean cake, and K2HPO4 which can be presented in terms of coded factors as in the following equation:
| (2) |
where Y is the inulinase activity (U/gds), X1, X2, X3, and X4 are NH4NO3, MnSO4·7H2O, Soya bean cake, and K2HPO4, respectively. ANOVA for the response surface is shown in Table 5. The Model F value of 7.37 implies the model is significant. Values of “Prob > F” less than 0.05 indicate that model terms are significant. Values greater than 0.1 indicate that the model terms are not significant. In the present work, all the linear, interactive effects of X1X3, X1X4, and square effects of X1, X2, X3, and X4 were significant for inulinase production. The coefficient of determination (R2) for inulinase activity was calculated as 0.8931, which is very close to 1 and can explain up to 89.31% variability of the response. The predicted R2 value of 0.6764 was in reasonable agreement with the adjusted R2 value of 0.7847. An adequate precision value greater than 4 is desirable. The adequate precision value of 8.76 indicates an adequate signal and suggests that the model can be used to navigate the design space.
Table 5.
Analysis of variance (ANOVA) for response surface quadratic model for the production of inulinase.
| Source | Coefficient factor | Sum of squares | DF | F | P > F |
|---|---|---|---|---|---|
| Model | 59.43 | 5756.33 | 14 | 7.37 | 0.0002 |
| X1 | −4.17 | 416.67 | 1 | 7.47 | 0.0154 |
| X2 | −4.25 | 433.50 | 1 | 7.78 | 0.0138 |
| X3 | 3.25 | 253.50 | 1 | 4.55 | 0.0499 |
| X4 | 3.17 | 240.67 | 1 | 4.32 | 0.0553 |
| X1∗X2 | −1.00 | 16.00 | 1 | 0.29 | 0.6000 |
| X1∗X3 | 4.50 | 324.00 | 1 | 5.81 | 0.0292 |
| X1∗X4 | −7.63 | 930.25 | 1 | 16.68 | 0.0010 |
| X2∗X3 | −1.75 | 49.00 | 1 | 0.88 | 0.3634 |
| X2∗X4 | −2.38 | 90.25 | 1 | 1.62 | 0.2226 |
| X3∗X4 | −0.38 | 2.25 | 1 | 0.040 | 0.8435 |
| X1∗X1 | −6.25 | 1050.11 | 1 | 18.83 | 0.0006 |
| X2∗X2 | −6.88 | 1272.96 | 1 | 22.83 | 0.0002 |
| X3∗X3 | −4.38 | 510.11 | 1 | 9.15 | 0.0085 |
| X4∗X4 | −7.13 | 1368.11 | 1 | 24.54 | 0.0002 |
| Residual | 836.33 | 15 | |||
| Lack of fit | 825.50 | 10 | 38.10 | 0.0004 | |
| Pure error | 10.83 | 5 | |||
| Cor total | 6592.67 | 29 |
Std. Dev.: 7.47; R2: 89.31%; Mean: 39.67; Adj R2: 78.47%; C.V.%: 18.78; Pred R2: 67.64%; Adeq Precision: 8.76.
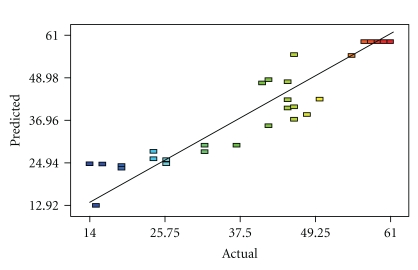
The above model can be used to predict the inulinase production within the limits of the experimental factors. Figure 2 shows that the actual response values agree well with the predicted response values.
Figure 2.
Predicted response versus actual value.
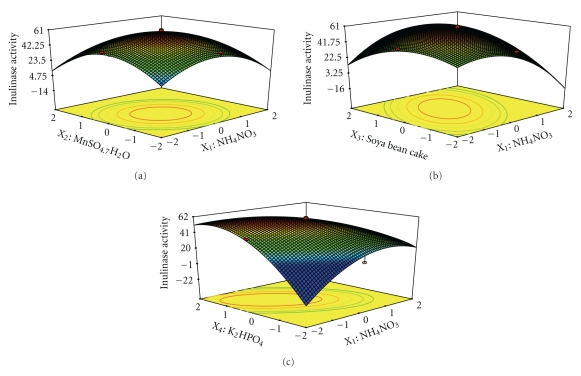
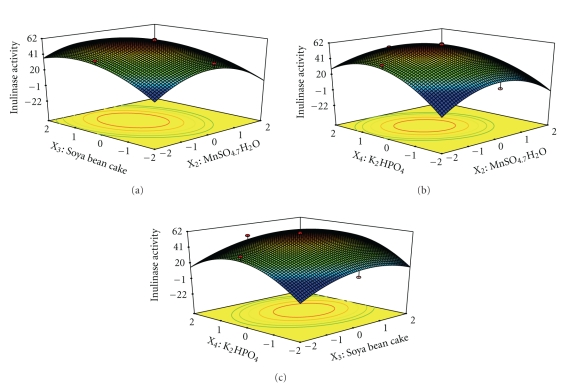
The interaction effects of variables on inulinase production were studied by plotting 3D surface curves against any two independent variables, while keeping another variable at its central (0) level. The 3D curves of the calculated response (Inulinase production) and contour plots from the interactions between the variables are shown in Figures 3(a)–3(c) and 4(a)–4(c). Figure 3(a) shows the dependency of inulinase on NH4NO3, and MnSO4·7H2O. The inulinase activity increases with increase in NH4NO3, and MnSO4·7H2O up to 6.63 and 26.16 mg/gds, respectively, and thereafter inulinase activity decreases with further increase in NH4NO3, and MnSO4·7H2O. The same trend was observed in Figures 3(b) and 3(c). Increase in Soya bean cake results in increase in inulinase activity up to 60.6 mg/gds. This is evident from Figures 3(b), 4(a), and 4(c). Figures 3(c), 4(b), and 4(c) shows the dependency of inulinase activity on K2HPO4. The effect of K2HPO4 on inulinase activity was similar to other variables. The optimum conditions for the maximum production of inulinase were determined by response surface analysis and also estimated by regression equation. The optimum conditions are NH4NO3—6.63 mg/gds, MnSO4·7H2O—26.16 mg/gds, Soya bean cake—60.6 mg/gds, and K2HPO4—5.24 mg/gds. The predicted results are shown in Table 4. The predicted values from the regression equation closely agreed with that obtained from experimental values. Validation of the experimental model was tested by carrying out the batch experiment under optimal operation conditions. Three repeated experiments were performed, and the results are compared. The inulinase activity obtained from experiments was very close to the actual response predicted by the regression model, which proved the validity of the model. At these optimized conditions, the maximum inulinase activity was found to be 76 U/gds.
Figure 3.
(a) Response surface plot for inulinase production from garlic by Streptomyces sp. in solid state fermentation as a function of NH4NO3 and MnSO4·7H2O. (b) Response surface plot for inulinase production from garlic by Streptomyces sp. in solid state fermentation as a function of NH4NO3 and Soya bean cake. (c) Response surface plot for inulinase production from garlic by Streptomyces sp. in solid state fermentation as a function of NH4NO3 and K2HPO4.
Figure 4.
(a) Response surface plot for inulinase production from garlic by Streptomyces sp. in solid state fermentation as a function of MnSO4·7H2O and Soya bean cake. (b) Response surface plot for inulinase production from garlic by Streptomyces sp. in solid state fermentation as a function of MnSO4·7H2O and K2HPO4. (c) Response surface plot for inulinase production from garlic by Streptomyces sp. in solid state fermentation as a function of Soya bean cake and K2HPO4 on inulinase activity.
4. Conclusions
In this work, Plackett-Burman design was used to test the relative importance of medium components on inulinase production. Among the variables, NH4NO3, MnSO4·7H2O, Soya bean cake, and K2HPO4 were found to be the most significant variables. From further optimization studies, the optimized values of the variables for inulinase production were as follows: NH4NO3—6.63 mg/gds, MnSO4·7H2O—26.16 mg/gds, Soya bean cake—60.6 mg/gds, and K2HPO4—5.24 mg/gds. This study showed that the garlic constitutes a good carbon source for the production of inulinase. Using the optimized conditions, the produced activity reaches 76 U/gds. The results show a close concordance between the expected and obtained activity level.
Acknowledgment
The authors wish to express their gratitude for the support extended by the authorities of Annamalai University, Annamalainagar, India in carrying out the research work in Environmental Engineering laboratory, Department of Chemical Engineering.
References
- 1.Sangeetha PT, Ramesh MN, Prapulla SG. Recent trends in the microbial production, analysis and application of Fructooligosaccharides. Trends in Food Science and Technology. 2005;16(10):442–457. [Google Scholar]
- 2.Vandamme EJ, Derycke DG. Microbial inulinases: fermentation process, properties, and applications. Advances in Applied Microbiology. 1983;29:139–176. doi: 10.1016/s0065-2164(08)70356-3. [DOI] [PubMed] [Google Scholar]
- 3.Cazetta ML, Martins PMM, Monti R, Contiero J. Yacon (Polymnia sanchifolia) extract as a substrate to produce inulinase by Kluyveromyces marxianus var. bulgaricus. Journal of Food Engineering. 2005;66(3):301–305. [Google Scholar]
- 4.Singh RS, Dhaliwal R, Puri M. Production of inulinase from Kluyveromyces marxianus YS-1 using root extract of Asparagus racemosus. Process Biochemistry. 2006;41(7):1703–1707. [Google Scholar]
- 5.Bajpai P, Margaritis A. Optimization studies for production of high fructose syrup from jerusalem artichoke using calcium alginate immobilized cells of Kluyveromyces marxianus. Process Biochemistry. 1986;21:16–18. [Google Scholar]
- 6.Manzoni M, Cavazzoni V. Hydrolysis of Topinambur (Jerusalem artichoke) fructans by extracellular inulinase of Kluyveromyces marxianus var. bulgaricus. Journal of Chemical Technology and Biotechnology. 1992;54(4):311–315. [Google Scholar]
- 7.Pandey A, Soccol CR, Selvakumar P, Soccol VT, Krieger N, Fontana JD. Recent developments in microbial inulinases its production, properties, and industrial applications. Applied Biochemistry and Biotechnology. 1999;81(1):35–52. doi: 10.1385/abab:81:1:35. [DOI] [PubMed] [Google Scholar]
- 8.Rocha JR, Catana R, Ferreira BS, Cabral JMS, Fernandes P. Design and characterisation of an enzyme system for inulin hydrolysis. Food Chemistry. 2006;95(1):77–82. [Google Scholar]
- 9.Gong F, Zhang T, Chi Z, Sheng J, Li J, Wang X. Purification and characterization of extracellular inulinase from a marine yeast Pichia guilliermondii and inulin hydrolysis by the purified inulinase. Biotechnology and Bioprocess Engineering. 2008;13(5):533–539. [Google Scholar]
- 10.Gill PK, Manhas RK, Singh P. Purification and properties of a heat-stable exoinulinase isoform from Aspergillus fumigatus. Bioresource Technology. 2006;97(7):894–902. doi: 10.1016/j.biortech.2005.04.034. [DOI] [PubMed] [Google Scholar]
- 11.Kim CH, Rhee SK. Fructose production from Jerusalem artichoke by inulinase immobilized on chitin. Biotechnology Letters. 1989;11(3):201–206. [Google Scholar]
- 12.Kalil SJ, Suzan R, Maugeri F, Rodrigues MI. Optimization of inulinase production by Kluyveromyces marxianus using factorial design. Applied Biochemistry and Biotechnology. 2001;94(3):257–264. doi: 10.1385/abab:94:3:257. [DOI] [PubMed] [Google Scholar]
- 13.Yépez Silva-Santisteban BO, Maugeri Filho F. Agitation, aeration and shear stress as key factors in inulinase production by Kluyveromyces marxianus. Enzyme and Microbial Technology. 2005;36(5-6):717–724. [Google Scholar]
- 14.Selvakumar P, Pandey A. Solid state fermentation for the synthesis of inulinase from Staphylococcus sp. and Kluyveromyces marxianus. Process Biochemistry. 1999;34(8):851–855. [Google Scholar]
- 15.Mazutti M, Bender JP, Treichel H, Luccio MD. Optimization of inulinase production by solid-state fermentation using sugarcane bagasse as substrate. Enzyme and Microbial Technology. 2006;39(1):56–59. [Google Scholar]
- 16.Bender JP, Mazutti MA, De Oliveira D, Di Luccio M, Treichel H. Inulinase production by Kluyveromyces marxianus NRRL Y-7571 using solid state fermentation. Applied Biochemistry and Biotechnology. 2006;132(1–3):951–958. doi: 10.1385/abab:132:1:951. [DOI] [PubMed] [Google Scholar]
- 17.Gill PK, Manhas RK, Singh P. Hydrolysis of inulin by immobilized thermostable extracellular exoinulinase from Aspergillus fumigatus. Journal of Food Engineering. 2006;76(3):369–375. [Google Scholar]
- 18.Pandey A. Solid state fermentation. Biochemical Engineering Journal. 2003;13:81–84. doi: 10.1016/s1369-703x(00)00065-6. [DOI] [PubMed] [Google Scholar]
- 19.Sharma AD, Kainth S, Gill PK. Inulinase production using garlic (Allium sativum) powder as a potential substrate in Streptomyces sp. Journal of Food Engineering. 2006;77(3):486–491. [Google Scholar]
- 20.Miller GL. Use of dinitrosalicylic acid reagent for determination of reducing sugar. Analytical Chemistry. 1959;31(3):426–428. [Google Scholar]
- 21.Uzunova K, Vassileva A, Ivanova V, Spasova D, Tonkova A. Thermostable exo-inulinase production by semicontinuous cultivation of membrane-immobilized Bacillus sp. 11 cells. Process Biochemistry. 2002;37(8):863–868. [Google Scholar]