Abstract
After entry into animal cells, most viruses hijack essential components involved in gene expression. This is the case of poliovirus, which abrogates cellular translation soon after virus internalization. Abrogation is achieved by cleavage of both eIF4GI and eIF4GII by the viral protease 2A. Apart from the interference of poliovirus with cellular protein synthesis, other gene expression steps such as RNA and protein trafficking between nucleus and cytoplasm are also altered. Poliovirus 2Apro is capable of hydrolyzing components of the nuclear pore, thus preventing an efficient antiviral response by the host cell. Here, we compare in detail poliovirus 2Apro with other viral proteins (from picornaviruses and unrelated families) as regard to their activity on key host factors that control gene expression. It is possible that future analyses to determine the cellular proteins targeted by 2Apro will uncover other cellular functions ablated by poliovirus infection. Further understanding of the cellular proteins hydrolyzed by 2Apro will add further insight into the molecular mechanism by which poliovirus and other viruses interact with the host cell.
1. Introduction
A great variety of animal viruses encode for proteases that accomplish crucial functions during the biological cycle of the virus [1]. Usually, the main function of these proteases is to proteolyze viral polypeptide precursors to render mature viral proteins that form part of viral capsids or participate in virus vegetative processes [2]. Although both DNA and RNA viruses can encode proteases, the proteolytic tailoring of polypeptide precursors is most common among viruses with positive single-stranded RNA genomes, such as picornaviruses, flaviviruses, caliciviruses, and retroviruses [3–7]. This mechanism of gene expression by proteolytic processing serves to compress the genetic information of viruses in the limited space provided by the genome. In this manner, viruses reduce the genetic space occupied by 5′ and 3′ untranslated regions (UTRs), the signals devoted for mRNA transcription and to initiate translation are minimal, such that, for instance, in the case of picornaviruses or flaviviruses, only one 5′ and 3′ UTR is necessary for viral replication, transcription, translation, and morphogenesis, despite the fact that several viral proteins are synthesized by the infected cells. In addition, a number of polypeptide precursors may exhibit functions that differ from those present in their mature products. In the case of poliovirus (PV), eleven mature proteins are produced from a single translation initiation event, and at least two precursors, 2BC and 3CD, accomplish functions which are not present in their mature proteins. Taking together all these considerations, the “proteolytic strategy” provides the small RNA viruses with an advantageous and efficient mechanism for distribution of the genome to accomplish all the viral biological functions with the smaller genetic space.
Apart from generation of active viral proteins that participate in capsid morphogenesis and genome replication, viral proteases may also target a number of cellular proteins. Proteolysis of these cellular substrates can very much affect a variety of cellular processes and play an important role in virus-induced cytopathogenesis [8, 9]. In this regard, productive poliovirus infection induces rapid morphological alterations in host-cell. Among them, the most prevalent is the accumulation of numerous membranous vesicles in the cytoplasm, derived from endoplasmic reticulum where the viral proteins 2C and 2BC play a central role [10]. In addition, cellular shape is modified upon viral replication giving rise to cell rounding, which is most probably induced by disorders in the cytoskeletal network [11]. Finally, chromatin condensates at late times postinfection, associated with the nuclear envelope except for sites where nuclear pores are placed [11]. Interestingly, individual expression of the viral proteases 2Apro and 3Cpro leads to the induction of most of these cytopathic effects, supporting the idea that these proteases actively contribute to the viral-induced morphological changes [12]. Indeed, long-term expression of either 2Apro or 3Cpro triggers the activation of caspases and, thus, cell death by apoptosis [11, 12], reflecting the strong cytotoxicity of both proteases. In addition to the cytopathic effects induced by 2Apro and 3Cpro, hydrolysis of host proteins may impact on other cellular functions such as the antiviral responses to virus infection. Activation of innate immunity pathways, as well as the establishment of an antiviral response, is absolutely dependent on signals traversing the nuclear membrane through the nuclear pore complex. Therefore, many viruses block cellular gene expression at different levels, that is, translation, transcription or protein and RNA trafficking between nucleus and cytoplasm. The blockade of active trafficking can inhibit the nuclear import of antiviral signals or prevent the export of cellular mRNAs detrimental to virus processes. All these effects can be achieved by hydrolysis of specific cellular proteins. The precise number of cellular proteins degraded by a viral protease, which is known as the “degradome,” still remains unknown for a given viral protease. Perhaps, one of the best-studied proteases in this respect is PV 2Apro. The discovery that PV 2Apro bisects the initiation factor of translation eIF4G leading to the regulation of translation in the infected cells has attracted much attention from many laboratories during the past three decades [13, 14]. More recently, 2Apro has been involved in the alteration of RNA and protein trafficking between the nucleus and the cytoplasm upon proteolysis of several nucleoporins [15–17]. The present paper focuses on the multifaceted activities of 2Apro and its regulation of different viral and cellular processes.
2. The Poliovirus Life Cycle: An Overview
PV is a prototype member of the Picornaviridae family that infects cells of human or simian origin cytolytically or persistently and is responsible for poliomyelitis in humans [18]. The RNA genome is housed in a naked capsid formed by 60 copies of each of the four structural proteins: VP1, VP2, VP3, and VP4. The infectious cycle commences by the attachment of a viral particle to cellular receptors present at the cell surface [19, 20]. This interaction leads to virion internalization and destabilization of the capsid, which adopts a less compact structure. Once the RNA is released in the cytoplasm, it interacts with the translational machinery, directing the synthesis of viral proteins during the early phase of infection.
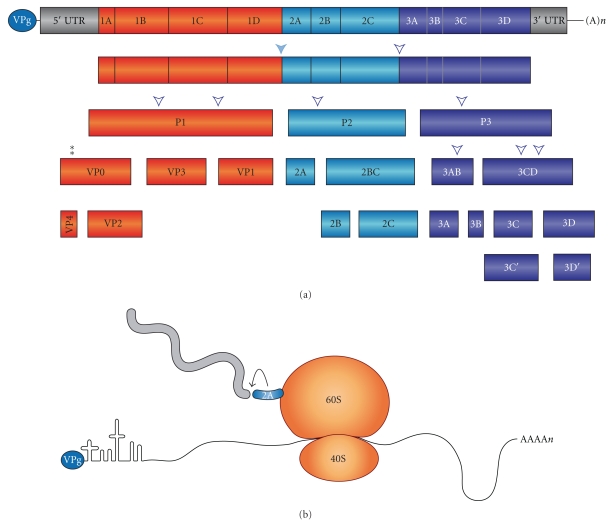
The PV genome is composed of a single-stranded RNA copy of positive polarity of about 7.4 Kb [21, 22]. This RNA molecule is uncapped and contains a poly(A) tail at its 3′ end and a single open reading frame, which encodes for a polyprotein of about two thousands amino acid residues. This polyprotein is proteolytically processed giving rise to the mature viral proteins [23] (Figure 1(a)). Three different cleavages can be distinguished on the viral polyprotein: (i) polysomal cleavages that are produced on the nascent polypeptide chain. The first of these cleavages is catalyzed by 2Apro at its amino terminus separating the P1 precursor that encodes for the structural proteins from the rest of nonstructural polypeptides (Figure 1(b)). The second cleavage still on polysomes is performed by 3Cpro, releasing the P2 precursor (2ABC) from P3 (3ABCD); (ii) cytoplasmic cleavages that are mostly exerted by 3Cpro and (iii) hydrolysis of VP0 (VP4–VP2), which is concomitant with the morphogenesis of virus particles [2, 23]. All these hydrolytic events lead to the formation of eleven mature proteins and several precursors such as P1, P2, P3, VP0, VP3, VP1, 2BC, 3AB, and 3CD. This last precursor, 3CD, can be used as substrate by 2Apro or 3Cpro. The alternative cleavage carried out by 2Apro renders the mature products 3C′ and 3D′, whereas 3Cpro generates the canonical proteins 3Cpro and 3Dpol. However, the biological significance of this alternative cleavage is obscure because PV mutated at 2Apro-cleavage site on 3CD does not exhibit defects in virus replication [24].
Figure 1.
(a) Structure of poliovirus genome and proteolytic processing of its polyprotein. The PV genome consists of a single-stranded, positive-sense polarity RNA molecule, which encodes a single polyprotein. The 5′ nontranslated region (NTR) is covalently linked to the viral protein VPg. The PV genome is polyadenylated (An) in its 3′ end. The polyprotein contains four structural (P1) and seven nonstructural (P2 and P3) proteins that are released from the polypeptide chain by proteolytic processing mediated by the viral-encoded proteinases 2Apro and 3Cpro /3CDpro. The intermediate products of processing 2BC, 3CD, and 3AB exhibit functions distinct from those of their respective final cleavage products. The alternative cleavage carried out by 2Apro rendering the mature products 3C′ and 3D′ is also shown. (b) Once the ribosome has synthesized the PV 2Apro sequence and continues translation on the P2 region, the autocatalytic activity of PV 2Apro is manifested by cleaving itself at its amino terminus still on the nascent polypeptide chain. This cleavage liberates the P1 precursor that will render the capsid proteins on subsequent proteolytic events catalyzed by 3Cpro or 3CDpro. Cleavage at the carboxy terminus of PV 2Apro on the P2 precursor is accomplished by PV 3Cpro, leaving free 2Apro and generating the 2BC precursor.
The nonstructural proteins that are generated participate in the replication of viral genomes [25, 26]. To this end, the positive RNA genome is recognized at its 3′ end by proteins of the replication complex to synthesize the complementary RNA strand of negative polarity. In this process, 3B protein, also known as VPg, acts as a primer to initiate viral RNA transcription [27]. This leads to the formation of a double-stranded RNA molecule, also known as the replicative form. The negative RNA synthesized serves in turn as a template to direct the synthesis of several copies of positive RNA, so this process leads to the production of several nascent RNA molecules with a VPg molecule bound to their 5′ ends on the negative RNA molecule forming a replicative intermediate. The positive RNA molecules synthesized may participate in three processes (i) to serve as templates for synthesizing more negative RNA molecules; (ii) as mRNAs that will be engaged in translation, and (iii) as genomes that will be encapsidated in new viral particles. In picornaviruses, the only type of mRNA molecule known is exactly the same as the genome.
Once the synthesis of several thousands of positive RNA molecules is performed, the late phase of translation takes place, giving rise also during this period to a great amount of viral proteins, some of which will participate in virus morphogenesis. This late phase of infection is preceded by the abrogation of cellular mRNA translation, such that only viral proteins are being synthesized late in the PV life cycle [14]. In the case of picornaviruses, transcription is dependent on continuous viral protein synthesis [28]. Thus, inhibition of viral mRNA translation provokes the sudden blockade of viral RNA synthesis. Moreover, translation is coupled to transcription, such that viral RNAs transfected into picornavirus-infected cells are not able to direct protein synthesis [29]. Therefore, these two processes of viral macromolecular biosynthesis are tightly coupled, making it difficult to determine exactly the function affected in some PV mutants. Notably, continuous lipid and cellular membrane synthesis is also necessary for PV RNA synthesis [30]. The morphogenesis of progeny virions in PV-infected cells is observed concomitantly with viral RNA translation and replication. The release of new viral particles takes place by cell lysis, due to membrane permeabilization that occurs at the late phase of infection [31]. Viroporin 2B and its precursor 2BC are responsible for this permeabilization upon the formation of pore channels in cellular membranes [32, 33].
PV has represented a useful model to gain insight into diverse aspects of molecular biology and gene expression. A number of discoveries concerning animal viruses with RNA genomes were initially made in PV. For example, the presence of uncapped mRNA, the sequencing and development of an infectious cDNA clone, the three-dimensional structure of a virus particle, the discovery of the IRES elements, the synthesis of an infectious virus in a cell-free system, the chemical synthesis of a complete viral genomes, among others, were initially reported in PV [34–38]. In addition, the first time that eIF4G was found proteolytically cleaved was in PV-infected cells [39].
3. Picornavirus Proteases: L, 2A and 3C
Picornaviruses encode different proteases depending on the virus species although it is common to all of them to encode 3Cpro and its precursor 3CDpro (Figure 2). In PV, both these exhibit protease activity, and they execute most of the hydrolytic events on the viral polyprotein [2, 23, 40]. Apart from these two proteases, 3Cpro and 3CDpro, picornaviruses also contain a 2A gene, whose product in some species exhibits proteolytic activity, as is the case for PV (Figure 2). 2Apro has a limited proteolytic effect on the polyprotein and its function is most probably one of altering cellular functions by the cleavage of a number of cellular proteins. In this regard, the best studied of these cleavages is the bisection of eIF4G (Figure 3) [14].
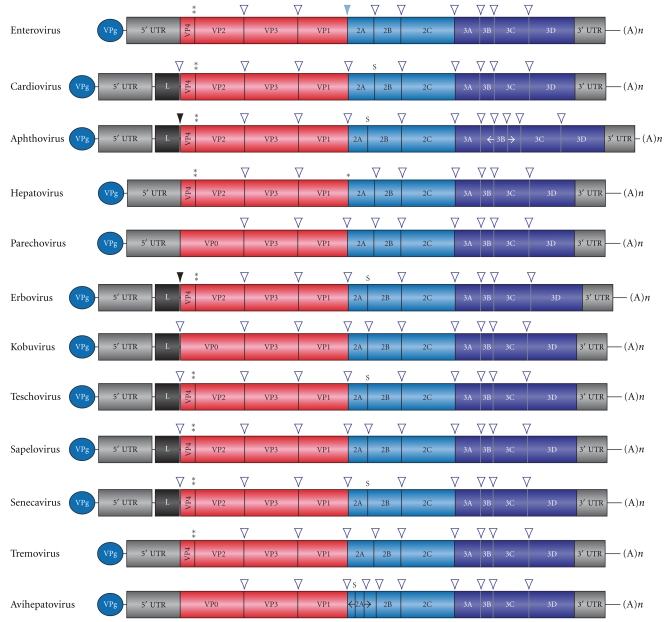
Figure 2.
Genomic structure of the different members of the Picornaviridae family. The single-stranded, positive-sense polarity RNA genome encodes a single polyprotein, which is different depending on family member [41]. Black arrow, Lpro cleavage site; two asterisk indicate the maturation cleavage between VP4 and VP2; blue arrow, enterovirus 2Apro cleavage site; an asterisk indicates the unknown cellular protease that achieves P1-2A cleavage in hepatovirus; empty arrow, 3Cpro cleavage sites; S, ribosome skipping at Asn-Pro-Gly-Pro site.
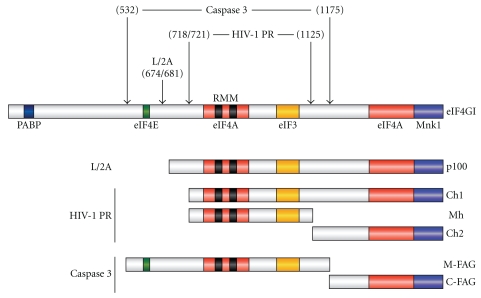
Figure 3.
Schematic representation of eIF4GI and cleavage sites. The boxes indicate binding sites for eIF4A, eIF4E, elF3, Mnk1 kinase and PABP. Arrows indicates the site of cleavage of eIF4GI by PV 2Apro, HIV-1 PR and caspase-3. Cleavage of eIF4GI by PV 2Apro generates a carboxi-terminal polypeptide named p100. These fragments are different from those produced after cleavage by HIV-1 PR and caspase-3.
The general organization of the picornavirus genomes is to encode for P1-P2-P3 precursors giving rise to 4-3-4 mature products. Some picornavirus species, in addition, encode a leader protein (L) placed before P1 (Figure 2) [22]. In the case of aphthoviruses, such as foot-and-mouth disease virus (FMDV), the L protein has proteolytic activity and it is known as Lpro [42, 43]. During polyprotein synthesis Lpro is the first protein synthesized and its autoproteolytic activity releases itself from the rest of the polypeptide chain. Thus, the only known hydrolysis executed by Lpro on the polyprotein is to hydrolyze between its carboxy terminus and the amino terminus of VP4 (Figure 2). Because Lpro does not play a direct role in viral replication [44] and its protease activity has a limited impact in the viral polyprotein, this protease may be involved in the interaction with the host-cell [45–47]. Indeed, Lpro also exhibits proteolytic activity on eIF4G acting at a position close to that of PV 2Apro (Figure 3) [48–50]. In the case of FMDV, the 2A protein is reduced to a small peptide of 18 residues that does not hydrolyze eIF4G; instead it induces the release of 2A from its carboxy terminus by a ribosomal skip mechanism [51, 52]. This model proposes that FMDV 2A modifies the activity of the ribosome to promote hydrolysis of the peptidyl(2A)-tRNA(Gly) ester linkage at the C-terminus of 2A, thereby releasing the polypeptide from the translational complex. However, not all L or 2A proteins from picornaviruses exhibit protease activity, since in the case of EMCV, which encodes both proteins, neither has been demonstrated to possess proteolytic activity [53].
All known proteases have been classified in four classes and many subgroups according to three parameters: (i) their catalytic center, (ii) their substrate specificity, and (iii) their three-dimensional structure. The classification of the different picornavirus proteases initially relied upon the effect of protease inhibitors. Compounds that blocked sulphydryl groups abrogated the proteolytic activity of 2Apro and 3Cpro, suggesting that the nucleophilic aminoacid in the active site was cysteine [54, 55]. However, another cysteine inhibitor such as E64 had no effect on these proteases, while Lpro activity was inhibited not only by sulphydryl-active compounds but also by E64 [56, 57]. These findings together with structural observations imply that Lpro belongs to the class of papain-like cysteine proteinases [43, 58–60]. Comparison of the structure of picornavirus proteases with prototypes of cellular ones revealed that both 2Apro and 3Cpro are similar in structure to the chymotrypsin like group [61–63]. Picornavirus 3Cpro reflects similarities to the staphylococcus aureus proteinase, whereas 2Apro is more akin to streptomyces griseus proteinase A.
4. The PV 2A. Structure-Function Relationship
PV 2Apro is a protein composed of 149 amino acids that belongs to the cysteine protease group [54]. PV 2Apro is autocatalytically processed at its amino terminus between the capsid protein VP1 and 2A [64] (see Figure 1(b)). The determinants of substrate specificity of picornaviral PV 2Apro have been investigated in detail by identification of cleavage sites by N-terminal Edman degradation, mutational analysis and using synthetic peptides as substrates [34, 65–67]. PV 2Apro can recognize a wide variety of amino acid residues at the P1 position. The determinants of substrate specificity for PV 2Apro lie at positions P4, P2, P1′, and P2′, which are preferentially occupied with Ile/Leu, Thr/Ser, Gly, and Pro, respectively. Moreover, the determinants of substrate specificity of HRV and coxsackievirus 2Apro are very similar to those found for PV 2Apro [65–67]. The yeast two-hybrid system has been used to identify the substrate sequence interacting with PV 2Apro. All the sequences identified contain the Leu-X-Thr-Z motif (X for any amino acid; Z for a hydrophobic residue) in positions from P4 to P1 suggesting the presence of a common interacting site on PV 2Apro substrates [68].
Several 2Apro variants have been generated in the entire PV genome or in the isolated 2A gene. Generation of PV 2Apro mutants was initially used to identify the Cys106, His18, and Asp35 as the residues that form part of the catalytic triad of 2Apro [69, 70]. These data have been confirmed in the structure of HRV2 2Apro [71]. The role of the conserved Cys and His residues in the structure-function relationship has also been studied by mutagenesis. The residues Cys55, Cys57, Cys115, and His117 play a critical role in the cis and trans proteolytic activity by maintaining 2Apro structure [72]. The structure of HRV2 2Apro shows that the Zn2+ ion is coordinated tetrahedrally by the side chains of these conserved Cys and His residues. This Zn2+ ion is tightly bound near to the C-terminal domain and may be important for the stability of the 2Apro [71, 73, 74].
The yeast Saccharomyces cerevisiae has been used as a system to obtain PV 2Apro variants [75]. The fact that this protease is very toxic for yeast has been exploited to generate 2Apro variants devoid of this cytotoxicity. Using this approach, several PV 2Apro unable to cleave eIF4G have been obtained. The characterization of these mutants revealed a region in 2Apro involved in the interaction with substrates but none of the mutations were found in the catalytic triad. A parallelism has been observed between the ability of these PV 2Apro variants to block protein synthesis and to cleave eIF4G [76]. PV mutants in 2A gene that lack trans but not cis proteolytic activity have been also identified. Normal processing of the viral polyprotein is observed upon infection with these PV variants, whereas eIF4G remains intact in these cells [77]. Interestingly, RNA replication of those mutant viruses is hampered, suggesting that there is a correlation between PV RNA replication and the trans activity of 2Apro. It is controversial whether PV 2Apro contributes directly to viral replication or not. Although for many years it was thought that PV 2Apro plays a direct role in PV replication, more recent studies have shown that a full-length dicistronic PV construct lacking 2Apro is capable to give rise to progeny viruses. Moreover, virus yields of PV variants lacking the P1 coding region is partially restored when P1 is expressed in trans, suggesting that cleavage of the viral polyprotein by PV 2Apro is not essential for viral replication. [78]. However, it is known that 2Apro is important for inducing the cytophatic effect and for avoiding the inhibition of PV replication in interferon (IFN) α treated cells [79]. In agreement with the idea that 2Apro participates in viral RNA replication, a fraction of this protease localizes in PV replicative foci although the majority of 2Apro is associated with the matrix structure in the cytoplasm of infected cells [80]. However, the presence of 2Apro in the proximity of replication complexes does not demonstrate that it participates directly in the replication process.
5. PV 2A Hijack Host Protein Synthesis Machinery by Cleaving eIF4G
5.1. Structure and Functioning of eIF4G
The process of translation can be divided in different steps: initiation, elongation, termination and ribosome recycling. The synthesis of cellular proteins is highly regulated, and in this sense, the most precisely controlled step is the initiation of translation (for a recent review, see [81, 82]). For most eukaryotic mRNAs, the initiation of translation commences with the recognition of the cap structure (7mGpppN) and the poly(A) tail by the heterotrimeric complex eIF4F and the poly(A)-binding protein (PABP), respectively, followed by the recruitment of the 43S preinitiation complex containing the 40S ribosomal subunit, the ternary complex Met-tRNAiMet-eIF2-GTP, and the eukaryotic initiation factors (eIFs), 1, 1A, 3, and 5. Then, the preinitiation complex scans along the 5′ untranslated region until an AUG initiation codon is encountered in a favourable context. The perfect complementarity between the AUG start codon and the anticodon of Met-tRNAi leads to the arrest of scanning and the hydrolysis of GTP in the ternary complex. The release of eIF2-GDP and other factors triggers the interaction of the preinitiation complex with the 60S ribosomal subunit to form the 80S initiation complex, proceeding to translation of the mRNA coding region.
A number of eukaryotic initiation factors participate in both mRNA binding and scanning of the 5′ UTR of the mRNA by the small ribosomal subunit. The cap-binding protein eIF4E, together with the DEAD-box helicase eIF4A and the translation initiation factor eIF4G, forms the protein complex eIF4F. The heterotrimeric complex eIF4F is required for recruiting the 43S preinitiation complex onto the cap structure located at the 5′ end of the mRNA [83]. In this sense, eIF4G, the larger polypeptide of eIF4F, functions as an adaptor molecule that bridges the mRNAs to ribosomes via interactions with factors eIF4E (which binds the 5′cap structure), PABP (which binds the poly(A) tail), and eIF3, which interacts with the 40S ribosome subunit (Figure 3) [81, 84–86]. In addition, eIF4G also contains binding sites for other polypeptides involved in translation, such as the RNA helicase eIF4A (Figure 3) [87], which is required to unwind the secondary structure within the mRNA 5′ leader sequence that would otherwise inhibit ribosome scanning. The simultaneous interaction of eIF4G with eIF4E and PABP promotes circularization of the mRNA in a closed loop that facilitates the initiation of new rounds of translation by the proximity of the 5′ and 3′ ends [88, 89]. Furthermore, eIF4G also interacts with the mitogen-activated protein kinase 1 (Mnk1) (Figure 3), which phosphorylates eIF4E, although the role of this phosphorylation in the initiation of translation in still unclear [90–94]. Two forms of eIF4G, known as eIF4GI and eIF4GII, have been identified in mammalian cells. Both forms show only 46% amino acid sequence identity but they are thought to be functionally interchangeable due to the high homology in key domains that interact with other factors [88]. Evidence obtained by specific depletion of each eIF4G form or differential cleavage of each of them by specific proteases (see below) points to the idea that both factors should be lacking for complete abolition of protein synthesis [95, 96]. eIF4GI is the dominant form in HeLa cells, in which the ratio between eIFGI and eIF4GII is 9 : 1 [97]. However, the specific role of each form of eIF4G in the initiation of translation remains unknown. It was proposed that both eIF4G forms are differentially regulated by different kinases, supporting the hypothesis that eIF4GI and eIF4GII could drive differentially translation initiation [98]. eIF4GI is phosphorylated in response to serum and in a rapamycin-dependent manner at Ser 1148, 1188, and 1232, although the role of these posttranslational modifications is still under investigation [99]. In addition, eIF4GI is phosphorylated by p21-activated protein kinase (Pak-2) that is induced under stress conditions. This phosphorylation takes place in the eIF4E-binding site of eIF4G and avoids the interaction between these two factors, inhibiting cap-dependent initiation of translation [100]. On the other hand, eIF4GII is phosphorylated during mitosis [101] by calmodulin-dependent kinase I at Ser1156 [102]. Therefore, activity of eIF4GI and eIF4GII might be tightly and reversibly regulated by phosphorylation under different physiological conditions. Nevertheless, this factor is also subjected to irreversible modifications such as caspase-mediated proteolysis, which is triggered during apoptosis and leads to shutoff of protein synthesis. Caspase-3 cleaves directly eIF4GI in positions 532 and 1175 removing PABP, Mnk1, and one eIF4A-binding domain from the eIF4GI core (Figure 3) [103, 104]. In contrast, eIF4GII is degraded during apoptosis with a delayed kinetics in relation to eIF4GI proteolysis, correlating with the shutoff of the protein synthesis [104].
Furthermore, many eIF4GI isoforms have been detected in HeLa cells and these are synthesized from several distinct mRNAs via alternative promoter usage and alternative splicing [105]. The largest is the eIF4GI-a isoform, which contains 1,600 residues, while the eIF4GI-b, -c, -d, and –e are shorter variants [106]. It has been described that the longer isoforms are more active in translation initiation, most probably because they contain the PABP-binding site [107].
5.2. Cleavage of eIF4G by PV 2Apro
Infection of cells with PV results in a rapid shutoff of host-cell protein synthesis, whereas viral mRNA translation takes place efficiently [108]. It was initially observed that the inhibition of host-cell translation in PV-infected cells correlated with the proteolysis of a component of the eIF4F complex with a molecular mass of about 220 kDa (later identified as eIF4G) [39, 109]. This cleavage is exerted by 2Apro and can be prevented by both insertion of mutations that abolish the protease activity and addition of 2A inhibitors [76, 110, 111]. Interestingly, this proteolysis is more effective when eIF4E is interacting with eIF4G, suggesting that PV 2Apro preferentially acts on the eIF4G pool involved in translation [112]. Cleavage of eIF4GI also occurs in cells infected with other picornaviruses such as HRV, coxsackievirus, and FMDV [48, 113, 114]. Interestingly, 2Apro from PV, HRV, and coxsackieviruses cleave eIF4GI at positions 681/682 (Figure 3), suggesting the conservation of the specificity of enterovirus 2A proteases for the substrate determinants present in eIF4GI [114, 115]. Cleavage at position 681/682 separates eIF4E- and eIF3-binding sites of eIF4GI, contained in N-terminal and C-terminal fragments respectively, thus decoupling mRNA and ribosome recruiting activities. The PV 2Apro cleaves eIF4GI directly and does not require any additional proteins for this process to occur [116, 117]. However, the fact that PV 2Apro is not copurified with eIF4GI fragments from PV-infected cell extracts suggest that 2Apro induced the activation of a host protease, which in turn cleaves eIF4G during PV infection [112]. In addition, it has been proposed that eIF3 and an unknown host-cell protein could act as cofactors for eIF4GI cleavage by PV 2Apro [118]. In this sense, Zamora and colleagues suggested that PV infection activates at least two host-cell proteases, which together with PV 2Apro, cleave eIF4GI [119]. Nevertheless, no additional evidence has been put forward to support this hypothesis and the identity of these host proteases has not yet been determined.
Many reports have demonstrated that the kinetics of protein synthesis shutoff and eIF4GI cleavage are not correlated in PV-infected cells [120–122]. These data clearly indicate that additional translation factors may be cleaved to achieve an efficient inhibition of cellular mRNA translation. In this regard, additional reports showed that eIF4GII is also proteolyzed by 2Apro during PV and HRV infections and that this cleavage is exerted between amino acids 699/700 leading to a proteolytic pattern similar to eIF4GI. Interestingly, eIF4GII is significantly more resistant to 2Apro-mediated cleavage than eIF4GI and the kinetics of protein synthesis shutoff close correlates with eIF4GII cleavage in PV- and RHV-infected cells [122, 123]. In those studies, Gradi and colleagues proposed that hydrolysis of both eIF4GI and eIF4GII is required for achieving PV- and HRV-mediated inhibition of host-cell mRNA translation and that the cleavage of eIF4GII is the rate-limiting step in the shutoff of host-cell translation after infection with those viruses [122, 123].
A variety of approaches have been devised to express PV 2Apro in order to cleave eIF4G in culture cells or in cell-free systems. Of these approaches, the most straightforward system has been the addition of the purified PV 2Apro, usually as a hybrid protein such as MBP-2Apro, to cell-free systems such as rabbit reticulocyte lysates (RRLs), HeLa and Krebs-2 extracts [76, 118, 124–127]. In this sense, the addition of about 1 to 5 μg MBP-2Apro suffices to hydrolyze eIF4G in those cell-free systems [76, 125–127]. An alternative method to cleave eIF4G in an in vitro system is the translation of an mRNA encoding PV 2Apro in translation competent extracts [50]. This assay has the advantage of providing genuine and freshly made PV 2Apro, leading to total cleavage of eIF4G in the test tube after several minutes of translation. Many different approaches have been explored in culture cells, the most popular being transfection of plasmids encoding PV 2Apro in different eukaryotic cell types [75, 76, 128–134]. Several plasmids have been utilized in this respect, and perhaps the most successful one is pTM1-2A, which is transfected in mammalian cells that transiently express T7 RNA polymerase by infection with a recombinant vaccinia T7 virus [76, 130, 131, 133, 134]. The amount of protease synthesized in this system is similar to that found in PV-infected cells at late times of infection, but these amounts are reached 1-2 hours after transfection. Similar results have been obtained in cells constitutively expressing T7 polymerase, which comprises a less pleiotropic system, because vaccinia virus proteins are not expressed (unpublished data). Since PV 2Apro targets a number of different cellular proteins, which affect several cellular functions depending on the amount of protease synthesized (see below), in some instances, it is useful to express 2Apro at low levels. We have explored many alternative methods trying to get a system that allows us to control the levels of PV 2Apro into the cells. Novoa and colleagues developed a protocol based on the addition of hybrid proteins bearing PV 2Apro. These recombinant proteins enter into the cytoplasm on cell membrane permeabilization by different methods such as addition of MBP-2Apro mixed with replicationally inactive chicken adenovirus particles [135]. Cleavage of eIF4G following these protocols takes place after incubation for 8–10 hours, suggesting that the amount of protease internalized is probably low but sufficient to hydrolyze eIF4GI in virtually all culture cells [136]. Probably one of the most attractive systems is a stable cell line that inducibly express PV 2Apro, obtained in two different laboratories including ours [137, 138]. In these cell lines, PV 2Apro is synthesized when tretracycline is removed from the culture medium, leading to low expression of PV 2Apro that induces efficient cleavage of eIF4G after 13 h post induction correlating with a potent inhibition of cellular translation. Finally, long term expression of PV 2Apro in Tet off cell lines triggers apoptosis [12, 137, 138]. The main drawback of this cell line is the low PV 2Apro escape under repression conditions that gives rise to a basal cytotoxicity. Probably, the most efficient method is electroporation of an mRNA encoding 2Apro under the control of EMCV leader sequence (IRES-2A) [95]. The biggest advantage of this method is the capacity to regulate levels of 2Apro expression by controlling the amounts of IRES-2A transfected. For example, electroporation of 9 μg of IRES-2A into ~1.5 · 106 HeLa cells leads to total cleavage of both eIF4GI and eIF4GII in only 2 h, resulting in an almost complete shutoff of cellular protein synthesis. In contrast, electroporation of low amounts of IRES-2A (1 μg) into HeLa cells induces efficient cleavage of eIF4GI, whereas eIF4GII remains largely intact. Therefore, 9-fold more IRES-2A mRNA is required to cleave eIF4GII compared to eIF4GI [95]. Based on the IRES-2A mRNA electroporation method we were able to induce the differential proteolysis of eIF4GI and eIF4GII in a time- and dose-dependent manner. Kinetics of protein synthesis shutoff and eIF4GII cleavage is closely correlated in HeLa cells, resembling what was found in PV-infected cells [122]. In agreement with what was observed with the addition of exogenous recombinant proteins [136], translation of de novo synthesized mRNAs showed higher susceptibility to low doses of PV 2Apro than mRNAs already engaged in translational machinery [95]. These results suggested a possible specific role of eIF4GI in the pioneer round of translation in agreement with a previous report [139]. However, specific ablation of eIF4GI using siRNAs induced a moderate inhibition of luciferase synthesis from de novo synthesized and preexisting mRNA (about 40% in both cases) [96]. These findings reported by Welnowska and colleagues indicated that the higher susceptibility of de novo synthesized mRNA translation to low doses of IRES-2A might be produced by an additional effect of PV 2Apro on another gene expression step. In this regard, further studies demonstrate that the stronger impact of 2Apro on de novo synthesized mRNAs is due to the concomitant inhibition of RNA nuclear export by Nucleoporin 98 cleavage, which is also achieved under these conditions (see below) [17]. Interestingly, cellular mRNAs are able to initiate translation after a polysome runoff with high salt treatment when eIF4GI is totally cleaved by PV 2Apro, whereas it is completely abolished when both forms of eIF4G are proteolyzed [95]. Taken together these set of data from cell expressing 2Apro [95, 136] as well as from PV-infected cells [120–122] we can conclude that complete shutoff of the protein synthesis induced by PV 2Apro is achieved when both eIF4GI and eIF4GII are completely cleaved. Therefore, when the levels of one of the two populations of eIF4G remain unaffected either because it is not cleaved by PV 2Apro [95] or it is not depleted by siRNAs [96], extensive host protein synthesis takes place.
The infection of PV and coxsackievirus also leads to hydrolysis of PABP [140, 141]. This cleavage is carried out by PV 3Cpro and coxsackievirus 2Apro and 3Cpro and it might actively contribute to the host translational shutoff induced by these viruses [142, 143]. In conclusion, the proteolysis of different components of the translation initiation machinery by picornavirus proteases can account for the shutoff of host translation induced after infection although the specific contribution of hydrolysis of eIF4GI, eIF4GII, and PABP remains still unclear.
5.3. Other Viral Proteases That Hydrolyze eIF4G
Infection of animal cells with FMDV also leads to proteolysis of eIF4G and to rapid inhibition of cellular translation [48]. The proteolysis of eIF4G is carried out by the two virally encoded proteases Lpro and 3Cpro [144, 145]. Lpro cleaves both eIF4GI and eIF4GII extremely rapidly at positions 674 (Figure 3) and 700, respectively, located seven and one amino acids upstream of the 2Apro cleavage sites on eIF4GI and eIF4GII [50, 146]. The cleavage of eIF4G by FMDV Lpro results in the rapid shutoff of host-cell protein synthesis [145]. Although the initial cleavage of eIF4GI can be carried out by FMDV Lpro in the absence of virus replication [145], a sequential cleavage of the C-terminal fragment of eIF4GI by FMDV 3Cpro also occurs in BHK cells at early stages of infection concomitant with the shutdown of viral translation. The 3Cpro cleavage site on eIF4GI has been located at position 712, 38 amino acids downstream of the Lpro cleavage site [147] although the role of this sequential cleavage is still unclear. The amino acid segment of eIF4G located between the Lpro and 3Cpro cleavage sites binds RNA and was suggested to be critical for mRNA scanning by the preinitiation complex [148]. Interestingly, this secondary cleavage does not occur in human cell lines due to an amino acid substitution at the cleavage site on eIF4GI [147].
Infection of cells with other picornaviruses such as cardioviruses (EMCV and mengovirus) leads to a shutoff of host-cell protein synthesis. However, eIF4G remains unaffected in these cells. These findings indicate that apart from eIF4G cleavage, there are other mechanisms that may block host translation by picornaviruses.
In addition to picornavirus Lpro and 2Apro, proteases from other viruses can also cleave eIF4G. The protease of human immunodeficiency virus type-1 (HIV-1) hydrolyzes eIF4GI during infection of human CD4+ cells [149]. The cleavage of eIF4GI takes place at positions 718, 721, and 1125, separating it in three domains (Figure 3) [149, 150]. Interestingly, HIV-1 protease efficiently cleaves eIF4GI, but not eIF4GII, both in cell-free systems and in mammalian cells [151]. The differential sensitivity of eIF4GI and eIF4GII to HIV-1 protease is more selective than that observed with picornaviral 2A proteases [122, 123]. HIV-1 protease also cleaves PABP at positions 237 and 477 separating the two first RNA-recognition motifs from the C-terminal domain of PABP [152]. Cleavage of eIF4GI and PABP by HIV-1 protease is sufficient to inhibit the translation of capped and polyadenylated mRNAs in cell-free systems, as well as in transfected cells [127, 151]. In contrast, IRES-driven translation is unaffected or even enhanced by HIV-1 PR after cleavage of both eIF4GI and PABP [127, 151]. Moreover, the translation of capped and polyadenylated HIV-1 genomic mRNA remains unaffected in HeLa extracts under these conditions suggesting that viral protein synthesis might persist at late phases of HIV-1 infection where those factors are cleaved [127]. In contrast, a previous report claimed that the hydrolysis of eIF4GI impaired the translation of both capped and IRES-driven mRNAs in reticulocyte lysate assays [150]. However, the different effect observed by these authors on IRES-driven translation can be due to differences already reported between HeLa extract and RRL [143, 153]. In addition, eIF4G cleavage is executed by proteases from other retrovirus species, such as HIV-2, simian immunodeficiency virus (SIV), human T-cell leukemia virus (HTLV-1), Moloney murine leukemia virus (MoMLV), and mouse mammary tumor virus (MMTV). These proteases hydrolyze eIF4GI and eIF4GII with different cleavage patterns and kinetics [134]. And indeed, several retroviruses, including HIV, SIV, and MoMLV, promote the translation of their gag gene products by internal ribosome entry, indicating that eIF4G cleavage could be compatible with viral protein synthesis in infected cells [154–156]. Furthermore, cleavage of eIF4GI and eIF4GII also occurs in feline calicivirus-infected cells although the cleavages occur at different sites to those observed for picornavirus proteases [157]. In addition, the 3C-like protease of two caliciviruses, like PV 3Cpro, cleaves PABP perhaps as a complementary strategy to inhibit cellular translation [158]. The fact that proteases from many picornaviruses, retroviruses and caliciviruses, target eIF4G, and, in some cases, PABP, strongly suggest that those viruses may share a common mechanism to regulate cellular and viral translation. However, further investigation is required to determine the specific contribution of eIF4GI, eIF4GII and PABP to the shutoff of host-cell translation and virus protein synthesis.
6. The Nuclear Pore and the Inhibition of Nucleus-Cytoplasm Trafficking by PV 2Apro
The biological cycle of picornaviruses is confined to the cytoplasm of infected cells. However, some of the PV proteins are able to target nuclear proteins such as transcription and splicing factors and proteins involved in nuclear-cytoplasmic trafficking. One of the best studied cases in this regard is the cleavage of nucleoporins (Nups), components of the nuclear pore complex (NPC), by PV and HRV 2Apro, which directly impacts on nuclear-cytoplasmic trafficking of proteins and RNAs [15, 16, 159]. Complementarily, EMCV L protein affects on the phosphorylation status of Nup62 and induces similar effects to those described for PV 2Apro in the transport of macromolecules through NPC [160]. Nevertheless, Nups are not only targets for picornavirus proteins but also for matrix (M) protein from Vesicular stomatitis virus (VSV) [161] and nonstructural protein 1 (NS1) from influenza virus [162], which also impair components of the nuclear export-import host machinery following analogous mechanisms. Taking into account that proteins from different positive and negative strand RNA viruses target Nups, we highlight in this paper NPC as a key target for viral proteins, although the possible role of NPC in virus biological cycle is still under intensive research.
6.1. Macromolecular Trafficking between Nucleus and Cytoplasm
Nucleus and cytoplasm are physically separated by a semipermeable barrier known as the nuclear envelope. Due to this compartmentalization, a large number of macromolecules traverse the nuclear envelope to reach their biological destination. For example, proteins are synthesized by ribosomes in the cytoplasm, but some of them such as polymerases, transcription factors, nucleosome components and splicing factors, have to traverse the NPC to reach the nucleoplasm. Conversely, all RNA species are transcribed and processed in the nucleus and later, most of mature RNAs are exported through the NPC to the cytoplasm, where their biological roles take place. Therefore, the regulation of RNA and protein trafficking between nucleus and cytoplasm directly impacts on gene expression [163, 164].
NPC forms large structures (~125 MDa) embedded in the nuclear envelope with a polarized eightfold symmetrical core. It is sandwiched by a cytoplasmic and nuclear ring, which projects eight filaments of about 50 nm into the cytoplasm and a basket-like structure of about 100 nm into the nucleoplasm [165, 166]. The NPC is composed of multiple copies (8, 16 or 32) of ~30 different proteins, called Nups, that are grouped in three major classes: (i) the phenylalanine-glycine (FG)-containing nucleoporins that actively work in the nuclear-cytoplasmic trafficking of macromolecules; (ii) the structural components, which lack FG-rich domains and (iii) the membrane integral proteins, which anchor the NPC to the nuclear envelope [163, 167]. Whereas the two last groups of nucleoporins play a role in the architecture and localization of the NPC, the FG-nucleoporins directly regulate the transport of RNAs and proteins through the NPC, and they are the main nucleoporin class targeted by viruses (Figure 4).
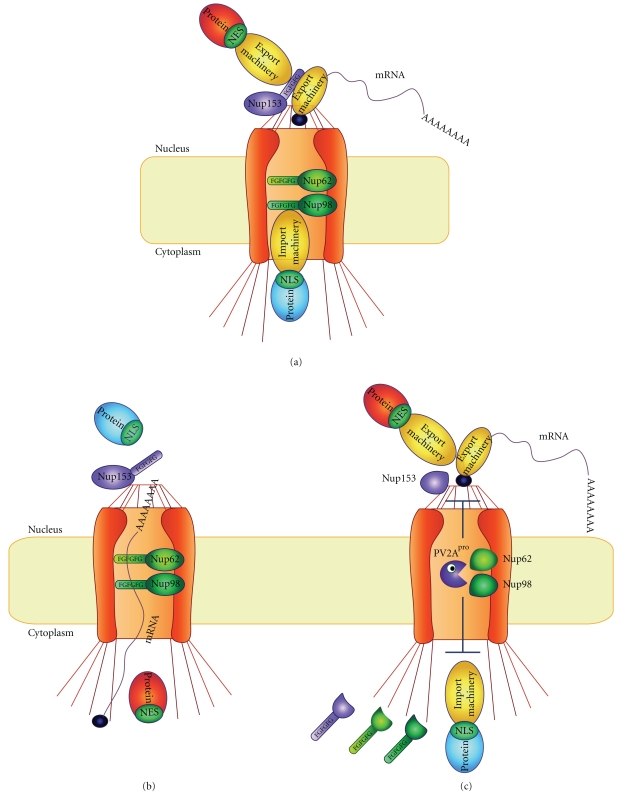
Figure 4.
Inhibition of nucleus-cytoplasm trafficking of macromolecules by PV 2Apro. (a and b) Proteins and RNAs (represented in the scheme by an mRNA) are imported and exported throughout the NPC by the mediation of FG nucleoporins (Nup98, Nup62, and Nup153) and import and export carriers. (c) However, cleavage of these nucleoporins by PV 2Apro induces the releasing of the FG domains impairing the interaction of these components of the NPC with the trafficking machinery.
Movement of ions, metabolites and other small molecules between nucleus and cytoplasm takes place by passive diffusion; however, transport cargos larger than 40 KDa require the participation of specific receptors and carriers [168, 169]. FG nucleoporins are placed on both cytoplasmic and nucleoplasmic sides of the NPC and play a central role in the active transport of macromolecules. The FG domains of nucleoporins are unfolded regions that participate in energy-independent transient interactions with the cargo receptors during the docking and translocation processes [170]. Nevertheless, the delivery of the cargo and some of the directional steps require hydrolysis of GTP. Trafficking of proteins through NPC is mediated by a family of conserved transport receptors named karyopherins, which recognize short peptides known as nuclear localization signal (NLS) and nuclear export signal (NES) [169, 171]. In addition, Karyopherins also recognize nucleotide sequences during the export of some classes of RNAs [163]. Due to their key role, Karyopherins involved in cargo import are known as importins and those involved in export are known as exportins. The RanGTP cycle also plays a central role in karyopherin activity and, therefore, in trafficking of macromolecules through the NPC. Importins bind the cargo in the cytoplasm and release it on binding to RanGTP in the nucleus. In contrast, exportins bind the cargo in the nucleus together with RanGTP; and then the Ran-associated GTP is hydrolyzed in the cytoplasm by RanGAP and cargo is liberated [163, 164].
Proteins containing constitutively active NLS are predominantly nuclear; but in some cases, the accessibility of NLS or the NLS itself is modified to selectively regulate the localization of the protein [172]. A similar regulatory mechanism is also exerted for control of NES activity [173]. For example, the NLS of the transcription factor NFκB is masked by the interaction with the inhibitor IκBα. However, IκBα is degraded on proinflammatory stimuli, exposing the NLS of NFκB for importin recognition. Therefore, under proinflammatory conditions, NFκB is imported to the nucleus, where it triggers a specific gene response [172]. In addition to “masking strategies,” phosphorylation, ubiquitination or methylation of NLS or NES also influences (negatively or positively) their recognition by importins or exportins. Therefore, trafficking of proteins between nucleus and cytoplasm could be finely regulated by posttranslational modifications in the NLS and NES [164, 174].
Conceptually, export of most of nuclear RNA follows a similar mechanism to that described above for protein trafficking. This process also involves cargo receptors, export factors, and nucleoporins to deliver mature RNA to the cytoplasm, but in this case, structure and function of nuclear export signals are not well understood. Aminoacylated tRNAs are necessary in the cytoplasm for protein synthesis, but tRNAs are transcribed in the nucleus. tRNA export to the cytoplasm is mediated by exportin-t, which belongs to the karyopherin family. Exportin-t forms a complex together with RanGTP in the nucleus, and once the exportin-t-RanGTP-tRNA complex reaches the cytoplasm, RanGAP induces the hydrolysis of the Ran-associated GTP and the release of the tRNA [175, 176]. It has been proposed that exportin-5 could act as an auxiliary protein in this process [177]. However, later reports in yeast have opened the possibility of alternative pathways for tRNA nuclear export [163].
Traffic of snRNA follows a complex mechanism involving adaptor proteins. snRNAs are transcribed by the RNA polymerase (Pol) II (with the exception of U6 snRNA which is produced by PolIII) and then they are capped but not polyadenylated. Cap-binding proteins (CBP) 80 and 20 interact with the cap of snRNAs in the nucleus and recruit an export adaptor known as PHAX [178]. This adaptor is phosphorylated and in this state, it is able to interact with CRM1, another member of karyopherin family. This interaction together with the joining of RanGTP is essential for snRNA trafficking [163]. Hydrolysis of RanGTP and dephosphorylation of PHAX lead to the release of the snRNA in the cytoplasm.
Ribosomal proteins are assembled together with the different rRNAs in the nucleolus following a complex process of maturation to give rise to the ribosomal subunits 40S and 60S. Although is known that preribosomal subunits are exported by separate routes that involve CRM1 and RanGTP, nowadays the exact mechanism followed by 40S preribosomal subunits to leave the nucleus remains unclear [163]. However, 60S preribosomal subunit relies on Nmd3 adaptor, which mediates the interaction with CRM1 [179, 180]. The release of the 60S preribosomal subunit requires two GTPase steps: (i) the hydrolysis of the Ran-associated GTP induces the liberation of CRM1; and (ii) the hydrolysis of GTP mediated by the cytoplasmic GTPase Lsg1, which induces the release of Nmd3 [181].
Finally, mRNAs compose the most heterogeneous group of RNAs, varying in length and structure. Thus, different export factors and adaptor proteins associate with each subpopulation of mRNAs [163]. mRNAs are transcribed by PolII, and, concomitantly with this process, a number of RNA-binding proteins assemble with them. These RNA-binding proteins exert different modifications in immature mRNAs such as polyadenylation, splicing and capping. In addition, export factors and adaptor proteins are also recruited to nascent pre-mRNAs, playing a further function in nuclear export. TREX (transcription-coupled export) complex is recruited to the 5′ end of nascent pre-mRNAs in a splicing-dependent manner by means of the interaction of one of its components, namely ALY/REF, with the subunit of the nuclear cap-binding complex CBP80 [182]. The TREX complex also recruits the conserved RNA-helicase UAP56 that is important for mRNP biogenesis [183]. TAP-p15, (also known as NXF1-NXT1) directly binds the mRNA immediately after splicing and actively participates in mRNA export. ALY/REF, TAP-p15 and UAP56 associate with exon junction complexes (EJC), which are deposited in a splicing-dependent manner at 20–24 nt of every exon-exon junction [184, 185]. This interaction network makes spliced mRNAs more susceptible to export and couple splicing and mRNA-trafficking. In fact, unspliced mRNAs are exported by an alternative and less efficient pathway that involves CRM1 and RanGTP. This alternative route is also followed by mRNAs encoding some protoncogenes and cytokines [186, 187]. It is important to mention that although mRNA can follow different nuclear export pathways, in all cases the interaction of export receptors with nucleoporins plays an essential role in the transport of the mRNPs throughout the NPC [170].
6.2. Inhibition of Nucleus-Cytoplasm Protein and RNA Trafficking by PV 2Apro
PV infection strongly impacts on host-cell protein localization, giving rise to an unusual cytoplasmic distribution of nuclear proteins [188]. This particular effect has been characterized by different laboratories for a number of nuclear factors involved in several cellular processes and containing different types of NLSs [189]. Nevertheless, not all nuclear proteins are re-localized after PV infection, evidencing the presence of a viral-specific mechanism affecting protein subcellular distribution [188]. Gustin and colleagues proposed the inhibition of nuclear protein import machinery as the cause of the cytoplasmic accumulation of nuclear proteins in PV-infected cells (Figure 4). They demonstrated that PV impairs protein trafficking across the NPC by expressing GFP proteins encoding classical or transporting NLSs in mock and infected cultured cells [15, 16, 159]. These recombinant proteins accumulate in the cytoplasm after PV infection, being almost completely depleted from the nucleus. Nevertheless, PV does not affect GFP distribution when the NLS is mutated or deleted, since the small size of GFP allows its inefficient efflux throughout the NPC [15]. Interestingly, cell-free nuclear import assays demonstrated that NLS-containing GFP is unable to traverse the NPC when cells are previously infected with PV [15]. In agreement with these findings, shuttling endogenous proteins, such as heterogeneous nuclear ribonucleoprotein (hnRNP) A1 and hnRNP K, are detected in the cytoplasm of infected cells from 3 hpi but are undetectable in the nucleus at 4.5 hpi. However, cytoplasmic accumulation of nuclear resident proteins such as hnRNP C requires longer times of infection [15]. These findings support the idea that the distribution of a protein that shuttles between nucleus and cytoplasm may be strongly altered by the disruption of protein trafficking pathways, as compared to nuclear resident proteins. However, not all the nuclear factors are redistributed to the cytoplasm of PV-infected cells. This is the case for SC35 (a serine/arginine-rich splicing factor), fibrillarin or TATA-binding protein (TBP), which remain in the nucleus for the duration of infection. The different behaviour of these groups of proteins could arise as a consequence of different turnover; thus, the distribution of highly stable nuclear proteins might be less affected by the inhibition of protein nuclear import than those proteins with low stability. Alternatively, PV might not impair all protein import pathways, and some might remain operative, thereby allowing the nuclear import of several families of nuclear proteins. Interestingly, some nuclear factors such as La antigen [190], PTB [191], Sam68 [192], or nucleolin [188] have been shown to interact with PV RNA or viral proteins. These proteins accumulate in the cytoplasm of PV-infected cells and, consequently, their availability for viral replication is increased [188, 193–195]. Cytoplasmic accumulation of nuclear factors might be produced mostly by the blockade of nuclear-cytoplasmic trafficking. However, loss of the NLS after the cleavage of PTB and La proteins by PV 3Cpro may also contribute to the subcellular relocalization of those proteins in PV-infected cells [193, 195]. Redistribution of nuclear proteins as well as impairment of cellular import machinery was also observed in cells infected with other picornaviruses such as HRV [159] or EMCV [160]. These findings indicate that most picornaviruses might share similar strategies to impair nuclear-cytoplasmic trafficking machinery.
An alternative hypothesis has been proposed as the cause of the redistribution of nuclear factors. Belov and colleagues reported that the nuclear envelope is permeabilized on PV infection, allowing nuclear proteins to diffuse across the nuclear membrane [196]. Indeed, electron microscopy revealed that PV 2Apro induces severe structural damage in NPC [196]. However, this hypothesis did not clarify why other proteins resident in the nucleus do not diffuse to the cytoplasm after PV infection. Both protein nuclear import and permeabilization of nuclear envelope could be integrated together as sequential steps in PV biological cycle. Protein import blockade is detected early after infection [16], correlating with the first modifications in the NPC (see below). However, prolonged expression of viral proteins might induce nuclear membrane leakiness, reflected by stronger alterations in NPC architecture [196]. Nevertheless, what might the biological relevance of these events be for a cytoplasmic virus? The most evident answer, which was extensively commented above, is that inhibition of nuclear protein import and further nuclear envelope leakiness might increase the presence of nuclear proteins in the cytoplasm of infected cells, which has been proposed in some cases to play a relevant role in PV replication. In addition, many transcription factors are arrested in the cytoplasm of uninfected cells (e.g., NF-κB, IRF7, and IRF3), but they are immediately activated and imported to the nucleus after proinflammatory extracellular signals or on the activation of intracellular sensors as a consequence of the viral replication. Once in the nucleus, these factors trigger the transcription of a set of genes involved in the antiviral response [197]. Inhibiting the import of these transcription factors, PV might prevent or, at least attenuate, the establishment of a hostile intracellular environment. Further effort will be made in the future to explore this attractive hypothesis.
Interestingly, Nup98, Nup153 and Nup62, components of the NPC belonging to FG Nup family, were found to be degraded in PV- as well as HRV-infected cells (Figure 4) [15, 16, 159]. These proteins are essential factors of the nuclear-cytoplasmic trafficking machinery since their N-terminal FG-rich domains serve as docking sites for soluble transport factors [163, 198]. Nup98, Nup153 and Nup62 are proteolyzed in PV-infected cells following different kinetics. Thus, Nup98 is the cleaved early after infection (from 1 hpi), whereas Nup153 and Nup62 are targeted at late times after infection (from 4 hpi) [15, 16]. In agreement with these findings, cleavage of Nup98 is induced even in presence of inhibitors of PV replication, suggesting that small amounts of viral proteins are sufficient for this proteolysis to occur. However, cleavage of Nup153, and Nup62 are efficiently prevented on arrest of viral replication, probably because they are only efficiently achieved when large amounts of viral proteins are produced [16]. Therefore, Nup98, Nup153, and Nup62 exhibit different susceptibilities to PV replication, thus PV might have a gradual impact on the nuclear-cytoplasmic trafficking machinery. Nup153 is also proteolyzed by caspase-3 and caspase-9 during apoptosis induction; however, the involvement of these cellular proteases in PV-induced Nup cleavage has been ruled out by different laboratories. First, Nup153 cleavage products generated upon caspase activation differ to those found in PV-infected cells [159]. In addition, Nup62 is cleaved in PV-infected cells, but it remains intact despite caspase activation [199]. Most importantly, PV-induced NPC structural damage takes place in cells lacking caspase 3 and 9 [196], and Nup153 and Nup62 are efficiently cleaved in PV-infected cells even in presence of the caspase inhibitor Z-VAD [159]. All together, these data support the idea that one or more viral proteins play a direct role in the cleavage of those Nups. In agreement with this hypothesis, PV-induced NPC damage is prevented by PV 2Apro inhibitors such as elastatinal, elastase, and MPCMK, suggesting an involvement of this viral protease in the alteration of NPC [196]. Indeed, individual expression of PV 2Apro in HeLa cells as well as addition of this protease to cell-free systems gives rise to Nup98, Nup62, and Nup153 cleavage [16, 17, 200].
In agreement with the data obtained from PV-infected cells, on PV 2Apro expression in HeLa cells, Nup98 is cleaved faster than Nup62 and Nup153, which suggests the presence of optimal cleavage sites in this protein [17]. Proteolysis of Nup98 in PV-infected cells as well as in cell-free systems generates two different cleavage products of around 50–65 KDa and 35 KDa [16]. There are two optimal cleavage sites in Nup98 for PV 2Apro located between aminoacids 373-374 and 551-552, containing Gly at P1′, Thr at P2, and Leu at P4. Hydrolysis at both sites results in N- and C-terminal products with predicted molecular masses of 37 and 53 or 55 and 35 KDa, in good agreement with the size of the peptides detected experimentally in PV-infected cells and 2A-treated HeLa extracts [16, 17]. An explanation for the delayed kinetics of Nup62 and Nup153 with respect to Nup98 is that optimal PV 2Apro cleavage sites were not found in these Nups (unpublished data). Recently, Park and colleagues have reported that PV 2Apro directly cleaves Nup62 at six different positions rendering multiple proteolytic products. These cleavage sites are located between aminoacids 103 and 298, thus releasing the FG-rich region from the protein core [200]. Functionally, loss of the FG-rich region might make Nup62 inactive for interaction with cargo receptors. This hypothetical mechanism of Nup62 functional decoupling could be extrapolated to Nup98 and Nup153 (Figure 4). However, it remains unknown whether PV 2Apro is able to directly cleave Nup98 and Nup153 or where cleavage might occur.
Nup98, Nup153 and Nup62 are also involved in RNA export from the nucleus and therefore, cleavage by PV 2Apro might also impact on this process (Figure 4). However, oligo d(T) hybridization studies showed that PV infection does not affect distribution of the polyadenylated mRNA bulk after 3 hpi [16]. A more detailed analysis revealed that expression of PV 2Apro in HeLa cells induces a number of disorders in RNA location. Nuclear export of cellular mRNAs is inhibited in 2Apro-expressing cells in a dose dependent manner concomitantly with Nup98, Nup62 and Nup153 cleavage [17]. Interestingly, mRNA export of constitutively expressed mRNAs such as β-actin is less affected than that of newly synthesized mRNAs. For example, tetracycline-induced luciferase mRNA was almost totally retained in the nucleus when these Nups are cleaved by PV 2Apro. This effect was also observed for endogenous mRNAs such as IL-6, c-myc or p53 mRNAs which are induced on PV 2Apro expression. Therefore, PV 2Apro could counteract the induction of proapoptotic (c-myc and p53) and proinflammatory (IL-6) responses by accumulating the c-myc, p53 and IL-6 mRNAs in the nucleus [17]. This export blockage may prevent the establishment of a host-cell response against PV infection. These findings could explain why PV 2Apro is essential for replication of PV in cells pre-treated with IFN-α [79]. Furthermore, impairment of mRNA export strongly alters the localization of mRNAs with high turnover as compared to constitutively expressed and highly stable mRNAs such as β-Actin [17]. As observed in PV-infected cells, oligo d(T) hybridization revealed that PV 2Apro expression hardly affects the distribution of the polyadenylated mRNA pool after short times of expression (8 h). However, nuclear accumulation of polyadenylated mRNA bulk is detected when cells are exposed to PV 2Apro for longer times (16 and 24 h). In this regard, the progression of the alterations of polyadenylated mRNA localization in 2Apro-expressing cells takes place as follows: (i) disruption of nuclear mRNA-containing foci (ii) appearance of mRNA-containing granules in the cytoplasm (most probably stress granules) and (iii) depletion of cytoplasmic mRNAs. These events were more clearly observed when high amounts of PV 2Apro are synthesized, reflecting that nuclear accumulation of mRNAs is a time- and dose-dependent process [17].
Nevertheless, PV 2Apro is not only able to block mRNA export, but also rRNA and snRNA transport. Both 18S rRNA and U2 snRNA accumulate in the nucleus of 2Apro-expressing cells in a dose-dependent manner. Most probably, cleavage of Nup98, Nup62 and Nup153 is involved in these effects, since rRNA, snRNA, and mRNA are exported using different cargo receptors and auxiliary proteins (see above) but all of them relay in Nup activity to traverse NPC [163]. Importantly, tRNAs (val-tRNA) are exported normally despite PV 2Apro expression, indicating that some RNA nuclear export pathways are not affected by this viral protease. In fact, Nup98, Nup62, and Nup153 are not directly involved in tRNA export [163], reinforcing the idea that nucleoporin cleavage plays a central role in the impairment of protein and RNA trafficking by PV (Figure 4).
Notably, IFN-γ induced a specific increase of Nup98 levels in HeLa cells that counteracts the inhibition of mRNA export by PV 2Apro [17, 201]. Collectively, these findings reflect the central role of Nup98 in PV infection and in antiviral response, since its overexpression by itself prevents, at least in part, blockade of nuclear RNA export. Therefore, secretion of IFN-γ by immune cells might allow the induction of antiviral response by neighbouring cells by increasing the levels of Nup98 in order to protect nuclear-cytoplasmic protein and RNA trafficking pathways. The physiological relevance of the crosstalk between Nup98-PV 2Apro in PV (and other viruses) infection might be studied in the future with cellular and animal systems.
6.3. Viral Proteins That Target the Nuclear Pore
As mentioned above, HRV also induces cleavage of Nup62, Nup153 and most probably Nup98, leading to the impairment of nuclear protein import [159]. Nevertheless, PV and HRV 2Apro exhibit high homology and both proteases are therefore expected to share common targets. In contrast, the 2A gene of cardioviruses encodes a short peptide with autoproteolytic activity but lacks trans-protease activity. However, EMCV and mengovirus are able to damage NPC [202]. As occurs in PV-infected cells, these cardioviruses induce both protein nuclear import inhibition and late membrane leakiness but they do not induce the cleavage of Nups [160, 202, 203]. Cardioviruses encode an additional protein known as L protein, which is highly cytopathic although it lacks protease activity (in contrast to aphthovirus Lpro). L protein contains a Zinc finger domain, and an acidic region, which is proposed to be phosphorylated in infected cells [204]. Individual expression of this protein in cultured cells or in cell-free systems induces several cellular disorders including the inhibition of protein nuclear import that resembles that observed in EMCV-infected cells [160, 203]. Several studies reported EMCV L protein mutations that resulted in defective virus growth phenotypes in cell culture [202, 204]. In particular, mutations in the zinc finger domain (Cys19Ala and Cys22Ala) or in the acidic region (Thr47Ala) partially avoid blockade of protein trafficking between the nucleus and the cytoplasm [202]. Taken together, these data support the involvement of cardiovirus L protein in NPC damaging and in the inhibition of protein nuclear import, inducing Nups phosphorylation rather than their cleavage. Indeed, Nup62 is quickly and strongly phosphorylated after EMCV infection (2 hpi), and it was clearly detected by conventional Western blotting. Nevertheless, analysis with Pro-Q diamond phosphoprotein stain revealed that Nup153 and Nup214 are also phosphorylated to a certain extent upon EMCV infection. Notably, Nups phosphorylation was avoided when Cys19Ala mutation was inserted in the L protein sequence, suggesting that the Zinc finger domain is essential for this posttranslational modification to occur [160]. However, the exact role of Nups phosphorylation in nuclear-cytoplasmic trafficking is still unknown. The idea that Nups phosphorylation could regulate protein import and RNA export as a switch that turns the different pathways on/off should be pursued in more detail. As mentioned above, Ran is essential for the regulation of most of the nuclear export and import pathways, because it acts as a cofactor modulating the affinity of importins and exportins for the cargo. The RanGTP cycle is described in detail in Section 6.1. It has been described that EMCV L directly interacts with Ran, and this interaction is abrogated by the insertion of C19A mutation in L [203]. However, the potential role of L/Ran interaction in the modulation of RanGTP cycle and its impact in nuclear import and export pathways have not yet been studied.
Alteration of nuclear-cytoplasmic trafficking is not only restricted to picornaviruses but has also been observed with negative strand viruses such as vesicular stomatitis virus (VSV) and influenza virus. Her and collaborators reported that RNA export and protein import are strongly inhibited by VSV matrix (M) protein, by microinjection of oocytes with radiolabeled RNAs and proteins. Radiolabeled tRNAs, mRNAs, U snRNAs, and rRNAs were injected directly into the nucleus of Xenopus laevis oocytes, and the subcellular localization of those RNAs was monitored by autoradiography. The conclusion of this work is that mRNA, U snRNA, and rRNA but not tRNA trafficking is blocked by VSV M protein [205], in agreement with our findings on 2Apro-expressing HeLa cells [17]. In addition, protein nuclear import was monitored by microinjection of radiolabeled proteins containing NLS in the oocytes cytoplasm. This assay revealed that VSV M protein abrogates protein nuclear import to the same extent as treatment with specific inhibitors of this pathway such as WGA [205]. These interesting findings support the idea that PV 2Apro and VSV M protein could target similar host proteins to impair macromolecule trafficking between nucleus and cytoplasm. In agreement with this possibility, it was found that the VSV M protein interacts with Nup98 [161], which is one of the primary targets of PV 2Apro [16]. The N-terminal domain of VSV M is sufficient to block RNA nuclear export and aa 52–54 may play an essential role in this blockade, because their mutations to Ala completely abrogate this inhibitory effect. Indeed, the N-terminal domain of M protein is involved in the interaction with Nup98 and, in particular, aa 52–54, because their mutation to Ala blocks the binding of VSV M to Nup98 [161]. In addition, binding of M to Nup98 requires active mRNA export pathways since, treatment with inhibitors such as WGA hampers this interaction. VSV M is also able to induce the accumulation of endogenous polyadenylated mRNAs in the nucleus of HeLa cells, and this effect is prevented again by mutations in aa 52–54 of M [161]. Furthermore, VSV M interacts with Rae1, which plays an essential role in mRNA nuclear export by its interaction with Nup98 and mRNPs. Overexpression of either Nup98 or Rae-1 prevents the nuclear accumulation of polyadenylated mRNAs, suggesting that both factors may play a role in the blockade of mRNA nuclear export by VSV M protein [206]. Interestingly, IFN-γ specifically increases the level of both Nup98 and Rae-1 and indicates a potential antiviral effect of these proteins. Indeed, overexpression of both proteins by IFN-γ treatment counteracts the inhibitory effects of VSV M protein on mRNA nuclear export, highlighting the possibility of a crosstalk between M and IFN-γ that might control the fate of the viral replication in infected animals [201, 206].
Influenza virus replication also impacts on nuclear-cytoplasmic trafficking and leads to the nuclear accumulation of host mRNAs [162, 207]. Influenza virus NS1 protein is a major virulence factor that is essential for pathogenesis, because it impairs innate and adaptive immunity by inhibiting host signal transduction and gene expression [208, 209]. NS1 forms a complex with NXF1/TAP, p15/NXT [162], Rae1, and E1B-AP5, which are components of the mRNA nuclear export machinery (see above). Individual expression of NS1 in 293T cells induces the accumulation of polyadenylated mRNAs in the nucleus, suggesting that the interaction of NS1 with these export factors yields an inactive complex for mRNA export. Influenza virus also induces a strong reduction of Nup98 steady-state levels although the viral mechanisms involved in this process are still unknown [162]. Expression of reporter luciferase mRNA synthesized from a nuclear plasmid is inhibited by NS1. However, this inhibition is overcome by overexpression of NXF1, p15, Rae-1 or Nup98, evidencing the role of NS1 interaction with these factors in the impairment of mRNA nuclear export [162]. Furthermore, mouse cells expressing low levels of Nup98 or/and Rae-1 show greater susceptibility to influenza infection, resulting in a significant increase in cell death and virus production. In addition, mRNAs encoding antiviral factors or immunomodulators such as IRF-1, MHC I and ICAM1 accumulated more in the nucleus of those cells than in cells expressing normal levels of Nup98 or Rae-1 [162]. All these data support the physiological role of NS1 interaction with RNA export factors as well as the reduction of Nup98 levels in influenza pathogenicity. Interestingly, VSV M, influenza NS1 and PV 2Apro expression gives rise to similar effects on mRNA trafficking. All these viral proteins target Nup98 and other components of the cellular machinery involved in nuclear-cytoplasmic trafficking.
7. Other Cellular Proteins Hydrolyzed by PV 2Apro: Hijacking the Cellular Splicing and Transcription Machinery
Survival of motor neurons (SMN) complex is composed by SMN and a class of proteins called Gemins, which localize in both cytoplasm and nucleoplasm [210, 211]. Gemin7 and Gemin8 constitute the core of the complex where the other Gemins associate by means of numerous protein-protein interactions from the periphery. The SMN complex is involved in the biogenesis of uridine-rich small nuclear ribonucleoprotein (U snRNP) in the cytoplasm and then the U snRNP carries out the splicing of pre-mRNAs in the nucleus [212–214]. The snRNPs are composed of the major U snRNAs U1, U2, U4, U5, and U6 as well as a group of seven proteins known as Sm ribonucleoproteins that collectively make up the extremely stable Sm core of the snRNP. Gemins (except Gemin-2) associate with Sm proteins to form a heptameric ring structure in the presence of U snRNAs [211]. After Sm core assembly, the U snRNPs are imported to the nucleus, localizing in foci known as Cajal Bodies, where further maturation processes take place [215]. Gemin-3, one of the main components of SMN complex, is cleaved in PV-infected HeLa cells leading to a 50 KDa cleavage product. Scission of Gemin-3 negatively impacts on the kinetics of Sm core assembly, which is prevented in presence of inhibitors of PV replication [216]. These results indicated that high levels of PV proteins are required for this process to occur, as is the case of Nup153, Nup62, and eIF4GII, [16, 122]. PV 2Apro is able to hydrolyze purified Gemin-3 in vitro, rendering a cleavage product similar to that found in PV-infected HeLa cells [216]. Only one potential 2Apro-cleavage site, between the amino acids Tyr462 and Gly463 (VHTYG), was found in this SMN complex component. Proteolysis of Gemin-3 at this position would render two cleavage products of about 50–30 KDa, in agreement with the polypeptide of about 50 KDa found in PV-infected and 2Apro-expressing cells. In addition, G463E mutation avoids direct hydrolysis of Gemin-3 exerted by PV 2Aproin vivo and in vitro [216]. Taken together, these findings support the notion that VHTYG is the cleavage site for PV 2Apro in Gemin-3. Although hydrolysis of Gemin-3 is exerted in cells transfected with plasmid encoding PV 2Apro, it does not take place when this protease is expressed from exogenous mRNAs; contrary to that found with eIF4GI, eIF4GII, Nup98, Nup153 and Nup62 [17] (and unpublished data). A probable explanation for this difference is that PV 2Apro is expressed at lower levels from transfected mRNAs than from plasmids [95] (Castello et al., 2006). Thus, Gemin-3 cleavage may be a very late event in PV-infected cells because it requires expression of high amounts of PV 2Apro. Gemin-3 hydrolysis may directly impact on pre-mRNA splicing since this event reduces the availability of SMN complexes, which is involved in U snRNPs biogenesis. Nevertheless, Alstead and colleagues could not detect any apparent effect of Gemin-3 proteolysis in splicing of cellular pre-mRNAs [216]. Therefore, the physiological relevance of Gemin-3 cleavage in PV biological cycle remains unknown. In addition to eIFs, Nups and proteins from SMN complex, PV 2Apro is able to cleave proteins involved in other cellular processes, such as transcription. TBP is cleaved by PV 2Apro between amino acids Tyr34 and Gly35 in vitro, although this cleavage only removes the first 34 aa located at the N-terminus and does not inhibit transcription carried out by RNA Polymerase II [217, 218]. These findings are in agreement with the fact that host mRNA transcription takes place in 2Apro-expressing cells when both translation and RNA nuclear export are inhibited, upon cleavage of eIF4G and Nups [17]. One attractive hypothesis is that PV 2Apro could cleave specific initiation factors affecting specific rather than general mRNA transcription in order to modulate host-cell response to viral infection. Further studies in this direction can be carried out using microarray platforms to detect precise alterations in cellular transcriptome after PV-infection or 2Apro expression. These studies could be complemented by screening for new host factors cleaved by PV 2Apro using different in silico and experimental approaches.
8. Conclusions and Future Prospects
The study of viral proteases is crucial to understand the mechanism used by animal viruses to replicate their genomes and to translate viral mRNAs at the molecular level. In addition, we wish to draw attention to the concept that viral proteases can be used as tools to reveal the exact functioning of their target cellular proteins. In this regard, PV 2Apro has been very useful for examining the requirements for eIF4G to translate different cellular or viral mRNAs. In addition to this, in this paper, we have highlighted the role of PV 2Apro not only in the processing of the poliovirus polyprotein but also in the interaction with the host-cell. Interestingly, a single viral protein is able to modulate many steps of gene expression in order to generate an optimal intracellular environment for the viral biological cycle. In particular, PV 2Apro cleaves cell proteins involved in transcription, pre-mRNA splicing, nucleus/cytoplasm transport and translation, in order to hijack those host functions and to concentrate the cellular resources on the production of the viral progeny. For example, PV 2Apro inactivates host translational machinery for capped cellular mRNAs by cleaving eIF4G, whereas viral protein synthesis takes place under those conditions by IRES-driven translation [14]. Because of the decrease of cellular mRNA translatability in PV-infected cells, host ribosomes are available for viral protein synthesis. Probably, the inhibition of host protein synthesis may prevent the production of antiviral proteins. Similarly, the blockade of nucleus/cytoplasm transport of macromolecules might isolate nuclear processes from cytoplasmic cellular ones, hampering the arrival of specific proinflammatory transcription factors to the nucleus and of mRNAs encoding proinflammatory, antiviral or proapoptotic proteins to the cytoplasm of infected cells. Finally, transcription and pre-mRNA splicing could be also modulated by PV 2Apro and might reduce the availability of mature mRNAs. Nevertheless, very little is known about the potential role of PV 2Apro in transcription and pre-mRNA splicing, and further studies of these steps of gene expression in PV-infected and 2A-expressing cells should be carried out. Making use of the “omics” technologies it would be possible to identify changes in the host transcriptome in those cells, allowing us to understand the readjustment of host gene expression to viral infection and to the cytotoxic effect of PV 2Apro. Gene ontology tools can be used to cluster the pathways (KEGG), molecular activities and biological functions of the genes which are transcriptionally up- or downregulated or not affected after these unfavourable stimuli. In silico analysis could be carried out in order to identify whether these gene clusters belong to particular networks controlled by specific transcription factors. This approach will give us additional information about potential host targets for PV 2Apro. Complementarily, deep sequence (RNAseq), specific microarrays types and conventional RT-PCR could be used to screen for nonspliced or abnormal spliced mRNA variants in PV-infected and 2Apro-expressing cells. Finally, microarrays can be employed to identify the cytoplasmic and nuclear transcriptome in those cells to determine whether mRNA nuclear export inhibition induced by PV 2Apro impacts on the distribution of the entire host mRNA bulk or in specific mRNA pools. Taken together, all this information may provide us with a general and deep vision of the modification induced by PV 2Apro on the different steps of mRNA metabolism. Additionally, the physiological role of Nups and Gemin-3 cleavage might be studied using different models. One possible and interesting approach is to engineer stable cell lines expressing noncleavable versions of Nup98, Nup62, Nup153 and/or Gemin-3 and then to analyze the fitness of PV in the different cell types, especially in presence of extracellular antiviral stimuli such as IFNs or interleukins, which will activate different epigenetic programs. These studies will provide essential information to help us understand the specific role that those host factors play in PV infection.
The total number of cellular proteins targeted by PV 2Apro (degradome) remains unknown, but it can be anticipated that with the expanding use of proteomic methodologies, this analysis will be known soon not only for PV 2Apro, but also for other viral proteases of interest. Furthermore, the analysis of the PV 2Apro-induced degradome in human cells will be of general interest for many researchers, including virologists and cellular biologists. This goal could be achieved combining in silico prediction of 2Apro cleavage sites and experimental tools such as proteomics. In the first case, Blom and collaborators developed a bioinformatics tool using neural network algorithms to predict cellular targets for picornavirus proteases [219]. This approach has been successfully used to predict the cleavage of dystrophin by coxsackievirus 2Apro [220] although most of the predicted human targets for rhinovirus and enterovirus 2Apro have not been proved yet. In addition, the algorithm did not predict the cleavage of cellular targets that have been later demonstrated to be proteolyzed by picornaviral 2Apro such as Nup98 and cytokeratin 8 [16, 17, 221]. Thus, it would be necessary to develop an improved algorithm able to find optimal cleavage sequences in the host proteome by implementing the proteolytic sites known for newly described 2Apro targets. Many parameters have to be taken into account, including the protein localization (cytoplasmic and nuclear protein will be considered, but not proteins resident in the lumen of other organelles such as RE or peroxisomes), the exposure of the cleavage site to the solvent (the sequence must be accessible to the protease), and the secondary structure in which the proteolytic site is included (optimally, unstructured regions). Potential targets could be ordered by their degree of homology with optimal cleavage sequences, as well as with the degree in which they fulfill the above prerequisites. On the other hand, novel PV 2Apro targets can be identified by proteomic tools such as two-dimensional differential gel electrophoresis (DIGE) or quantitative proteomics such as stable isotope labeling by amino acids in cell culture (SILAC) coupled to monodimensional electrophoresis. Following these two methods, it will be feasible to identify proteins with reduced levels on 2Apro expression and, in addition, to detect the cleavage products that will appear as lower size peptides. By the uncovering of novel 2Apro targets we will be able to map the cellular networks impacted by PV 2Apro and to integrate them in the context of PV infection. In fact, the role of 2Apro hijacking host processes could be potentially expanded to other cellular pathways with direct impact on control of viral infection. Such knowledge will provide more insight into our understanding of the cytopathogenicity of viral proteases at the molecular level.
Acknowledgments
Our group is supported by a DGICYT Grant (no. BFU2009-07352). The Institutional Grant awarded to the Centro de Biología Molecular “Severo Ochoa” by the Fundación Ramón Areces is also acknowledged. A. Castello is a beneficiary of Marie Curie IEF fellowship (Seventh Framework Programme). Alfredo Castelló and Enrique Álvarez contributed equally to this work.
References
- 1.Tong L. Viral proteases. Chemical Reviews. 2002;102(12):4609–4626. doi: 10.1021/cr010184f. [DOI] [PubMed] [Google Scholar]
- 2.Skern T, Hampölz B, Guarné A, et al. Structure and function of picornavirus proteinases. In: Semler BL, Wimmer E, editors. Molecular Biology of Picornaviruses. Washington, DC, USA: ASM Press; 2002. pp. 199–212. [Google Scholar]
- 3.Frankel AD, Young JAT. HIV-1: fifteen proteins and an RNA. Annual Review of Biochemistry. 1998;67:1–25. doi: 10.1146/annurev.biochem.67.1.1. [DOI] [PubMed] [Google Scholar]
- 4.Seipelt J, Guarné A, Bergmann E, et al. The structures of picornaviral proteinases. Virus Research. 1999;62(2):159–168. doi: 10.1016/s0168-1702(99)00043-x. [DOI] [PubMed] [Google Scholar]
- 5.Clarke IN, Lambden PR. Organization and expression of calicivirus genes. Journal of Infectious Diseases. 2000;181(supplement 2):S309–S316. doi: 10.1086/315575. [DOI] [PubMed] [Google Scholar]
- 6.Flügel RM, Pfrepper KI. Proteolytic processing of foamy virus Gag and Pol proteins. Current Topics in Microbiology and Immunology. 2003;277:63–88. doi: 10.1007/978-3-642-55701-9_3. [DOI] [PubMed] [Google Scholar]
- 7.Davidson AD. Chapter 2. New insights into flavivirus nonstructural protein 5. Advances in Virus Research. 2009;74:41–101. doi: 10.1016/S0065-3527(09)74002-3. [DOI] [PubMed] [Google Scholar]
- 8.Blondel B, Colbère-Garapin F, Couderc T, Wirotius A, Guivel-Benhassine F. Poliovirus, pathogenesis of poliomyelitis, and apoptosis. Current Topics in Microbiology and Immunology. 2005;289:25–56. doi: 10.1007/3-540-27320-4_2. [DOI] [PubMed] [Google Scholar]
- 9.Blondel B, Couderc T, Simonin Y, Gosselin AS, Guivel-Benhassine F. Poliovirus and apoptosis. Progress in Molecular and Subcellular Biology. 2004;36:151–169. doi: 10.1007/978-3-540-74264-7_8. [DOI] [PubMed] [Google Scholar]
- 10.Aldabe R, Carrasco L. Induction of membrane proliferation by poliovirus proteins 2C and 2BC. Biochemical and Biophysical Research Communications. 1995;206(1):64–76. doi: 10.1006/bbrc.1995.1010. [DOI] [PubMed] [Google Scholar]
- 11.Agol VI, Belov GA, Bienz K, et al. Two types of death of poliovirus-infected cells: caspase involvement in the apoptosis but not cytopathic effect. Virology. 1998;252(2):343–353. doi: 10.1006/viro.1998.9438. [DOI] [PubMed] [Google Scholar]
- 12.Calandria C, Irurzun A, Barco A, Carrasco L. Individual expression of poliovirus 2A and 3C induces activation of caspase-3 and PARP cleavage in HeLa cells. Virus Research. 2004;104(1):39–49. doi: 10.1016/j.virusres.2004.02.042. [DOI] [PubMed] [Google Scholar]
- 13.Belsham GJ, Sonenberg N. Picornavirus RNA translation: roles for cellular proteins. Trends in Microbiology. 2000;8(7):330–335. doi: 10.1016/s0966-842x(00)01788-1. [DOI] [PubMed] [Google Scholar]
- 14.Lloyd RE. Translational control by viral proteinases. Virus Research. 2006;119(1):76–88. doi: 10.1016/j.virusres.2005.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gustin KE, Sarnow P. Effects of poliovirus infection on nucleo-cytoplasmic trafficking and nuclear pore complex composition. EMBO Journal. 2001;20(1-2):240–249. doi: 10.1093/emboj/20.1.240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Park N, Katikaneni P, Skern T, Gustin KE. Differential targeting of nuclear pore complex proteins in poliovirus-infected cells. Journal of Virology. 2008;82(4):1647–1655. doi: 10.1128/JVI.01670-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Castelló A, Izquierdo JM, Welnowska E, Carrasco L. RNA nuclear export is blocked by poliovirus 2A protease and is concomitant with nucleoporin cleavage. Journal of Cell Science. 2009;122(20):3799–3809. doi: 10.1242/jcs.055988. [DOI] [PubMed] [Google Scholar]
- 18.Nathanson N. The pathogenesis of poliomyelitis: what we don't know. Advances in Virus Research. 2008;71:1–50. doi: 10.1016/S0065-3527(08)00001-8. [DOI] [PubMed] [Google Scholar]
- 19.Hogle JM. Poliovirus cell entry: common structural themes in viral cell entry pathways. Annual Review of Microbiology. 2002;56:677–702. doi: 10.1146/annurev.micro.56.012302.160757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Racaniello VR. One hundred years of poliovirus pathogenesis. Virology. 2006;344(1):9–16. doi: 10.1016/j.virol.2005.09.015. [DOI] [PubMed] [Google Scholar]
- 21.Wimmer E, Hellen CUT, Cao X. Genetics of poliovirus. Annual Review of Genetics. 1993;27:353–436. doi: 10.1146/annurev.ge.27.120193.002033. [DOI] [PubMed] [Google Scholar]
- 22.Agol VA. Picornavirus genome: an overview. In: Semler BL, Wimmer E, editors. Molecular Biology of Picornaviruses. Washington, DC, USA: ASM Press; 2002. pp. 127–148. [Google Scholar]
- 23.Leong LEC, Cornell CT, Semler BL. Processing determinants and functions of cleavage products of picornavirus polyproteins. In: Semler BL, Wimmer E, editors. Molecular Biology of Picornaviruses. Washington, DC, USA: ASM Press; 2002. pp. 187–198. [Google Scholar]
- 24.Lee CK, Wimmer E. Proteolytic processing of poliovirus polyprotein: elimination of 2A(pro)-mediated, alternative cleavage of polypeptide 3CD by in vitro mutagenesis. Virology. 1988;166(2):405–414. doi: 10.1016/0042-6822(88)90511-9. [DOI] [PubMed] [Google Scholar]
- 25.Cameron CE, Gohara DW, Arnold JJ. Poliovirus RNA-dependent RNA polymerase (3Dpol): structure, function, and mechanism. In: Semler BL, Wimmer E, editors. Molecular Biology of Picornaviruses. Washington, DC, USA: ASM Press; 2002. pp. 255–268. [Google Scholar]
- 26.Paul AV. Possible Unifying mechanism of picornavirus genome replication. In: Semler BL, Wimmer E, editors. Molecular Biology of Picornaviruses. Washington, DC, USA: ASM Press; 2002. pp. 227–246. [Google Scholar]
- 27.Paul AV, Van Boom JH, Filippov D, Wimmer E. Protein-primed RNA synthesis by purified poliovirus RNA polymerase. Nature. 1998;393(6682):280–284. doi: 10.1038/30529. [DOI] [PubMed] [Google Scholar]
- 28.Almela MJ, Gonzalez ME, Carrasco L. Inhibitors of poliovirus uncoating efficiently block the early membrane permeabilization induced by virus particles. Journal of Virology. 1991;65(5):2572–2577. doi: 10.1128/jvi.65.5.2572-2577.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sanz MA, Castelló A, Carrasco L. Viral translation is coupled to transcription in sindbis virus-infected cells. Journal of Virology. 2007;81(13):7061–7068. doi: 10.1128/JVI.02529-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Guinea R, Carrasco L. Phospholipid biosynthesis and poliovirus genome replication, two coupled phenomena. EMBO Journal. 1990;9(6):2011–2016. doi: 10.1002/j.1460-2075.1990.tb08329.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Carrasco L, Guinea R, Irurzun A, Barco A. Effects of viral replication on cellular membrane metabolism and function. In: Semler BL, Wimmer E, editors. Molecular Biology of Picornaviruses. Washington, DC, USA: ASM Press; 2002. pp. 337–356. [Google Scholar]
- 32.Agirre A, Barco A, Carrasco L, Nieva JL. Viroporin-mediated membrane permeabilization: pore formation by nonstructural poliovirus 2B protein. Journal of Biological Chemistry. 2002;277(43):40434–40441. doi: 10.1074/jbc.M205393200. [DOI] [PubMed] [Google Scholar]
- 33.Gonzalez ME, Carrasco L. Viroporins. FEBS Letters. 2003;552(1):28–34. doi: 10.1016/s0014-5793(03)00780-4. [DOI] [PubMed] [Google Scholar]
- 34.Kitamura N, Semler BL, Rothberg PG, et al. Primary structure, gene organization and polypeptide expression of poliovirus RNA. Nature. 1981;291(5816):547–553. doi: 10.1038/291547a0. [DOI] [PubMed] [Google Scholar]
- 35.Hogle JM, Chow M, Filman DJ. Three-dimensional structure of poliovirus at 2.9 Å resolution. Science. 1985;229(4720):1358–1365. doi: 10.1126/science.2994218. [DOI] [PubMed] [Google Scholar]
- 36.Pelletier J, Sonenberg N. Internal initiation of translation of eukaryotic mRNA directed by a sequence derived from poliovirus RNA. Nature. 1988;334(6180):320–325. doi: 10.1038/334320a0. [DOI] [PubMed] [Google Scholar]
- 37.Molla A, Paul AV, Wimmer E. Cell-free, de novo synthesis of poliovirus. Science. 1991;254(5038):1647–1651. doi: 10.1126/science.1661029. [DOI] [PubMed] [Google Scholar]
- 38.Wimmer E, Mueller S, Tumpey TM, Taubenberger JK. Synthetic viruses: a new opportunity to understand and prevent viral disease. Nature Biotechnology. 2009;27(12):1163–1172. doi: 10.1038/nbt.1593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Etchison D, Milburn SC, Edery I, Sonenberg N, Hershey JW. Inhibition of HeLa cell protein synthesis following poliovirus infection correlates with the proteolysis of a 220,000-dalton polypeptide associated with eucaryotc initiation factor 3 and a cap binding protein complex. Journal of Biological Chemistry. 1982;257(24):14806–14810. [PubMed] [Google Scholar]
- 40.Ryan MD, Flint M. Virus-encoded proteinases of the picornavirus super-group. Journal of General Virology. 1997;78(4):699–723. doi: 10.1099/0022-1317-78-4-699. [DOI] [PubMed] [Google Scholar]
- 41.Palmenberg AC, Neubauer D, Skern T. Genome organization and encoded proteins. In: Eherenfeld E, Domingo E, Roos R, editors. The Picornaviruses. Washington, DC, USA: ASM Press; 2010. pp. 3–17. [Google Scholar]
- 42.Guarné A, Tormo J, Kirchweger R, Pfistermueller D, Fita I, Skern T. Structure of the foot-and-mouth disease virus leader protease: a papain-like fold adapted for self-processing and eIF4G recognition. EMBO Journal. 1998;17(24):7469–7479. doi: 10.1093/emboj/17.24.7469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Guarné A, Hampoelz B, Glaser W, et al. Structural and biochemical features distinguish the foot-and-mouth disease virus leader proteinase from other papain-like enzymes. Journal of Molecular Biology. 2000;302(5):1227–1240. doi: 10.1006/jmbi.2000.4115. [DOI] [PubMed] [Google Scholar]
- 44.Piccone ME, Rieder E, Mason PW, Grubman MJ. The foot-and-mouth disease virus leader proteinase gene is not required for viral replication. Journal of Virology. 1995;69(9):5376–5382. doi: 10.1128/jvi.69.9.5376-5382.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.De Los Santos T, De Avila Botton S, Weiblen R, Grubman MJ. The leader proteinase of foot-and-mouth disease virus inhibits the induction of beta interferon mRNA and blocks the host innate immune response. Journal of Virology. 2006;80(4):1906–1914. doi: 10.1128/JVI.80.4.1906-1914.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.De Los Santos T, Segundo FDS, Zhu J, Koster M, Dias CCA, Grubman MJ. A conserved domain in the leader proteinase of foot-and-mouth disease virus is required for proper subcellular localization and function. Journal of Virology. 2009;83(4):1800–1810. doi: 10.1128/JVI.02112-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hato SV, Sorgeloos F, Ricour C, et al. Differential IFN-α/β production suppressing capacities of the leader proteins of mengovirus and foot-and-mouth disease virus. Cellular Microbiology. 2010;12(3):310–317. doi: 10.1111/j.1462-5822.2009.01395.x. [DOI] [PubMed] [Google Scholar]
- 48.Devaney MA, Vakharia VN, Lloyd RE, Ehrenfeld E, Grubman MJ. Leader protein of foot-and-mouth disease virus is required for cleavage of the p220 component of the cap-binding protein complex. Journal of Virology. 1988;62(11):4407–4409. doi: 10.1128/jvi.62.11.4407-4409.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lloyd RE, Grubman MJ, Ehrenfeld E. Relationship of p220 cleavage during picornavirus infection to 2A proteinase sequencing. Journal of Virology. 1988;62(11):4216–4223. doi: 10.1128/jvi.62.11.4216-4223.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Glaser W, Skern T. Extremely efficient cleavage of eIF4G by picornaviral proteinases L and 2A in vitro. FEBS Letters. 2000;480(2-3):151–155. doi: 10.1016/s0014-5793(00)01928-1. [DOI] [PubMed] [Google Scholar]
- 51.Ryan MD, Drew J. Foot-and-mouth disease virus 2A oligopeptide mediated cleavage of an artificial polyprotein. EMBO Journal. 1994;13(4):928–933. doi: 10.1002/j.1460-2075.1994.tb06337.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Donnelly MLL, Luke G, Mehrotra A, et al. Analysis of the aphthovirus 2A/2B polyprotein 'cleavage' mechanism indicates not a proteolytic reaction, but a novel translational effect: a putative ribosomal 'skip'. Journal of General Virology. 2001;82(5):1013–1025. doi: 10.1099/0022-1317-82-5-1013. [DOI] [PubMed] [Google Scholar]
- 53.Donnelly MLL, Gani D, Flint M, Monaghan S, Ryan MD. The cleavage activities of aphthovirus and cardiovirus 2A proteins. Journal of General Virology. 1997;78(1):13–21. doi: 10.1099/0022-1317-78-1-13. [DOI] [PubMed] [Google Scholar]
- 54.Konig H, Rosenwirth B. Purification and partial characterization of poliovirus protease 2A by means of a functional assay. Journal of Virology. 1988;62(4):1243–1250. doi: 10.1128/jvi.62.4.1243-1250.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Nicklin MJH, Harris KS, Pallai PV, Wimmer E. Poliovirus proteinase 3C: large-scale expression, purification, and specific cleavage activity on natural and synthetic substrates in vitro. Journal of Virology. 1988;62(12):4586–4593. doi: 10.1128/jvi.62.12.4586-4593.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Burroughs JN, Sangar DV, Clarke BE, et al. Multiple proteases in foot-and-mouth disease virus replication. Journal of Virology. 1984;50(3):878–883. doi: 10.1128/jvi.50.3.878-883.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kleina LG, Grubman MJ. Antiviral effects of a thiol protease inhibitor on foot-and-mouth disease virus. Journal of Virology. 1992;66(12):7168–7175. doi: 10.1128/jvi.66.12.7168-7175.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Gorbalenya AE, Koonin EV, Lai MMC. Putative papain-related thiol proteases of positive-strand RNA viruses. Identification of rubi- and aphthovirus proteases and delineation of a novel conserved domain associated with proteases of rubi, α- and coronaviruses. FEBS Letters. 1991;288(1-2):201–205. doi: 10.1016/0014-5793(91)81034-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Piccone ME, Zellner M, Kumosinski TF, Mason PW, Grubman MJ. Identification of the active-site residues of the L proteinase of foot- and-mouth disease virus. Journal of Virology. 1995;69(8):4950–4956. doi: 10.1128/jvi.69.8.4950-4956.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Roberts PJ, Belsham GJ. Identification of critical amino acids within the foot-and-mouth disease virus leader protein, a cysteine protease. Virology. 1995;213(1):140–146. doi: 10.1006/viro.1995.1554. [DOI] [PubMed] [Google Scholar]
- 61.Blinov VM, Gorbalenia AE, Donchenko AP. Structural similarity of poliovirus cysteine proteinase P3-7c and cellular serine proteinase of trypsin. Doklady Akademii Nauk SSSR. 1984;279(2):502–505. [PubMed] [Google Scholar]
- 62.Gorbalenya AE, Donchenko AP, Blinov VM, Koonin EV. Cysteine proteases of positive strand RNA viruses and chymotrypsin-like serin proteases: a distinct protein superfamily with a common structural fold. FEBS Letters. 1989;243(2):103–114. doi: 10.1016/0014-5793(89)80109-7. [DOI] [PubMed] [Google Scholar]
- 63.Bazan JF, Fletterick RJ. Viral cysteine proteases are homologous to the trypsin-like family of serine proteases: structural and functional implications. Proceedings of the National Academy of Sciences of the United States of America. 1988;85(21):7872–7876. doi: 10.1073/pnas.85.21.7872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Toyoda H, Nicklin MJH, Murray MG, et al. A second virus-encoded proteinase involved in proteolytic processing of poliovirus polyprotein. Cell. 1986;45(5):761–770. doi: 10.1016/0092-8674(86)90790-7. [DOI] [PubMed] [Google Scholar]
- 65.Skern T, Sommergruber W, Auer H, et al. Substrate requirements of a human rhinoviral 2A proteinase. Virology. 1991;181(1):46–54. doi: 10.1016/0042-6822(91)90468-q. [DOI] [PubMed] [Google Scholar]
- 66.Hellen CUT, Lee CK, Wimmer E. Determinants of substrate recognition by poliovirus 2A proteinase. Journal of Virology. 1992;66(6):3330–3338. doi: 10.1128/jvi.66.6.3330-3338.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Sommergruber W, Ahorn H, Zophel A, et al. Cleavage specificity on synthetic peptide substrates of human rhinovirus 2 proteinase 2A. Journal of Biological Chemistry. 1992;267(31):22639–22644. [PubMed] [Google Scholar]
- 68.Ventoso I, Barco A, Carrasco L. Genetic selection of poliovirus 2A(pro)-binding peptides. Journal of Virology. 1999;73(1):814–818. doi: 10.1128/jvi.73.1.814-818.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Yu SF, Lloyd RE. Identification of essential amino acid residues in the functional activity of poliovirus 2A protease. Virology. 1991;182(2):615–625. doi: 10.1016/0042-6822(91)90602-8. [DOI] [PubMed] [Google Scholar]
- 70.Sommergruber W, Seipelt J, Fessl F, Skern T, Liebig HD, Casari G. Mutational analyses support a model for the HRV2 2A proteinase. Virology. 1997;234(2):203–214. doi: 10.1006/viro.1997.8595. [DOI] [PubMed] [Google Scholar]
- 71.Petersen JFW, Cherney MM, Liebig HD, Skern T, Kuechler E, James MNG. The structure of the 2A proteinase from a common cold virus: a proteinase responsible for the shut-off of host-cell protein synthesis. EMBO Journal. 1999;18(20):5463–5475. doi: 10.1093/emboj/18.20.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Yu SF, Lloyd RE. Characterization of the roles of conserved cysteine and histidine residues in poliovirus 2A protease. Virology. 1992;186(2):725–735. doi: 10.1016/0042-6822(92)90039-r. [DOI] [PubMed] [Google Scholar]
- 73.Sommergruber W, Casari G, Fessl F, Seipelt J, Skern T. The 2A proteinase of human rhinovirus is a zinc containing enzyme. Virology. 1994;204(2):815–818. doi: 10.1006/viro.1994.1599. [DOI] [PubMed] [Google Scholar]
- 74.Voss T, Meyer R, Sommergruber W. Spectroscopic characterization of rhinoviral protease 2A: Zn is essential for the structural integrity. Protein Science. 1995;4(12):2526–2531. doi: 10.1002/pro.5560041209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Barco A, Ventoso I, Carrasco L. The yeast Saccharomyces cerevisiae as a genetic system for obtaining variants of poliovirus protease 2A. Journal of Biological Chemistry. 1997;272(19):12683–12691. doi: 10.1074/jbc.272.19.12683. [DOI] [PubMed] [Google Scholar]
- 76.Ventoso I, Barco A, Carrasco L. Mutational analysis of poliovirus 2A(pro): distinct inhibitory functions of 2A(pro) on translation and transcription. Journal of Biological Chemistry. 1998;273(43):27960–27967. doi: 10.1074/jbc.273.43.27960. [DOI] [PubMed] [Google Scholar]
- 77.Yu SF, Benton P, Bovee M, Sessions J, Lloyd RE. Defective RNA replication by poliovirus mutants deficient in 2A protease cleavage activity. Journal of Virology. 1995;69(1):247–252. doi: 10.1128/jvi.69.1.247-252.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Igarashi H, Yoshino Y, Miyazawa M, Horie H, Ohka S, Nomoto A. 2A protease is not a prerequisite for poliovirus replication. Journal of Virology. 2010;84(12):5947–5957. doi: 10.1128/JVI.02575-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Morrison JM, Racaniello VR. Proteinase 2A is essential for enterovirus replication in type I interferon-treated cells. Journal of Virology. 2009;83(9):4412–4422. doi: 10.1128/JVI.02177-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Teterina NL, Levenson EA, Ehrenfeld E. Viable polioviruses that encode 2A proteins with fluorescent protein tags. Journal of Virology. 2010;84(3):1477–1488. doi: 10.1128/JVI.01578-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Sonenberg N, Hinnebusch AG. Regulation of translation initiation in eukaryotes: mechanisms and biological targets. Cell. 2009;136(4):731–745. doi: 10.1016/j.cell.2009.01.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Jackson RJ, Hellen CUT, Pestova TV. The mechanism of eukaryotic translation initiation and principles of its regulation. Nature Reviews Molecular Cell Biology. 2010;11(2):113–127. doi: 10.1038/nrm2838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Gingras AC, Raught B, Sonenberg N. eIF4 initiation factors: effectors of mRNA recruitment to ribosomes and regulators of translation. Annual Review of Biochemistry. 1999;68:913–963. doi: 10.1146/annurev.biochem.68.1.913. [DOI] [PubMed] [Google Scholar]
- 84.Gebauer F, Hentze MW. Molecular mechanisms of translational control. Nature Reviews Molecular Cell Biology. 2004;5(10):827–835. doi: 10.1038/nrm1488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Hentze MW. eIF4G: a multipurpose ribosome adapter? Science. 1997;275(5299):500–501. doi: 10.1126/science.275.5299.500. [DOI] [PubMed] [Google Scholar]
- 86.Morley SJ, Curtis PS, Pain VM. elF4G: translation’s mystery factor begins to yield its secrets. RNA. 1997;3(10):1085–1104. [PMC free article] [PubMed] [Google Scholar]
- 87.Imataka H, Sonenberg N. Human eukaryotic translation initiation factor 4G (eIF4G) possesses two separate and independent binding sites for eIF4A. Molecular and Cellular Biology. 1997;17(12):6940–6947. doi: 10.1128/mcb.17.12.6940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Imataka H, Gradi A, Sonenberg N. A newly identified N-terminal amino acid sequence of human eIF4G binds poly(A)-binding protein and functions in poly(A)-dependent translation. EMBO Journal. 1998;17(24):7480–7489. doi: 10.1093/emboj/17.24.7480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Wells SE, Hillner PE, Vale RD, Sachs AB. Circularization of mRNA by eukaryotic translation initiation factors. Molecular Cell. 1998;2(1):135–140. doi: 10.1016/s1097-2765(00)80122-7. [DOI] [PubMed] [Google Scholar]
- 90.Fukunaga R, Hunter T. MNK1, a new MAP kinase-activated protein kinase, isolated by a novel expression screening method for identifying protein kinase substrates. EMBO Journal. 1997;16(8):1921–1933. doi: 10.1093/emboj/16.8.1921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Waskiewicz AJ, Flynn A, Proud CG, Cooper JA. Mitogen-activated protein kinases activate the serine/threonine kinases Mnk1 and Mnk2. EMBO Journal. 1997;16(8):1909–1920. doi: 10.1093/emboj/16.8.1909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Pyronnet S, Imataka H, Gingras AC, Fukunaga R, Hunter T, Sonenberg N. Human eukaryotic translation initiation factor 4G (eIF4G) recruits Mnk1 to phosphorylate eIF4E. EMBO Journal. 1999;18(1):270–279. doi: 10.1093/emboj/18.1.270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Mohr I. Phosphorylation and dephosphorylation events that regulate viral mRNA translation. Virus Research. 2006;119(1):89–99. doi: 10.1016/j.virusres.2005.10.009. [DOI] [PubMed] [Google Scholar]
- 94.Richter JD, Sonenberg N. Regulation of cap-dependent translation by eIF4E inhibitory proteins. Nature. 2005;433(7025):477–480. doi: 10.1038/nature03205. [DOI] [PubMed] [Google Scholar]
- 95.Castello A, Alvarez E, Carrasco L. Differential cleavage of eIF4GI and eIF4GII in mammalian cells: effects on translation. Journal of Biological Chemistry. 2006;281(44):33206–33216. doi: 10.1074/jbc.M604340200. [DOI] [PubMed] [Google Scholar]
- 96.Welnowska E, Castelló A, Moral P, Carrasco L. Translation of mRNAs from vesicular stomatitis virus and vaccinia virus is differentially blocked in cells with depletion of eIF4GI and/or eIF4GII. Journal of Molecular Biology. 2009;394(3):506–521. doi: 10.1016/j.jmb.2009.09.036. [DOI] [PubMed] [Google Scholar]
- 97.Marissen WE, Lloyd RE. Eukaryotic translation initiation factor 4G is targeted for proteolytic cleavage by caspase 3 during inhibition of translation in apoptotic cells. Molecular and Cellular Biology. 1998;18(12):7565–7574. doi: 10.1128/mcb.18.12.7565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Caron S, Charon M, Cramer E, Sonenberg N, Dusanter-Fourt I. Selective modification of eukaryotic initiation factor 4F (eIF4F) at the onset of cell differentiation: recruitment of eIF4GII and long-lasting phosphorylation of eIF4E. Molecular and Cellular Biology. 2004;24(11):4920–4928. doi: 10.1128/MCB.24.11.4920-4928.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Raught B, Gingras AC, Gygi SP, et al. Serum-stimulated, rapamycin-sensitive phosphorylation sites in the eukaryotic: translation initiation factor 4GI. EMBO Journal. 2000;19(3):434–444. doi: 10.1093/emboj/19.3.434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Ling J, Morley SJ, Traugh JA. Inhibition of cap-dependent translation via phosphorylation of eIF4G by protein kinase Pak2. EMBO Journal. 2005;24(23):4094–4105. doi: 10.1038/sj.emboj.7600868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Pyronnet S, Dostie J, Sonenberg N. Suppression of cap-dependent translation in mitosis. Genes and Development. 2001;15(16):2083–2093. doi: 10.1101/gad.889201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Qin H, Raught B, Sonenberg N, Goldstein EG, Edelman AM. Phosphorylation screening identifies translational initiation factor 4GII as an intracellular target of Ca2+/calmodulin-dependent protein kinase I. Journal of Biological Chemistry. 2003;278(49):48570–48579. doi: 10.1074/jbc.M308781200. [DOI] [PubMed] [Google Scholar]
- 103.Bushell M, Poncet D, Marissen WE, et al. Cleavage of polypeptide chain initiation factor eIF4GI during apoptosis in lymphoma cells: characterisation of an internal fragment generated by caspase-3-mediated cleavage. Cell Death and Differentiation. 2000;7(7):628–636. doi: 10.1038/sj.cdd.4400699. [DOI] [PubMed] [Google Scholar]
- 104.Marissen WE, Gradi A, Sonenberg N, Lloyd RE. Cleavage of eukaryotic translation initiation factor 4GII correlates with translation inhibition during apoptosis. Cell Death and Differentiation. 2000;7(12):1234–1243. doi: 10.1038/sj.cdd.4400750. [DOI] [PubMed] [Google Scholar]
- 105.Byrd MP, Zamora M, Lloyd RE. Translation of eukaryotic translation initiation factor 4GI (eIF4GI) proceeds from multiple mRNAs containing a novel cap-dependent internal ribosome entry site (IRES) that is active during poliovirus infection. Journal of Biological Chemistry. 2005;280(19):18610–18622. doi: 10.1074/jbc.M414014200. [DOI] [PubMed] [Google Scholar]
- 106.Byrd MP, Zamora M, Lloyd RE. Generation of multiple isoforms of eukaryotic translation initiation factor 4GI by use of alternate translation initiation codons. Molecular and Cellular Biology. 2002;22(13):4499–4511. doi: 10.1128/MCB.22.13.4499-4511.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Coldwell MJ, Morley SJ. Specific isoforms of translation initiation factor 4GI show differences in translational activity. Molecular and Cellular Biology. 2006;26(22):8448–8460. doi: 10.1128/MCB.01248-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Leibowitz R, Penman S. Regulation of protein synthesis in HeLa cells. 3. Inhibition during poliovirus infection. Journal of Virology. 1971;8(5):661–668. doi: 10.1128/jvi.8.5.661-668.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Helentjaris T, Ehrenfeld E. Control of protein synthesis in extracts from poliovirus-infected cells. I. mRNA discrimination by crude initiation factors. Journal of Virology. 1978;26(2):510–521. doi: 10.1128/jvi.26.2.510-521.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Krausslich HG, Nicklin MJH, Toyoda H, Etchison D, Wimmer E. Poliovirus proteinase 2A induces cleavage of eucaryotic initiation factor 4f polypeptide p220. Journal of Virology. 1987;61(9):2711–2718. doi: 10.1128/jvi.61.9.2711-2718.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Ventoso I, Carrasco L. A poliovirus 2A(pro) mutant unable to cleave 3CD shows inefficient viral protein synthesis and transactivation defects. Journal of Virology. 1995;69(10):6280–6288. doi: 10.1128/jvi.69.10.6280-6288.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Bovee ML, Lamphear BJ, Rhoads RE, Lloyd RE. Direct cleavage of elF4G by poliovirus 2A protease is inefficient in vitro. Virology. 1998;245(2):241–249. doi: 10.1006/viro.1998.9172. [DOI] [PubMed] [Google Scholar]
- 113.Ethison D, Fout S. Human rhinovirus 14 infection of HeLa cells results in the proteolytic cleavage of the p220 cap-binding complex subunit and inactivates globin mRNA translation in vitro. Journal of Virology. 1985;54(2):634–638. doi: 10.1128/jvi.54.2.634-638.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Lamphear BJ, Yan R, Yang F, et al. Mapping the cleavage site in protein synthesis initiation factor eIF-4γ of the 2A proteases from human Coxsackievirus and rhinovirus. Journal of Biological Chemistry. 1993;268(26):19200–19203. [PubMed] [Google Scholar]
- 115.Sommergruber W, Ahorn H, Klump H, et al. 2A proteinases of coxsackie- and rhinovirus cleave peptides derived from eIF-4 gamma via a common recognition motif. Virology. 1994;198:741–745. doi: 10.1006/viro.1994.1089. [DOI] [PubMed] [Google Scholar]
- 116.Haghighat A, Svitkin Y, Novoa I, Kuechler E, Skern T, Sonenberg N. The eIF4G-eIF4E complex is the target for direct cleavage by the rhinovirus 2A proteinase. Journal of Virology. 1996;70(12):8444–8450. doi: 10.1128/jvi.70.12.8444-8450.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Ventoso I, MacMillan SE, Hershey JWB, Carrasco L. Poliovirus 2A proteinase cleaves directly the eIF-4G subunit of eIF-4F complex. FEBS Letters. 1998;435(1):79–83. doi: 10.1016/s0014-5793(98)01027-8. [DOI] [PubMed] [Google Scholar]
- 118.Wyckoff EE, Lloyd RE, Ehrenfeld E. Relationship of eukaryotic initiation factor 3 to poliovirus-induced p220 cleavage activity. Journal of Virology. 1992;66(5):2943–2951. doi: 10.1128/jvi.66.5.2943-2951.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Zamora M, Marissen WE, Lloyd RE. Multiple eIF4GI-specific protease activities present in uninfected and poliovirus-infected cells. Journal of Virology. 2002;76(1):165–177. doi: 10.1128/JVI.76.1.165-177.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Perez L, Carrasco L. Lack of direct correlation between p220 cleavage and the shut-off of host translation after poliovirus infection. Virology. 1992;189(1):178–186. doi: 10.1016/0042-6822(92)90693-j. [DOI] [PubMed] [Google Scholar]
- 121.Irurzun A, Sanchez-Palomino S, Novoa I, Carrasco L. Monensin and nigericin prevent the inhibition of host translation by poliovirus, without affecting p220 cleavage. Journal of Virology. 1995;69(12):7453–7460. doi: 10.1128/jvi.69.12.7453-7460.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Gradi A, Svitkin YV, Imataka H, Sonenberg N. Proteolysis of human eukaryotic translation initiation factor eIF4GII, but not EIF4GI, coincides with the shutoff of host protein synthesis after poliovirus infection. Proceedings of the National Academy of Sciences of the United States of America. 1998;95(19):11089–11094. doi: 10.1073/pnas.95.19.11089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Svitkin YV, Gradi A, Imataka H, Morino S, Sonenberg N. Eukaryotic initiation factor 4GII (eIF4GII), but not eIF4GI, cleavage correlates with inhibition of host cell protein synthesis after human rhinovirus infection. Journal of Virology. 1999;73(4):3467–3472. doi: 10.1128/jvi.73.4.3467-3472.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Martinez-Abarca F, Alonso MA, Carrasco L. High level expression in Escherichia coli cells and purification of poliovirus protein 2A(pro) Journal of General Virology. 1993;74(12):2645–2652. doi: 10.1099/0022-1317-74-12-2645. [DOI] [PubMed] [Google Scholar]
- 125.Novoa I, Martínez-Abarca F, Fortes P, Ortín J, Carrasco L. Cleavage of p220 by purified poliovirus 2A(pro) in cell-free systems: effects on translation of capped and uncapped mRNAs. Biochemistry. 1997;36(25):7802–7809. doi: 10.1021/bi9631172. [DOI] [PubMed] [Google Scholar]
- 126.Svitkin YV, Imataka H, Khaleghpour K, Kahvejian A, Liebig HD, Sonenberg N. Poly(A)-binding protein interaction with elF4G stimulates picornavirus IRES-dependent translation. RNA. 2001;7(12):1743–1752. [PMC free article] [PubMed] [Google Scholar]
- 127.Castelló A, Franco D, Moral-López P, et al. HIV-1 protease inhibits cap-and poly(A)-dependent translation upon eIF4GI and PABP cleavage. PLoS One. 2009;4(11) doi: 10.1371/journal.pone.0007997. Article ID e7997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Sun XH, Baltimore D. Human immunodeficiency virus tat-activated expression of poliovirus protein 2A inhibits mRNA translation. Proceedings of the National Academy of Sciences of the United States of America. 1989;86(7):2143–2146. doi: 10.1073/pnas.86.7.2143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Davies MV, Pelletier J, Meerovitch K, Sonenberg N, Kaufman RJ. The effect of poliovirus proteinase 2A(pro) expression on cellular metabolism. Inhibition of DNA replication, RNA polymerase II transcription, and translation. Journal of Biological Chemistry. 1991;266(22):14714–14720. [PubMed] [Google Scholar]
- 130.Aldabe R, Feduchi E, Novoa I, Carrasco L. Expression of poliovirus 2A(pro) in mammalian cells: effects on translation. FEBS Letters. 1995;377(1):1–5. doi: 10.1016/0014-5793(95)01269-9. [DOI] [PubMed] [Google Scholar]
- 131.Aldabe R, Feduchi E, Novoa I, Carrasco L. Efficient cleavage of p220 by poliovirus 2A(pro) expression in mammalian cells: effects on vaccinia virus. Biochemical and Biophysical Research Communications. 1995;215(3):928–936. doi: 10.1006/bbrc.1995.2553. [DOI] [PubMed] [Google Scholar]
- 132.Novoa I, Feduchi E, Carrasco L. Hybrid proteins between pseudomonas exotoxin a and poliovirus protease 2A(pro) FEBS Letters. 1994;355(1):45–48. doi: 10.1016/0014-5793(94)01157-5. [DOI] [PubMed] [Google Scholar]
- 133.Feduchi E, Aldabe R, Novoa I, Carrasco L. Effects of poliovirus 2A(pro) on vaccinia virus gene expression. European Journal of Biochemistry. 1995;234(3):849–854. doi: 10.1111/j.1432-1033.1995.849_a.x. [DOI] [PubMed] [Google Scholar]
- 134.Alvarez E, Menéndez-Arias L, Carrasco L. The eukaryotic translation initiation factor 4GI is cleaved by different retroviral proteases. Journal of Virology. 2003;77(23):12392–12400. doi: 10.1128/JVI.77.23.12392-12400.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Novoa I, Cotten M, Carrasco L. Hybrid proteins between pseudomonas aeruginosa exotoxin A and poliovirus 2Apro cleave p220 in HeLa cells. Journal of Virology. 1996;70(5):3319–3324. doi: 10.1128/jvi.70.5.3319-3324.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Novoa I, Carrasco L. Cleavage of eukaryotic translation initiation factor 4G by exogenously added hybrid proteins containing poliovirus 2A(pro) in HeLa cells: effects on gene expression. Molecular and Cellular Biology. 1999;19(4):2445–2454. doi: 10.1128/mcb.19.4.2445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Barco A, Feduchi E, Carrasco L. A stable HeLa cell line that inducibly expresses poliovirus 2A(pro): effects on cellular and viral gene expression. Journal of Virology. 2000;74(5):2383–2392. doi: 10.1128/jvi.74.5.2383-2392.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Goldstaub D, Gradi A, Bercovitch Z, et al. Poliovirus 2A protease induces apoptotic cell death. Molecular and Cellular Biology. 2000;20(4):1271–1277. doi: 10.1128/mcb.20.4.1271-1277.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Lejeune F, Ranganathan AC, Maquat LE. eIF4G is required for the pioneer round of translation in mammalian cells. Nature Structural and Molecular Biology. 2004;11(10):992–1000. doi: 10.1038/nsmb824. [DOI] [PubMed] [Google Scholar]
- 140.Joachims M, Van Breugel PC, Lloyd RE. Cleavage of poly(A)-binding protein by enterovirus proteases concurrent with inhibition of translation in vitro. Journal of Virology. 1999;73(1):718–727. doi: 10.1128/jvi.73.1.718-727.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Kerekatte V, Keiper BD, Badorff C, Cat A, Knowlton KU, Rhoads RE. Cleavage of poly(A)-binding protein by coxsackievirus 2A protease in vitro and in vivo: another mechanism for host protein synthesis shutoff? Journal of Virology. 1999;73(1):709–717. doi: 10.1128/jvi.73.1.709-717.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Kuyumcu-Martinez NM, Joachims M, Lloyd RE. Efficient cleavage of ribosome-associated poly(A)-binding protein by enterovirus 3C protease. Journal of Virology. 2002;76(5):2062–2074. doi: 10.1128/jvi.76.5.2062-2074.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Kuyumcu-Martinez NM, Van Eden ME, Younan P, Lloyd RE. Cleavage of poly(A)-binding protein by poliovirus 3C protease inhibits host cell translation: a novel mechanism for host translation shutoff. Molecular and Cellular Biology. 2004;24(4):1779–1790. doi: 10.1128/MCB.24.4.1779-1790.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Kirchweger R, Ziegler E, Lamphear BJ, et al. Foot-and-mouth disease virus leader proteinase: purification of the Lb form and determination of its cleavage site on eIF-4γ. Journal of Virology. 1994;68(9):5677–5684. doi: 10.1128/jvi.68.9.5677-5684.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Belsham GJ, McInerney GM, Ross-Smith N. Foot-and-mouth disease virus 3C protease induces cleavage of translation initiation factors eIF4A and eIF4G within infected cells. Journal of Virology. 2000;74(1):272–280. doi: 10.1128/jvi.74.1.272-280.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Gradi A, Foeger N, Strong R, et al. Cleavage of eukaryotic translation initiation factor 4GII within foot-and-mouth disease virus-infected cells: identification of the L-protease cleavage site in vitro. Journal of Virology. 2004;78(7):3271–3278. doi: 10.1128/JVI.78.7.3271-3278.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Strong R, Belsham GJ. Sequential modification of translation initiation factor elF4Gl by two different foot-and-mouth disease virus proteases within infected baby hamster kidney cells: identification of the 3C cleavage site. Journal of General Virology. 2004;85(10):2953–2962. doi: 10.1099/vir.0.80254-0. [DOI] [PubMed] [Google Scholar]
- 148.Prevot D, Darlix JL, Ohlmann T. Conducting the initiation of protein synthesis: the role of eIF4G. Biology of the Cell. 2003;95(3-4):141–156. doi: 10.1016/s0248-4900(03)00031-5. [DOI] [PubMed] [Google Scholar]
- 149.Ventoso I, Blanco R, Perales C, Carrasco L. HIV-1 protease cleaves eukaryotic initiation factor 4G and inhibits cap-dependent translation. Proceedings of the National Academy of Sciences of the United States of America. 2001;98(23):12966–12971. doi: 10.1073/pnas.231343498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Ohlmann T, Prevot D, Décimo D, et al. In vitro cleavage of eIF4GI but not eIF4GII by HIV-1 protease and its effects on translation in the rabbit reticulocyte lysate system. Journal of Molecular Biology. 2002;318(1):9–20. doi: 10.1016/S0022-2836(02)00070-0. [DOI] [PubMed] [Google Scholar]
- 151.Perales C, Carrasco L, Ventoso I. Cleavage of eIF4G by HIV-1 protease: effects on translation. FEBS Letters. 2003;533(1-3):89–94. doi: 10.1016/s0014-5793(02)03764-x. [DOI] [PubMed] [Google Scholar]
- 152.Alvarez E, Castelló A, Menéndez-Arias L, Carrasco L. HIV protease cleaves poly(A)-binding protein. Biochemical Journal. 2006;396(2):219–226. doi: 10.1042/BJ20060108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Michel YM, Poncet D, Piron M, Kean KM, Borman AM. Cap-poly(A) synergy in mammalian cell-free extracts. Investigation of the requirements for poly(A)-mediated stimulation of translation initiation. Journal of Biological Chemistry. 2000;275(41):32268–32276. doi: 10.1074/jbc.M004304200. [DOI] [PubMed] [Google Scholar]
- 154.López-Lastra M, Gabus C, Darlix JL. Characterization of an internal ribosomal entry segment within the 5’ leader of avian reticuloendotheliosis virus type A RNA and development of novel MLV-REV-Based retroviral vectors. Human Gene Therapy. 1997;8(16):1855–1865. doi: 10.1089/hum.1997.8.16-1855. [DOI] [PubMed] [Google Scholar]
- 155.Ohlmann T, Lopez-Lastra M, Darlix JL. An internal ribosome entry segment promotes translation of the simian immunodeficiency virus genomic RNA. Journal of Biological Chemistry. 2000;275(16):11899–11906. doi: 10.1074/jbc.275.16.11899. [DOI] [PubMed] [Google Scholar]
- 156.Brasey A, Lopez-Lastra M, Ohlmann T, et al. The leader of human immunodeficiency virus type 1 genomic RNA harbors an internal ribosome entry segment that is active during the G/M phase of the cell cycle. Journal of Virology. 2003;77(7):3939–3949. doi: 10.1128/JVI.77.7.3939-3949.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Willcocks MM, Carter MJ, Roberts LO. Cleavage of eukaryotic initiation factor elF4G and inhibition of host-cell protein synthesis during feline calicivirus infection. Journal of General Virology. 2004;85(5):1125–1130. doi: 10.1099/vir.0.19564-0. [DOI] [PubMed] [Google Scholar]
- 158.Kuyumcu-Martinez M, Belliot G, Sosnovtsev SV, Chang KO, Green KY, Lloyd RE. Calicivirus 3C-like proteinase inhibits cellular translation by cleavage of poly(A)-binding protein. Journal of Virology. 2004;78(15):8172–8182. doi: 10.1128/JVI.78.15.8172-8182.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Gustin KE, Sarnow P. Inhibition of nuclear import and alteration of nuclear pore complex composition by rhinovirus. Journal of Virology. 2002;76(17):8787–8796. doi: 10.1128/JVI.76.17.8787-8796.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Porter FW, Palmenberg AC. Leader-induced phosphorylation of nucleoporins correlates with nuclear trafficking inhibition by cardioviruses. Journal of Virology. 2009;83(4):1941–1951. doi: 10.1128/JVI.01752-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Von Kobbe C, Van Deursen JMA, Rodrigues JP, et al. Vesicular stomatitis virus matrix protein inhibits host cell gene expression by targeting the nucleoporin Nup98. Molecular Cell. 2000;6(5):1243–1252. doi: 10.1016/s1097-2765(00)00120-9. [DOI] [PubMed] [Google Scholar]
- 162.Satterly N, Tsai PL, Van Deursen J, et al. Influenza virus targets the mRNA export machinery and the nuclear pore complex. Proceedings of the National Academy of Sciences of the United States of America. 2007;104(6):1853–1858. doi: 10.1073/pnas.0610977104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Köhler A, Hurt E. Exporting RNA from the nucleus to the cytoplasm. Nature Reviews Molecular Cell Biology. 2007;8(10):761–773. doi: 10.1038/nrm2255. [DOI] [PubMed] [Google Scholar]
- 164.Terry LJ, Shows EB, Wente SR. Crossing the nuclear envelope: hierarchical regulation of nucleocytoplasmic transport. Science. 2007;318(5855):1412–1416. doi: 10.1126/science.1142204. [DOI] [PubMed] [Google Scholar]
- 165.Rout MP, Aitchison JD, Suprapto A, Hjertaas K, Zhao Y, Chait BT. The yeast nuclear pore complex: composition, architecture, transport mechanism. Journal of Cell Biology. 2000;148(4):635–651. doi: 10.1083/jcb.148.4.635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Cronshaw JM, Krutchinsky AN, Zhang W, Chait BT, Matunis MLJ. Proteomic analysis of the mammalian nuclear pore complex. Journal of Cell Biology. 2002;158(5):915–927. doi: 10.1083/jcb.200206106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Wälde S, Kehlenbach RH. The Part and the Whole: functions of nucleoporins in nucleocytoplasmic transport. Trends in Cell Biology. 2010;20:461–469. doi: 10.1016/j.tcb.2010.05.001. [DOI] [PubMed] [Google Scholar]
- 168.Panté N, Kann M. Nuclear pore complex is able to transport macromolecules with diameters of about 39 nm. Molecular Biology of the Cell. 2002;13(2):425–434. doi: 10.1091/mbc.01-06-0308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Fried H, Kutay U. Nucleocytoplasmic transport: taking an inventory. Cellular and Molecular Life Sciences. 2003;60(8):1659–1688. doi: 10.1007/s00018-003-3070-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Strambio-De-Castillia C, Niepel M, Rout MP. The nuclear pore complex: bridging nuclear transport and gene regulation. Nature Reviews Molecular Cell Biology. 2010;11(7):490–501. doi: 10.1038/nrm2928. [DOI] [PubMed] [Google Scholar]
- 171.Friedrich B, Quensel C, Sommer T, Hartmann E, Köhler M. Nuclear localization signal and protein context both mediate importin α specificity of nuclear import substrates. Molecular and Cellular Biology. 2006;26(23):8697–8709. doi: 10.1128/MCB.00708-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Poon IKH, Jans DA. Regulation of nuclear transport: central role in development and transformation? Traffic. 2005;6(3):173–186. doi: 10.1111/j.1600-0854.2005.00268.x. [DOI] [PubMed] [Google Scholar]
- 173.Kanai M, Hanashiro K, Kim SH, et al. Inhibition of Crm1-p53 interaction and nuclear export of p53 by poly(ADP-ribosyl)ation. Nature Cell Biology. 2007;9(10):1175–1183. doi: 10.1038/ncb1638. [DOI] [PubMed] [Google Scholar]
- 174.Smith WA, Schurter BT, Wong-Staal F, David M. Arginine methylation of RNA helicase A determines its subcellular localization. Journal of Biological Chemistry. 2004;279(22):22795–22798. doi: 10.1074/jbc.C300512200. [DOI] [PubMed] [Google Scholar]
- 175.Kutay U, Lipowsky G, Izaurralde E, et al. Identification of a tRNA-specific nuclear export receptor. Molecular Cell. 1998;1(3):359–369. doi: 10.1016/s1097-2765(00)80036-2. [DOI] [PubMed] [Google Scholar]
- 176.Arts GJ, Fornerod M, Mattaj IW. Identification of a nuclear export receptor for tRNA. Current Biology. 1998;8(6):305–314. doi: 10.1016/s0960-9822(98)70130-7. [DOI] [PubMed] [Google Scholar]
- 177.Calado A, Treichel N, Müller EC, Otto A, Kutay U. Exportin-5-mediated nuclear export of eukaryotic elongation factor 1A and tRNA. EMBO Journal. 2002;21(22):6216–6224. doi: 10.1093/emboj/cdf620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Ohno M, Segref A, Bachi A, Wilm M, Mattaj IW. PHAX, a mediator of U snRNA nuclear export whose activity is regulated by phosphorylation. Cell. 2000;101(2):187–198. doi: 10.1016/S0092-8674(00)80829-6. [DOI] [PubMed] [Google Scholar]
- 179.Ho JHN, Kallstrom G, Johnson AW. Nmd3p is a Crm1p-dependent adapter protein for nuclear export of the large ribosomal subunit. Journal of Cell Biology. 2000;151(5):1057–1066. doi: 10.1083/jcb.151.5.1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Nissan TA, Baßler J, Petfalski E, Tollervey D, Hurt E. 60S pre-ribosome formation viewed from assembly in the nucleolus until export to the cytoplasm. EMBO Journal. 2002;21(20):5539–5547. doi: 10.1093/emboj/cdf547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Hedges J, West M, Johnson AW. Release of the export adapter, Nmd3p, from the 60S ribosomal subunit requires Rpl10p and the cytoplasmic GTPase Lsg1p. EMBO Journal. 2005;24(3):567–579. doi: 10.1038/sj.emboj.7600547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Cheng H, Dufu K, Lee CS, Hsu JL, Dias A, Reed R. Human mRNA export machinery recruited to the 5' end of mRNA. Cell. 2006;127(7):1389–1400. doi: 10.1016/j.cell.2006.10.044. [DOI] [PubMed] [Google Scholar]
- 183.Strasser K, Hurt E. Splicing factor Sub2p is required for nuclear mRNA export through its interaction with Yra1p. Nature. 2001;413(6856):648–652. doi: 10.1038/35098113. [DOI] [PubMed] [Google Scholar]
- 184.Le Hir H, Gatfield D, Izaurralde E, Moore MJ. The exon-exon junction complex provides a binding platform for factors involved in mRNA export and nonsense-mediated mRNA decay. EMBO Journal. 2001;20(17):4987–4997. doi: 10.1093/emboj/20.17.4987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Kataoka N, Diem MD, Kim VN, Yong J, Dreyfuss G. Magoh, a human homolog of Drosophila mago nashi protein, is a component of the splicing-dependent exon-exon junction complex. EMBO Journal. 2001;20(22):6424–6433. doi: 10.1093/emboj/20.22.6424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Clouse KN, Luo MJ, Zhou Z, Reed R. A Ran-independent pathway for export of spliced mRNA. Nature Cell Biology. 2001;3(1):97–99. doi: 10.1038/35050625. [DOI] [PubMed] [Google Scholar]
- 187.Gallouzi IE, Steitz JA. Delineation of mRNA export pathways by the use of cell-permeable peptides. Science. 2001;294(5548):1895–1901. doi: 10.1126/science.1064693. [DOI] [PubMed] [Google Scholar]
- 188.Waggoner S, Sarnow P. Viral ribonucleoprotein complex formation and nucleolar-cytoplasmic relocalization of nucleolin in poliovirus-infected cells. Journal of Virology. 1998;72(8):6699–6709. doi: 10.1128/jvi.72.8.6699-6709.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Gustin KE. Inhibition of nucleo-cytoplasmic trafficking by RNA viruses: targeting the nuclear pore complex. Virus Research. 2003;95(1-2):35–44. doi: 10.1016/S0168-1702(03)00165-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Meerovitch K, Pelletier J, Sonenberg N. A cellular protein that binds to the 5’-noncoding region of poliovirus RNA: implications for internal translation initiation. Genes & Development. 1989;3(7):1026–1034. doi: 10.1101/gad.3.7.1026. [DOI] [PubMed] [Google Scholar]
- 191.Hellen CUT, Witherell GW, Schmid M, et al. A cytoplasmic 57-kDa protein that is required for translation of picornavirus RNA by internal ribosomal entry is identical to the nuclear pyrimidine tract-binding protein. Proceedings of the National Academy of Sciences of the United States of America. 1993;90(16):7642–7646. doi: 10.1073/pnas.90.16.7642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Mcbride AE, Schlegel A, Kirkegaard K. Human protein Sam68 relocalization and interaction with poliovirus RNA polymerase in infected cells. Proceedings of the National Academy of Sciences of the United States of America. 1996;93(6):2296–2301. doi: 10.1073/pnas.93.6.2296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Shiroki K, Isoyama T, Kuge S, et al. Intracellular redistribution of truncated La protein produced by poliovirus 3C(pro)-mediated cleavage. Journal of Virology. 1999;73(3):2193–2200. doi: 10.1128/jvi.73.3.2193-2200.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.McBride AE, Taylor SJ, Shalloway D, Kirkegaard K. KH domain integrity is required for wild-type localization of Sam68. Experimental Cell Research. 1998;241(1):84–95. doi: 10.1006/excr.1998.4047. [DOI] [PubMed] [Google Scholar]
- 195.Back SH, Kim YK, Kim WJ, et al. Translation of polioviral mRNA is inhibited by cleavage of polypyrimidine tract-binding proteins executed by polioviral 3C(pro) Journal of Virology. 2002;76(5):2529–2542. doi: 10.1128/jvi.76.5.2529-2542.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Belov GA, Lidsky PV, Mikitas OV, et al. Bidirectional increase in permeability of nuclear envelope upon poliovirus infection and accompanying alterations of nuclear pores. Journal of Virology. 2004;78(18):10166–10177. doi: 10.1128/JVI.78.18.10166-10177.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Ranjan P, Bowzard JB, Schwerzmann JW, Jeisy-Scott V, Fujita T, Sambhara S. Cytoplasmic nucleic acid sensors in antiviral immunity. Trends in Molecular Medicine. 2009;15(8):359–368. doi: 10.1016/j.molmed.2009.06.003. [DOI] [PubMed] [Google Scholar]
- 198.Ball JR, Ullman KS. Versatility at the nuclear pore complex: lessons learned from the nucleoporin Nup153. Chromosoma. 2005;114(5):319–330. doi: 10.1007/s00412-005-0019-3. [DOI] [PubMed] [Google Scholar]
- 199.Buendia B, Santa-Maria A, Courvalin JC. Caspase-dependent proteolysis of integral and peripheral proteins of nuclear membranes and nuclear pore complex proteins during apoptosis. Journal of Cell Science. 1999;112(11):1743–1753. doi: 10.1242/jcs.112.11.1743. [DOI] [PubMed] [Google Scholar]
- 200.Park N, Skern T, Gustin KE. Specific cleavage of the nuclear pore complex protein Nup62 by a viral protease. Journal of Biological Chemistry. 2010;285(37):28796–28805. doi: 10.1074/jbc.M110.143404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Enninga J, Levy DE, Blobel G, Fontoura BMA. Role of nucleoporin induction in releasing an mRNA nuclear export block. Science. 2002;295(5559):1523–1525. doi: 10.1126/science.1067861. [DOI] [PubMed] [Google Scholar]
- 202.Lidsky PV, Hato S, Bardina MV, et al. Nucleocytoplasmic traffic disorder induced by cardioviruses. Journal of Virology. 2006;80(6):2705–2717. doi: 10.1128/JVI.80.6.2705-2717.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Porter FW, Bochkov YA, Albee AJ, Wiese C, Palmenberg AC. A picornavirus protein interacts with Ran-GTPase and disrupts nucleocytoplasmic transport. Proceedings of the National Academy of Sciences of the United States of America. 2006;103(33):12417–12422. doi: 10.1073/pnas.0605375103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Dvorak CMT, Hall DJ, Hill M, et al. Leader protein of encephalomyocarditis virus binds zinc, is phosphorylated during viral infection, and affects the efficiency of genome translation. Virology. 2001;290(2):261–271. doi: 10.1006/viro.2001.1193. [DOI] [PubMed] [Google Scholar]
- 205.Her LS, Lund E, Dahlberg JE. Inhibition of ran guanosine triphosphatase-dependent nuclear transport by the matrix protein of vesicular stomatitis virus. Science. 1997;276(5320):1845–1848. doi: 10.1126/science.276.5320.1845. [DOI] [PubMed] [Google Scholar]
- 206.Faria PA, Chakraborty P, Levay A, et al. VSV disrupts the Rae1/mrnp41 mRNA nuclear export pathway. Molecular Cell. 2005;17(1):93–102. doi: 10.1016/j.molcel.2004.11.023. [DOI] [PubMed] [Google Scholar]
- 207.Fortes P, Beloso A, Ortin J. Influenza virus NS1 protein inhibits pre-mRNA splicing and blocks mRNA nucleocytoplasmic transport. EMBO Journal. 1994;13(3):704–712. doi: 10.1002/j.1460-2075.1994.tb06310.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Pichlmair A, Schulz O, Tan CP, et al. RIG-I-mediated antiviral responses to single-stranded RNA bearing 5′-phosphates. Science. 2006;314(5801):997–1001. doi: 10.1126/science.1132998. [DOI] [PubMed] [Google Scholar]
- 209.Mibayashi M, Martínez-Sobrido L, Loo YM, Cárdenas WB, Gale M, Jr., García-Sastre A. Inhibition of retinoic acid-inducible gene I-mediated induction of beta interferon by the NS1 protein of influenza A virus. Journal of Virology. 2007;81(2):514–524. doi: 10.1128/JVI.01265-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Carissimi C, Saieva L, Baccon J, et al. Gemin8 is a novel component of the survival motor neuron complex and functions in small nuclear ribonucleoprotein assembly. Journal of Biological Chemistry. 2006;281(12):8126–8134. doi: 10.1074/jbc.M512243200. [DOI] [PubMed] [Google Scholar]
- 211.Carissimi C, Saieva L, Gabanella F, Pellizzoni L. Gemin8 is required for the architecture and function of the survival motor neuron complex. Journal of Biological Chemistry. 2006;281(48):37009–37016. doi: 10.1074/jbc.M607505200. [DOI] [PubMed] [Google Scholar]
- 212.Meister G, Bühler D, Pillai R, Lottspeich F, Fischer U. A multiprotein complex mediates the ATP-dependent assembly of spliceosomal U snRNPs. Nature Cell Biology. 2001;3(11):945–949. doi: 10.1038/ncb1101-945. [DOI] [PubMed] [Google Scholar]
- 213.Meister G, Hannus S, Plöttner O, et al. SMNrp is an essential pre-mRNA splicing factor required for the formation of the mature spliceosome. EMBO Journal. 2001;20(9):2304–2314. doi: 10.1093/emboj/20.9.2304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Golembe TJ, Yong J, Dreyfuss G. Specific sequence features, recognized by the SMN complex, identify snRNAs and determine their fate as snRNPs. Molecular and Cellular Biology. 2005;25(24):10989–11004. doi: 10.1128/MCB.25.24.10989-11004.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Kiss T. Biogenesis of small nuclear RNPs. Journal of Cell Science. 2004;117(25):5949–5951. doi: 10.1242/jcs.01487. [DOI] [PubMed] [Google Scholar]
- 216.Almstead LL, Sarnow P. Inhibition of U snRNP assembly by a virus-encoded proteinase. Genes and Development. 2007;21(9):1086–1097. doi: 10.1101/gad.1535607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Yalamanchili P, Banerjee R, Dasgupta A. Poliovirus-encoded protease 2A(Pro) cleaves the TATA-binding protein but does not inhibit host cell RNA polymerase II transcription in vitro. Journal of Virology. 1997;71(9):6881–6886. doi: 10.1128/jvi.71.9.6881-6886.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Weidman MK, Sharma R, Raychaudhuri S, Kundu P, Tsai W, Dasgupta A. The interaction of cytoplasmic RNA viruses with the nucleus. Virus Research. 2003;95(1-2):75–85. doi: 10.1016/s0168-1702(03)00164-3. [DOI] [PubMed] [Google Scholar]
- 219.Blom N, Hansen J, Blaas D, Brunak S. Cleavage site analysis in picornaviral polyproteins: discovering cellular targets by neural networks. Protein Science. 1996;5(11):2203–2216. doi: 10.1002/pro.5560051107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Badorff C, Lee GH, Lamphear BJ, et al. Enteroviral protease 2A cleaves dystrophin: evidence of cytoskeletal disruption in an acquired cardiomyopathy. Nature Medicine. 1999;5(3):320–326. doi: 10.1038/6543. [DOI] [PubMed] [Google Scholar]
- 221.Seipelt J, Liebig HD, Sommergruber W, Gerner C, Kuechler E. 2A proteinase of human rhinovirus cleaves cytokeratin 8 in infected HeLa cells. Journal of Biological Chemistry. 2000;275(26):20084–20089. doi: 10.1074/jbc.275.26.20084. [DOI] [PubMed] [Google Scholar]