Abstract
A circular cryptic plasmid named pPAGA (2,734 bp) was isolated from Pantoea agglomerans strain EGE6 (an endophytic bacterial isolate from eucalyptus). Sequence analysis revealed that the plasmid has a G+C content of 51% and contains four potential ORFs, 238(A), 250(B), 131(C), and 129(D) amino acids in length without homology to known proteins. The shuttle vector pLGM1 was constructed by combining the pPAGA plasmid with pGFPmut3.0 (which harbors a gene encoding green fluorescent protein, GFP), and the resulting construct was used to over-express GFP in E. coli and P. agglomerans cells. GFP production was used to monitor the colonization of strain EGE6gfp in various plant tissues by fluorescence microscopy. Analysis of EGE6gfp colonization showed that 14 days after inoculation, the strain occupied the inner tissue of Eucalyptus grandis roots, preferentially colonizing the xylem vessels of the host plants.
Keywords: green fluorescent protein, bacteria, host plant, Eucalyptus
Introduction
Endophytic bacteria are microorganisms that can reside inside a host plant without triggering harmful reactions and/or induce the development of external structures like rhizobial nodules (Azevedo and Araújo, 2007). Due to the ability of endophytes to proliferate inside plant tissue, they are more likely to interact closely with the host than are rhizosphere bacteria (Reinhold-Hurek and Hurek, 1998). These characteristics make bacterial endophytes excellent candidates for the development of sustainable crop production strategies (Sturz et al. 2000).
Eucalyptus is one of the most important crops in Brazil and is used for the production of cellulose and paper (Sociedade Brasileira de Silvicultura). Previous studies have described the genus Pantoea (Gammaproteobacteria) as an important group of bacteria endophytically colonizing plants (Araújo et al., 2001, 2002; Duan et al., 2007; Procópio et al., 2009). The roles of such endophytic bacteria inside the host plant include nitrogen fixation, production of phytohormones, solubilization of phosphates, and induction of systemic resistance (Sturz et al. 2000; Feng et al., 2006; Ortmann and Moerschbacher, 2006: Son et al., 2006).
The combination of its endophytic nature and the high occurrence of the genus Pantoea in plants generate an opportunity to address the increasing interest in genetically modified endophytes (GMEs) (Andreote et al., 2004). The use of GMEs is based on the introduction of heterologous genes into endophytic bacteria to confer new characteristics that may be useful in monitoring plant colonization and, features within the inner tissue of the host plant. Here we describe the characterization and cloning of a cryptic plasmid found in strains of Pantoea agglomerans isolated from eucalyptus. Moreover, we also describe the construction of a shuttle vector carrying the gene gfp, which was efficiently used to monitor the bacterial colonization of eucalyptus plants.
Materials and Methods
Bacterial strains
The bacterial strain EGE6 (P. agglomerans) was previously isolated from Eucalyptus grandis (Procópio REL, PhD Thesis, University of São Paulo, 2004), and its phylogenetic affiliation was assessed by sequencing the 16S rDNA gene. Genomic DNA from the strain was extracted from 1 mL of an overnight culture as described by Sambrook et al. (1989). The 16S rRNA gene was amplified by PCR using primers (27f and 1378r) and protocols described by Weisburg et al. (1991). The PCR product was purified using a GFX PCR DNA (Amersham Biosciences) and Gel Band Purification kit (Amersham Biosciences), and the resulting sample was sequenced using the 1378R primer on an automated sequencer (Applied Biosystems 3100). The resulting chromatogram was analyzed for sequence quality using Phred/Phrap, and only bases with quality values above 20 were used for phylogenetic analysis (490 bp in total). The final sequence was compared to the database from the GenBank by a non-redundant BLASTn search (nr/nt). Additionally, the phylogenetic analysis of the obtained sequence was performed using the ARB software package (Department of Microbiology, Technical University of Munich, Munich, Germany). The nucleotide sequence obtained in this study was deposited at GenBank under the accession code (FN868159).
Nucleotide sequencing and analysis
Plasmid DNA in P. agglomerans strains was isolated by alkaline lysis, as described by Sambrook et al. (1998). A total of approximately 2.7 kb of the pPAGA plasmid was analyzed by restriction mapping, and a unique site for restriction with the endonuclease EcoRI was found. The pPAGA plasmid was then cloned into the vector pUC18 (in Escherichia coli strain DH5α) using the EcoRI site and sequenced by primer walking using the DNA sequencer ABI model 3100 with the Big Dye terminator kit (Applied Biosystems, Foster City, CA), according to the manufacturer's instructions.
The quality of obtained sequences was assessed as previously described for the strain identification, and resulting sequences were assembled using Consed, compounding the final sequence of the pPAGA plasmid. The final sequence was also compared with the GenBank database (with an nr/nt search). Estimation of GC content was made at the EMBL-EBI website. Additionally, analysis of open reading frames (ORFs) and restriction maps was performed using the NEB cutter 2.0 program, and promoter sequences were predicted using the “Neural Network Promoter Prediction” program of the University of California, Berkeley. The nucleotide sequence of pPAGA has been assigned GenBank accession number (FN868248).
Expression vector construction and transformation of the EGE6 strain
A constitutive ribosomal RNA promoter was first cloned into the vector pGFPmut3.1, driving a strong and continuous expression of the gene gfp. The 340-bp region containing the RNA promoter was amplified by PCR with specific primers for the ribosomal RNA promoter from the E. coli genome (Shen et al., 1982). Second, the pGFPmut3.1 vector harboring the promoter region was linearized with the endonuclease EcoRI for ligation with EcoRI-digested pPAGA. This ligation generated the endophyte vector pLGM1, which expresses the gfp gene constitutively. Such ligation did not interfere with any ORF present in the original pPAGA plasmid, maintaining the plasmid function as in the original strain.
The pLGM1 vector was then introduced into EGE6 cells by electroporation (2.5 kV, 200 Ω, 25 μF) as described by Andreote et al. (2004), and the recombinant P. agglomerans strain was named EGE6gfp. Under ultraviolet light, EGE6gfp cells display an intense fluorescent green color, evidencing the vector induced production of GFP protein within the bacteria.
E. grandis cultivation and inoculation with EGE6gfp
Eucalyptus seedlings were inoculated with a suspension of EGE6gfp cells. To generate the cell suspension, EGE6gfp was cultured in 5 mL of liquid LB medium supplemented with ampicillin (100 μg/mL) for 5 h at 150 rpm. Cells were harvested by centrifugation (5,000 g for 5 min), washed and inoculated into new liquid LB medium without antibiotics. Following culture for 10 h at 150 rpm, cells were harvested and rinsed twice with 10 mM potassium phosphate buffer (pH 7). The final suspension was prepared in sterilized distilled water at a final concentration of 106 cells per ml (as determined turbidimetrically and confirmed by plating counts).
Eucalyptus seedlings used in this study were 40 days old and had an average height of 25 cm. Seedlings were obtained by seed cultivation in vermiculite supplied with water. Seedlings were inoculated with 1 mL of bacterial suspension administered to the rhizospheres. To achieve a proper inoculation, the bacterial cell suspension was carefully introduced 1 cm below the vermiculite surface using a pipette tip. This process prevented the contamination of sampled aliquots with aboveground E. grandis tissue. Plants were grown at 28 °C with a 14 h photoperiod in a controlled environmental chamber for 14 days. At day 14 after inoculation, the plants were examined with an epifluorescence microscope (Zeiss Axiophot-2) and photographed with an automatic photograph system as described by Lacava et al. (2007).
Results
Sequencing analysis of the pPAGA plasmid
Restriction analysis of pPAGA revealed a unique site for restriction with the endonuclease EcoRI. This site was used for the insertion of the cryptic plasmid into the pUC18 cloning vector, allowing for its sequencing by primer walking. This procedure resulted in the determination of the sequence of 2,734 base pairs making up the pPAGA plasmid.
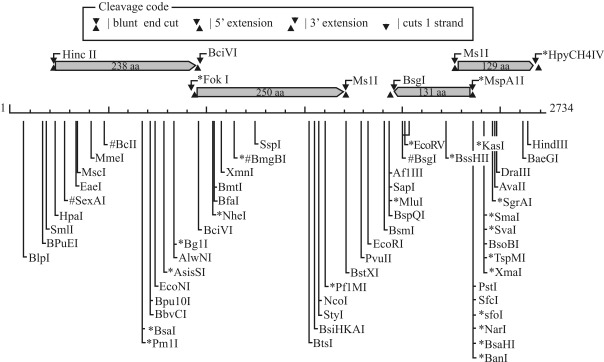
The GC content of pPAGA is 51.71%, which is within the range of GC contents previously reported in the genomic analysis of E. coli (50.8%, Blattner et al., 1997) and Erwinia carotovora (51.0%, Bell et al., 2004). Sequence analysis showed the presence of four putative ORFs in pPAGA, each of which would encode a protein of more than 100 amino acids (aa) (Figure 1).
Figure 1.
Physical map and genetic organization of the cryptic plasmid pPAGA, indicating the locations and orientations of the four described putative ORFs. The most relevant restriction sites are also indicated.
A putative operon was also found, comprising ORFs 1 and 2, which are separated by only three nucleotides. orf1 putatively encodes the largest polypeptide (238 aa), extending from nucleotides 232 to 948. The sequence of this ORF displays high coverage (up to 98%) and similarity (> 50%) values with hypothetical proteins from distinct bacterial species such as Solibacter, Methylobacterium, Bacillus, Beijerinckia, and Burkholderia. orf2 spans nucleotides 951 to 1703 and putatively encodes a protein of 250 aa. Within this ORF, it was possible to observe the presence of a highly conserved domain named DUF2382, which is found in many bacteria but is of unknown function. These results regarding ORFs 1 and 2 indicate that such proteins may have common roles in different bacterial groups. Their presence on a plasmid, which can be easily exchanged in the environment among bacterial cells not related phylogenetically, may explain the diversity of species carrying similar proteins.
Both other ORFs were minor, encoding peptides of 131 and 129 aa, possibly related to the presence of truncated genes in the plasmid. orf3 (131 aa) shows low similarity (30% to 50%) with proteins involved in type IV secretion system in Cupriavidus taiwanensis (Betaproteo-bacteria), while orf4 (129 aa) presents also low levels of similarity with enzymes involved in the promotion of oxidative processes in bacterial cells. In general, there was low resolution in the BLAST analysis of these ORFs, indicating that there is limited knowledge about plasmids found in endophytic bacteria.
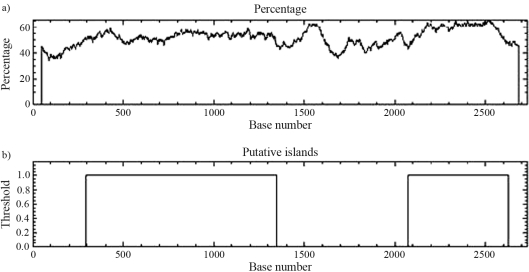
Despite the presence of these ORFs, no similarity was found between the plasmid sequence and sequences reported as origins of plasmid replication (ori regions). However, analysis of the DNA sequence revealed two AT-rich regions in the plasmid (Figure 2). These regions are described in the literature as being related to a replication origin (ori) region, due to the intrinsic capacity of such a sequence to promote double helix dissociation (Ioannidis et al., 2007). The existence of islands within the plasmid sequence was also indicated, reinforcing its origin from gene transfer and recombination with other DNA sequences.
Figure 2.
GC content analysis of the pPAGA plasmid extracted from strain EGE6, a eucalyptus plant endophyte. The GC content (%) along the sequence is represented (a) along with regions possibly constituting islands within the plasmid (b).
The attempt to find putative promoter regions within the sequence of the pPAGA plasmid resulted in the identification of nine regions possibly involved in the promotion of gene expression with scores higher than 0.90 (scale from 0.0 to 1.0) (Table 1). Five of these regions were found in the forward orientation, along with three ORFs, and four putative promoters were observed in the reverse orientation, where only one ORF is located. The location of two putative promoter regions (PrFw1 and PrFw2) upstream of ORFs 1 and 2 corroborates the possibility of an operon being formed by these two ORFs and also suggests that the regulation of such an operon would be modulated by more than one distinct factor. Considering the location of other putative promoter regions, it is also possible to attribute the regulation of orf4 to PrFw4 and PrFw5 and the possible control of orf3 to PrRv4. The other putative promoter regions were not adjacent to any ORF but may be involved in the regulation of plasmid replication regions.
Table 1.
List of putative promoter regions found within the sequence of the cryptic plasmid pPAGA obtained from cells of the endophytic Pantoea agglomerans strain EGE6.
| Strand | Putative promoter name | Sequence location | Putative promoter sequence | Promoter score | Possible ORF regulation |
|---|---|---|---|---|---|
| Forward | PrFw1 | 86–131 | CTTTTACTTAAATTGCCTGCATTTTTCTGTATTTTTCTATCTTTTGTAGT | 0.99 | orf1, orf2 |
| PrFw2 | 94–139 | TAAATTGCCTGCATTTTTCTGTATTTTTCTATCTTTTGTAGTTCTTTGTG | 0.90 | orf1, orf2 | |
| PrFw3 | 1813–1858 | TCATGGTTTTAAGAATTCACCAGACATGATACAACTCCCTGCATGTCGAA | 0.99 | ND | |
| PrFw4 | 2066–2110 | AGATGGCTGAAAGCCGACACTGATGAGAATAATATCCAATGGATCTGAAA | 0.98 | orf4 | |
| PrFw5 | 2246–2291 | AGTGTGATGACAGCCACCGGATGCTGACATTTCATACCGATGTCCGCAGC | 0.93 | orf4 | |
| Reverse | PrRv1 | 2130-2085 | CAGGCGGTTGCTTCAAACGTTTTCAGATCCATTGGATATTATTCTCATCA | 0.98 | ND |
| PrRv2 | 1742-1697 | CTGTAATCGTATTTACTCAAAATGTTGCATCCACCATCATGGCATTACGG | 0.91 | ND | |
| PrRv3 | 1421-1376 | TCGCTGGCCTCTTTACGCAGCACCACTTCTTCAATAATTTCCGCAGTCTT | 0.93 | ND | |
| PrRv4 | 246-201 | TCAGGAGGTTGATTGTTCATGGTTAACCTGCCTGTAATACAGAGCGATCA | 0.94 | orf3 | |
ND - ORF regulation was not indicated based on the location within the pPAGA plasmid sequence.
Construction of an expression vector based on pPAGA
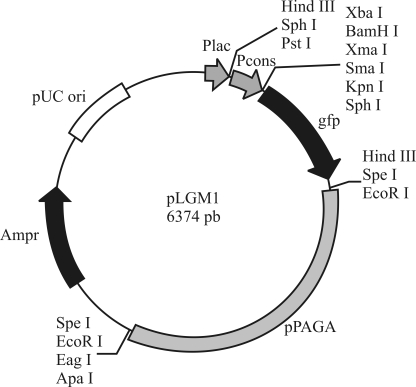
The relatively small size (2,734 bp) of the plasmid pPAGA makes it a candidate for the development of a shuttle vector for endophytic P. agglomerans and related species. Such a strategy followed by introducing pPAGA sequence into the pUC18 cloning vector along with a 212-bp fragment from the E. coli rRNA promoter controlling the gene responsible for the synthesis of Gfp (Figure 3). A chimeric plasmid was generated, containing the cryptic plasmid, the pUC18 vector, and a reporter gene. Re-introduction of the new vector, designated pLGM1, into P. agglomerans cells resulted in an excellent means for visualizing the cells under ultraviolet light. Due to the use of the pPAGA backbone, the final vector was stable in EGE6 cells, where it showed high levels of GFP production.
Figure 3.
Schematic representation of the shuttle vector pLGM1. The cloning sites and promoter are indicated.
Endophytic colonization by P. agglomerans EGE6gfp
The colonization of P. agglomerans in eucalyptus seedlings was analyzed by the introduction of the expression vector pLGM1 into cells of the strain EGE6, generating the genetically modified endophytic strain EGE6gfp. Image analysis of cells from strain EGE6gfp showed that these bacteria have the capacity to enter root tissue and establish colonization in the inner tissue of the host plant (Figure 4). Based on such imaging, it is also possible to suggest that these bacteria inhabit the vascular tissue of eucalyptus seedlings, preferentially xylem cells.
Figure 4.
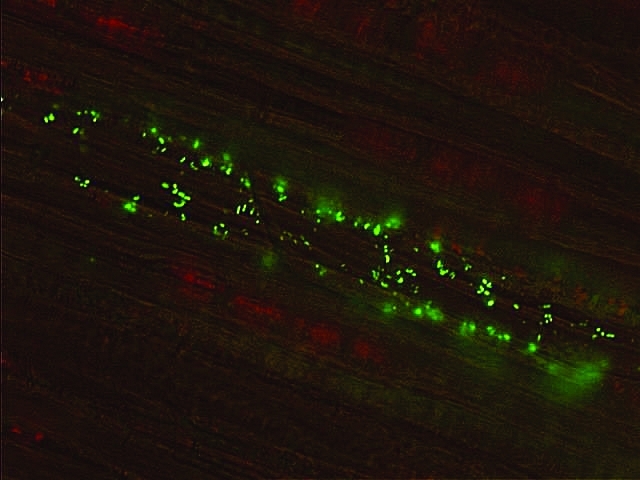
Fluorescence microscopy image of the endophytic P. agglomerans EGE6gfp colonizing the root tissue of E. grandis. The fluorescent green area indicates the presence of bacteria within the plant, colonizing the region occupied by the vascular system (xylem). The plant provides a support for the colonizing bacteria (red and brown background).
Discussion
In this study we describe the characterization of a cryptic plasmid named pPAGA isolated from Pantoea agglomerans strain EGE6, an endophytic bacterial isolate from eucalyptus. Bacteria of the species P. agglomerans have been considered one of the most important groups in terms of endophytic interaction with plants (Torres et al., 2008). Strains of this species have been found in studies using several plant species, such as rice (Verma et al., 2004), citrus (Araújo et al., 2001), and eucalyptus (Ferreira et al., 2008). In some of these studies, the authors have demonstrated plant colonization by the addition of reporter genes into the bacterial cells (Sabaratnam and Beattie, 2003; Verma et al., 2004; Duan et al., 2007). Previous work in eucalyptus has shown P. agglomerans to be a seed endophyte, suggesting that endophytic P. agglomerans can be transmitted vertically from seeds to seedlings in Eucalyptus (Ferreira et al., 2008). It has also been suggested that P. agglomerans can be transported through xylem vessels or through the colonization of intercellular spaces in root and aerial tissues (Compant et al., 2005). Verma et al. (2004) suggested that genus Pantoea is an aggressive endophytic colonizer of deep-water rice. The authors made this conclusion based on a competition experiment, in which another endophytic strain of the genus Ochrobactrum showed very little colonization in the presence of Pantoea sp.
The strain EGE6, used in this study, was previously described (Procópio REL, PhD Thesis, University of São Paulo, 2004) as harboring a cryptic plasmid, named pPAGA, which was characterized and sequenced in this study. A similar approach was recently described by Andreote et al. (2008), who also found a cryptic plasmid, pPA3.0, within cells of endophytic P. agglomerans isolated from citrus plants. In a comparison with the findings obtained by Andreote et al. (2008), the identified plasmids did not present similarities. Plasmid sizes vary by approximately 200 pb (pPA3.0 is 2.9 kb, while pPAGA is 2.7 kb in length), and are divergent regarding the genetic information they carry. While the plasmid described in citrus endophytes primarily presents small ORFs, with the biggest one encoding a peptide of 199 aa, the pPAGA plasmid harbors several ORFs coding for peptides of around 230 aa. Based on both analyses, it is possible to suggest that endophytic P. agglomerans species are important keepers of cryptic plasmids, possibly providing for the efficient exchange of genetic material with other endophytic bacteria within plants. Further studies are necessary to determine the essentiality of these ORFs for endophytic behavior and development within plants.
There was variation in the GC content of the cryptic plasmid, suggesting the presence of an origin of replication without high similarity to any previously described ori regions and also the existence of regions that were incorporated into pPAGA by horizontal gene transfer and recombination. Horizontal transfer likely occurs in soil (Syvanen and Kado, 1998; Gebhard and Smalla, 1999;), where these bacteria can live part of their life, but it still represents a phenomenon to be explored in endophytic communities. One could suggest that the putative genes orf1 and orf2, found under the regulation of the same promoter regions, are essential for the endophytic characteristics of P. agglomerans and, therefore, are refractory to transmission by horizontal gene transfer within the host plant. However, this hypothesis must be tested further to be properly affirmed.
Similar to results described by Andreote et al. (2008), we used the cryptic plasmid backbone to develop a shuttle vector that is able to carry and express exogenous genes within the host plant. This approach was based on other studies, in which several shuttle vectors were constructed using replication regions from small cryptic plasmids (An and Miyamoto, 2006; Matsui et al., 2007; Sangrador-Vegas et al., 2007). The selected reporter gene, from pGFPmut3.1, was a variant gfp gene with higher fluorescence and reduced half-life (Andersen et al., 1998, 2001). The efficient introduction of such a gene in endophytes and the further visualization of cells expressing the exogenous gene within plant tissue support the possibility of introducing new characteristics into endophytes. The genetic modification of bacteria is useful for the expression of genes that can benefit the host plant.
We also observed preferential occupation of the plant vessels by the genetically modified endophyte EGE6gfp, suggesting that this bacterium can invade internal root tissues of eucalyptus seedlings by passing epidermal and cortical cells and permeating the central cylinder. Such results corroborate the data described by Ferreira et al. (2008), suggesting that if bacteria related to P. agglomerans are transferred by seeds, it will result in the preferential spreading of bacterial cells along the aerial portion of the plant through xylem vessels once the plant begins to develop.
In this study we caracterized a cryptic plasmid found in endophytic P. agglomerans isolated from eucalyptus. In addition, we used this plasmid to create a shuttle vector, which can provide a means for introducing genes into plant cells. This study will support further research aiming to manipulate endophytic bacteria for the benefit of healthy plants.
Acknowledgments
This work was supported by a grant from Suzano de Papel e Celulose S.A. We thank FAPESP for providing a fellowship to R.E.L.P. (Grant no. 00/07053-2).
Footnotes
Associate Editor: Luis Carlos de Souza Ferreira
References
- An HY, Miyamoto T. Cloning and sequencing of plasmid pLC494 isolated from human intestinal Lactobacillus casei: Construction of an E. coli-Lactobacillus shuttle vector. Plasmid. 2006;55:128–134. doi: 10.1016/j.plasmid.2005.08.005. [DOI] [PubMed] [Google Scholar]
- Andersen JB, Sternberg C, Poulsen LK, Bjørn SP, Givskov M, Molin S. New unstable variants of green fluorescent protein for studies of transient gene expression in bacteria. Appl Environ Microbiol. 1998;64:2240–2246. doi: 10.1128/aem.64.6.2240-2246.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersen JB, Heydorn A, Hentzer M, Eberl L, Geisenberger O, Christensen BB, Molin SR, Givskov M. Gfp-based N-acyl homoserine-lactone sensor systems for detection of bacterial communication. Appl Environ Microbiol. 2001;67:575–585. doi: 10.1128/AEM.67.2.575-585.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andreote FD, Gullo MJM, Lima AOD, Maccheroni W, Jr, Azevedo JL, Araujo WL. Impact of genetically modified Enterobacter cloacae on indigenous endophytic community of Citrus sinensis seedlings. J Microbiol. 2004;42:169–173. [PubMed] [Google Scholar]
- Andreote FD, Rossetto PB, Souza LCA, Marcon J, Maccheroni W, Jr, Azevedo JL, Araujo WL. Endophytic population of Pantoea agglomerans in citrus plants and development of a cloning vector for endophytes. J Basic Microbiol. 2008;48:338–346. doi: 10.1002/jobm.200700341. [DOI] [PubMed] [Google Scholar]
- Araújo WL, Maccheroni W, Jr, Aguilar-Vildoso CI, Barroso PAV, Saridakis HO, Azevedo JL. Variability and interactions between endophytic bacteria and fungi isolated from leaf tissues of citrus rootstocks. Can J Microbiol. 2001;47:229–236. doi: 10.1139/w00-146. [DOI] [PubMed] [Google Scholar]
- Araújo WL, Marcon J, Maccheroni W, Junior, Elsas JD, van Vuurde JWL, Azevedo JL. Diversity of endophytic bacterial populations and their interaction with Xylella fastidiosa in citrus plants. Appl Environ Microbiol. 2002;68:4906–4914. doi: 10.1128/AEM.68.10.4906-4914.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azevedo JL, Araújo WL. Diversity and applications of endophytic fungi isolated from tropical plants. In: Ganguli BN, Desmukh SK, editors. Fungi: Multifaceted Microbes. Anamaya Publishers; New Delhi: 2007. pp. 189–207. [Google Scholar]
- Bell KS, Sebaihia M, Pritchard L, Holden MTG, Hyman LJ, Holeva MC, Thomson NR, Bentley SD, Churcher LJC, Mungall K, et al. Genome sequence of the enterobacterial phytopathogen Erwinia carotovora subsp. atroseptica and characterization of virulence factors. Proc Natl Acad Sci USA. 2004;101:11105–11110. doi: 10.1073/pnas.0402424101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blattner FR, Plunkett G, III, Bloch CA, Perna NT, Burland V, Riley M, Collado-Vides J, Glasner JD, Rode CK, Mayhew GF, et al. The complete genome sequence of Escherichia coli K-12. Science. 1997;277:1453–1474. doi: 10.1126/science.277.5331.1453. [DOI] [PubMed] [Google Scholar]
- Compant S, Reiter B, Sessitsch A, Nowak J, Clement C, Barka EA. Endophytic colonization of Vitis vinifera L. by plant growth-promoting bacterium Burkholderia sp. strain PsJN. Appl Environ Microbiol. 2005;71:1685–1693. doi: 10.1128/AEM.71.4.1685-1693.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan J, Yi T, Lu Z, Shen D, Feng Y. Rice endophyte Pantoea agglomerans YS19 forms multicellular symplasmata via cell aggregation. FEMS Microbiol Lett. 2007;270:220–226. doi: 10.1111/j.1574-6968.2007.00677.x. [DOI] [PubMed] [Google Scholar]
- Feng Y, Shen D, Song W. Rice endophyte Pantoea agglomerans YS19 promotes host plant growth and affects allocations of host photosynthates. J Appl Microbiol. 2006;100:938–945. doi: 10.1111/j.1365-2672.2006.02843.x. [DOI] [PubMed] [Google Scholar]
- Ferreira A, Quecine MC, Laçava PT, Azevedo JL, Araújo WL. Diversity of endophytic bacteria from Eucalyptus species seeds and colonization of seedlings by Pantoea agglomerans. FEMS Microbiol Lett. 2008;287:8–14. doi: 10.1111/j.1574-6968.2008.01258.x. [DOI] [PubMed] [Google Scholar]
- Gebhard F, Smalla K. Monitoring field releases of genetically modified sugar beets for persistence of transgenics plant DNA and horizontal transfer. FEMS Microbiol Ecol. 1999;28:261–272. [Google Scholar]
- Ioannidis P, Hotopp JCD, Sapountzis P, Siozios S, Tsiamis G, Bordenstein SR, Baldo L, Werren JH, Bourtzis K. New criteria for selecting the origin of DNA replication in Wolbachia and closely related bacteria. BMC Genomics. 2007;8:1–15. doi: 10.1186/1471-2164-8-182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lacava PT, Araujo WL, Azevedo JL. Evaluation of endophytic colonization of Citrus sinensis and Catharanthus roseus seedlings by endophytic bacteria. J Microbiol. 2007;45:11–14. [PubMed] [Google Scholar]
- Matsui T, Saeki H, Shinzato N, Matsuda H. Analysis of the 7.6-kb cryptic plasmid pNC500 from Rhodococcus rhodochrous B-276 and construction of Rhodococcus-E. coli shuttle vector. Appl Microbiol Biotech. 2007;74:169–175. doi: 10.1007/s00253-006-0660-7. [DOI] [PubMed] [Google Scholar]
- Ortmann I, Moerschbacher BM. Spent growth medium of Pantoea agglomerans primes wheat suspension cells for augmented accumulation of hydrogen peroxide and enhanced peroxidase activity upon elicitation. Planta. 2006;224:963–970. doi: 10.1007/s00425-006-0271-7. [DOI] [PubMed] [Google Scholar]
- Procópio REL, Araújo WL, Maccheroni W, Jr, Azevedo JL. Characterization of an endophytic bacterial community associated with Eucalyptus spp. Genet Mol Res. 2009;8:1408–1422. doi: 10.4238/vol8-4gmr691. [DOI] [PubMed] [Google Scholar]
- Reinhold-Hurek B, Hurek T. Interactions of gramineous plants with Azoarcus spp. and other diazotrophs: Identification, localization, and perspectives to study their function. Crit Rev Plant Sci. 1998;17:29–54. [Google Scholar]
- Sabaratnam S, Beattie GA. Differences between Pseudomonas syringae pv. syringae B728a and Pantoea agglomerans BRT98 in epiphytic and endophytic colonization of leaves. Appl Environ Microbiol. 2003;69:1220–1228. doi: 10.1128/AEM.69.2.1220-1228.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EFE, Maniatis T. Molecular Cloning: A Laboratory Manual. 2nd edition. Cold Spring Harbor Laboratory Press; New York: 1998. [Google Scholar]
- Sangrador-Vegas A, Stanton C, van Sinderen D, Fitzgerald GF, Ross RP. Characterization of plasmid pASV479 from Bifidobacterium pseudolongum subsp. globosum and its use for expression vector construction. Plasmid. 2007;58:140–147. doi: 10.1016/j.plasmid.2007.02.004. [DOI] [PubMed] [Google Scholar]
- Shen WF, Squires C, Squires CL. Nucleotide sequence of the rrnG ribosomal RNA promoter region of Escherichia coli. Nucleic Acids Res. 1982;10:3303–3313. doi: 10.1093/nar/10.10.3303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Syvanen M, Kado CI. Horizontal Gene Transfer. Chapman & Hall; London: 1998. p. 474. [Google Scholar]
- Son HJ, Park GT, Cha MS, Heo MS. Solubilization of insoluble inorganic phosphates by a novel salt- and pH-tolerant Pantoea agglomerans R-42 isolated from soybean rhizosphere. Biores Techn. 2006;97:204–210. doi: 10.1016/j.biortech.2005.02.021. [DOI] [PubMed] [Google Scholar]
- Sturz AV, Christie BR, Nowak J. Bacterial endophytes: Potential role in developing sustainable systems of crop production. Crit Rev Plant Sci. 2000;19:1–30. [Google Scholar]
- Torres AR, Araujo WL, Cursino L, Hungria M, Plotegher F, Mostasso FL, Azevedo JL. Diversity of endophytic enterobacteria associated with different host plants. J Microbiol. 2008;46:373–379. doi: 10.1007/s12275-007-0165-9. [DOI] [PubMed] [Google Scholar]
- Verma SC, Singh A, Chowdhury SP, Tripathi AK. Endophytic colonization ability of two deep-water rice endophytes, Pantoea sp. and Ochrobactrum sp. using green fluorescent protein reporter. Biotech Lett. 2004;26:425–429. doi: 10.1023/b:bile.0000018263.94440.ab. [DOI] [PubMed] [Google Scholar]
- Weisburg WG, Barns SM, Pelletier DA, Lane DJ. 16S ribosomal DNA amplification for phylogenetic study. J Bacteriol. 1991;173:697–703. doi: 10.1128/jb.173.2.697-703.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
Internet Resources
- EMBL-EBI website, http://www.ebi.ac.uk/Tools (March 2009).
- NEB cutter 2.0 program, http://tools.neb.com/NEBcutter2/index.php (February 2010).
- Neural Network Promoter Prediction, program of the University of California. Berkeley: http://www.fruitfly.org/seq_tools/promoter.html (February 2010). [Google Scholar]
- Sociedade Brasileira de Silvicultura, http://www.sbs.org.br (June 2010).