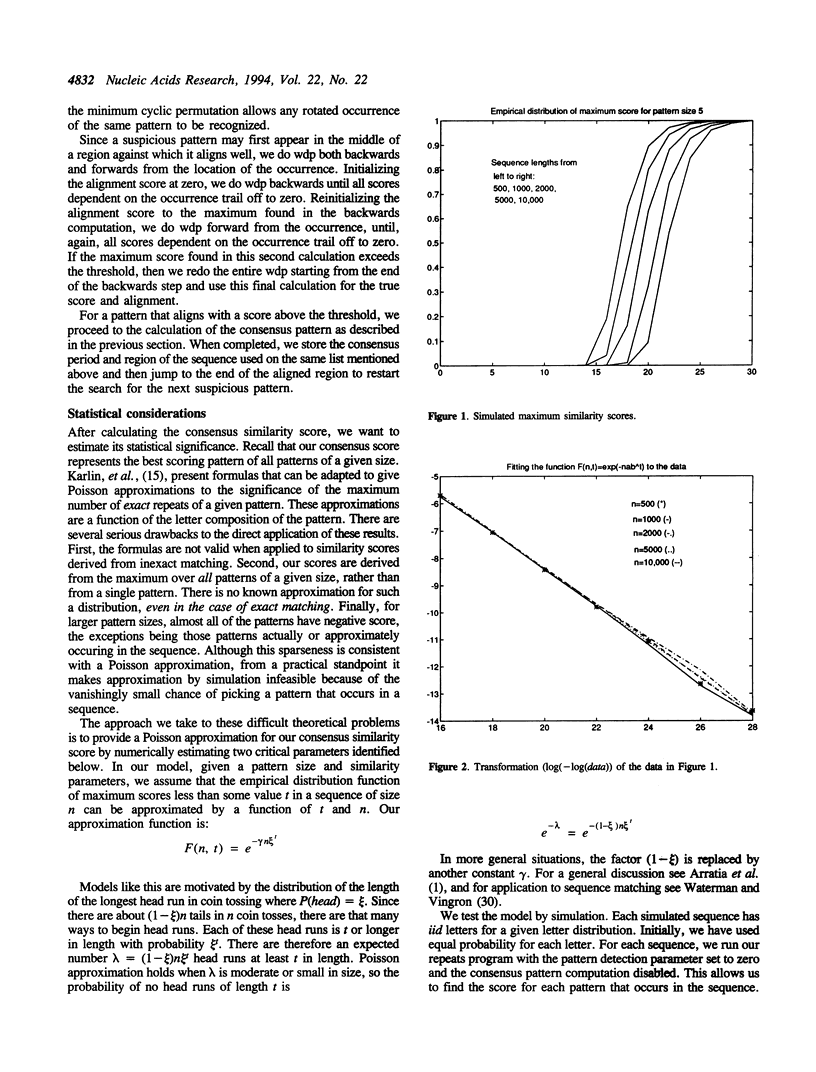
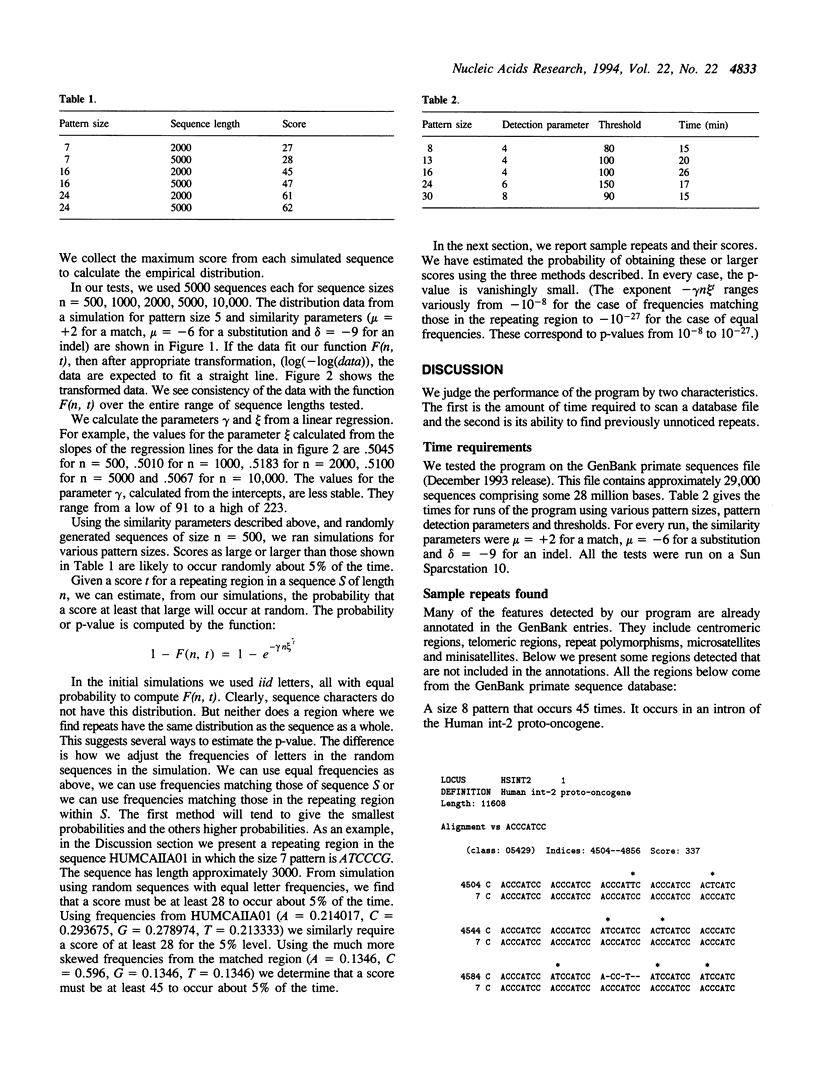
Abstract
A significant portion of DNA consists of repeating patterns of various sizes, from very small (one, two and three nucleotides) to very large (over 300 nucleotides). Although the functions of these repeating regions are not well understood, they appear important for understanding the expression, regulation and evolution of DNA. For example, increases in the number of trinucleotide repeats have been associated with human genetic disease, including Fragile-X mental retardation and Huntington's disease. Repeats are also useful as a tool in mapping and identifying DNA; the number of copies of a particular pattern at a site is often variable among individuals (polymorphic) and is therefore helpful in locating genes via linkage studies and also in providing DNA fingerprints of individuals. The number of repeating regions is unknown as is the distribution of pattern sizes. It would be useful to search for such regions in the DNA database in order that they may be studied more fully. The DNA database currently consists of approximately 150 million basepairs and is growing exponentially. Therefore, any program to look for repeats must be efficient and fast. In this paper, we present some new techniques that are useful in recognizing repeating patterns and describe a new program for rapidly detecting repeat regions in the DNA database where the basic unit of the repeat has size up to 32 nucleotides. It is our hope that the examples in this paper will illustrate the unrealized diversity of repeats in DNA and that the program we have developed will be a useful tool for locating new and interesting repeats.
Full text
PDF






Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Edwards A., Hammond H. A., Jin L., Caskey C. T., Chakraborty R. Genetic variation at five trimeric and tetrameric tandem repeat loci in four human population groups. Genomics. 1992 Feb;12(2):241–253. doi: 10.1016/0888-7543(92)90371-x. [DOI] [PubMed] [Google Scholar]
- Fu Y. H., Pizzuti A., Fenwick R. G., Jr, King J., Rajnarayan S., Dunne P. W., Dubel J., Nasser G. A., Ashizawa T., de Jong P. An unstable triplet repeat in a gene related to myotonic muscular dystrophy. Science. 1992 Mar 6;255(5049):1256–1258. doi: 10.1126/science.1546326. [DOI] [PubMed] [Google Scholar]
- Gall J. G., Atherton D. D. Satellite DNA sequences in Drosophila virilis. J Mol Biol. 1974 Jan 5;85(4):633–664. doi: 10.1016/0022-2836(74)90321-0. [DOI] [PubMed] [Google Scholar]
- Gotoh O. Optimal sequence alignment allowing for long gaps. Bull Math Biol. 1990;52(3):359–373. doi: 10.1007/BF02458577. [DOI] [PubMed] [Google Scholar]
- Hamada H., Seidman M., Howard B. H., Gorman C. M. Enhanced gene expression by the poly(dT-dG).poly(dC-dA) sequence. Mol Cell Biol. 1984 Dec;4(12):2622–2630. doi: 10.1128/mcb.4.12.2622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hellman L., Steen M. L., Sundvall M., Pettersson U. A rapidly evolving region in the immunoglobulin heavy chain loci of rat and mouse: postulated role of (dC-dA)n.(dG-dT)n sequences. Gene. 1988 Aug 15;68(1):93–100. doi: 10.1016/0378-1119(88)90602-6. [DOI] [PubMed] [Google Scholar]
- La Spada A. R., Wilson E. M., Lubahn D. B., Harding A. E., Fischbeck K. H. Androgen receptor gene mutations in X-linked spinal and bulbar muscular atrophy. Nature. 1991 Jul 4;352(6330):77–79. doi: 10.1038/352077a0. [DOI] [PubMed] [Google Scholar]
- Milosavljević A., Jurka J. Discovering simple DNA sequences by the algorithmic significance method. Comput Appl Biosci. 1993 Aug;9(4):407–411. doi: 10.1093/bioinformatics/9.4.407. [DOI] [PubMed] [Google Scholar]
- Myers E. W., Miller W. Optimal alignments in linear space. Comput Appl Biosci. 1988 Mar;4(1):11–17. doi: 10.1093/bioinformatics/4.1.11. [DOI] [PubMed] [Google Scholar]
- Needleman S. B., Wunsch C. D. A general method applicable to the search for similarities in the amino acid sequence of two proteins. J Mol Biol. 1970 Mar;48(3):443–453. doi: 10.1016/0022-2836(70)90057-4. [DOI] [PubMed] [Google Scholar]
- Pardue M. L., Lowenhaupt K., Rich A., Nordheim A. (dC-dA)n.(dG-dT)n sequences have evolutionarily conserved chromosomal locations in Drosophila with implications for roles in chromosome structure and function. EMBO J. 1987 Jun;6(6):1781–1789. doi: 10.1002/j.1460-2075.1987.tb02431.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith G. P. Evolution of repeated DNA sequences by unequal crossover. Science. 1976 Feb 13;191(4227):528–535. doi: 10.1126/science.1251186. [DOI] [PubMed] [Google Scholar]
- Smith T. F., Waterman M. S. Identification of common molecular subsequences. J Mol Biol. 1981 Mar 25;147(1):195–197. doi: 10.1016/0022-2836(81)90087-5. [DOI] [PubMed] [Google Scholar]
- Stallings R. L., Ford A. F., Nelson D., Torney D. C., Hildebrand C. E., Moyzis R. K. Evolution and distribution of (GT)n repetitive sequences in mammalian genomes. Genomics. 1991 Jul;10(3):807–815. doi: 10.1016/0888-7543(91)90467-s. [DOI] [PubMed] [Google Scholar]
- Strand M., Prolla T. A., Liskay R. M., Petes T. D. Destabilization of tracts of simple repetitive DNA in yeast by mutations affecting DNA mismatch repair. Nature. 1993 Sep 16;365(6443):274–276. doi: 10.1038/365274a0. [DOI] [PubMed] [Google Scholar]
- Streisinger G., Okada Y., Emrich J., Newton J., Tsugita A., Terzaghi E., Inouye M. Frameshift mutations and the genetic code. This paper is dedicated to Professor Theodosius Dobzhansky on the occasion of his 66th birthday. Cold Spring Harb Symp Quant Biol. 1966;31:77–84. doi: 10.1101/sqb.1966.031.01.014. [DOI] [PubMed] [Google Scholar]
- Sutherland G. R., Richards R. I. Anticipation legitimized: unstable DNA to the rescue. Am J Hum Genet. 1992 Jul;51(1):7–9. [PMC free article] [PubMed] [Google Scholar]
- Verkerk A. J., Pieretti M., Sutcliffe J. S., Fu Y. H., Kuhl D. P., Pizzuti A., Reiner O., Richards S., Victoria M. F., Zhang F. P. Identification of a gene (FMR-1) containing a CGG repeat coincident with a breakpoint cluster region exhibiting length variation in fragile X syndrome. Cell. 1991 May 31;65(5):905–914. doi: 10.1016/0092-8674(91)90397-h. [DOI] [PubMed] [Google Scholar]
- Weber J. L., May P. E. Abundant class of human DNA polymorphisms which can be typed using the polymerase chain reaction. Am J Hum Genet. 1989 Mar;44(3):388–396. [PMC free article] [PubMed] [Google Scholar]
- de Bustros A., Nelkin B. D., Silverman A., Ehrlich G., Poiesz B., Baylin S. B. The short arm of chromosome 11 is a "hot spot" for hypermethylation in human neoplasia. Proc Natl Acad Sci U S A. 1988 Aug;85(15):5693–5697. doi: 10.1073/pnas.85.15.5693. [DOI] [PMC free article] [PubMed] [Google Scholar]



