Abstract
Similarities between New World and Old World vultures have been interpreted to reflect a close relationship and to suggest the inclusion of both in Accipitridae (Falconiformes). However, deeper analyses indicated that the placement of the New World vultures (cathartids) in this Order is uncertain. Chromosome analysis has shown that cathartids retained a karyotype similar to the putative avian ancestor. In order to verify the occurrence of intrachromosomal rearrangements in cathartids, we hybridized whole chromosome probes of two species (Gallus gallus and Leucopternis albicollis) onto metaphases of Cathartes aura. The results showed that not only were the syntenic groups conserved between Gallus and C. aura, but probably also the general gene order, suggesting that New World vultures share chromosomal symplesiomorphies with most bird lineages.
Keywords: Cathartes, FISH, Gallus, Leucopternis, whole-chromosome probes
Vultures are large, carrion-eating birds with hooked bills, featherless heads, and soaring food-search behavior. Similarities in external appearance and lifestyle between New World (“cathartid”) and Old World (“accipitrid”) vulture species usually have been interpreted to reflect a close phylogenetic relationship, such that both groups traditionally have been included in the Falconiformes (Avise et al., 1994). However, deeper analyses have shown that cathartid vultures are anatomically very distinct and it is uncertain whether they belong in the order Falconiformes (Amadon, 1977). DNA-DNA hybridization (Sibley et al., 1988) and mitochondrial cytochrome b sequencing (Avise et al., 1994) were used to address a controversial suggestion that New World vultures are more closely related to storks than to Old World vultures. This would indicate that the morphological and behavioral similarities between New World and Old World vultures probably represent convergent adaptations associated with carrion-feeding, rather than proximity of descent. According to Seibold and Helbig (1995), New World vultures are not birds of prey, but the phylogenetic information available was insufficient to determine if they are more closely related to storks or to Accipitriformes.
Karyological data also support the exclusion of Cathartidae from the clade of birds of prey: New World vultures have conserved karyotypes similar to the putative complement of the Avian ancestor, with 2n = 80 and two distinct groups of chromosomes – macro and microchromosomes. In fact, six of the seven species of Cathartidae have already been analyzed, at least by conventional staining. They showed very similar karyotypes with 2n = 80 (Takagi and Sazaki, 1974; de Boer, 1975, 1976; Raudsepp et al., 2002; Tagliarini et al., 2007, 2009). Banding analyses showed that the few differences were due to distinct amounts and distribution of heterochromatic blocks in Cathartes aura, C. burrovianus and Coragyps atratus (Tagliarini et al., 2009). Chromosome painting has demonstrated a complete retention of the syntenic groups proposed as present in the putative bird ancestral karyotype. This conclusion was based on chromosome painting of metaphases of the Californian Condor, Gymnogyps californianus, with the six largest chromosomes of Gallus gallus (Raudsepp et al., 2002).
In this work, we performed fluorescent in situ hybridization (FISH) with whole chromosome probes of ten autosome pairs and the Z chromosome of Gallus gallus onto Turkey vulture (Cathartes aura) chromosomes. Our aim was to evaluate if the retention of the syntenic groups observed in Gymnogyps californianus is also present in species of Cathartidae with more derived karyotypes.
Metaphase chromosomes were obtained from a male of Cathartes aura using tissue culture from feather pulp (Sasaki et al., 1968, with modifications). Chromosome preparations were obtained by standard arrest with colcemid (Gibco), hypotonic treatment with 0.075 M KCl, and cell fixation with methanol:acetic acid (3: 1). The chromosome preparations were dropped onto glass slides and air-dried. For diploid number (2n) counting and karyotyping the chromosomes were stained with Giemsa (5%) and G-banded.
Chromosome specific paints of Gallus gallus (chromosomes GGA 1–10 and GGA Z) and of Leucopternis albicollis (LAL 2, 3, 4, 6, 7, 9, 13, 15, 17, 18, 20 and 26, which are region-specific to GGA chromosomes and altogether cover pairs 1, 2 and 3). Both sets of probes were generated from flow sorted chromosomes, which were amplified and labelled with biotin (Roche) or fluorescein (Roche) by degenerate oligonucleotide primed PCR (DOP-PCR). Single and dual color experiments were performed, according to the protocol of de Oliveira et al. (2005). Images of Giemsa-stained metaphases and of FISH experiments were captured with a cooled CCD camera coupled to a Zeiss Axiophot microscope. Camera control, digital image acquisition and pseudocolour assignment were performed using the Smartcapture VP 1.4 software (DigitalScientific, Cambridge, UK).
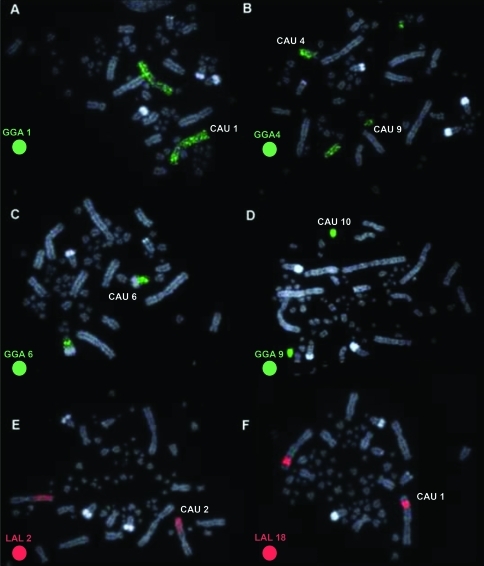
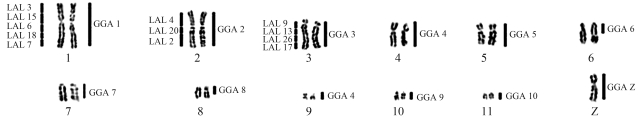
The diploid number of Cathartes aura (CAU) was 2n = 80 and the chromosomal morphology was similar to those previously reported for other Cathartidae species. Each of the chromosome-specific painting probes GGA1-10 and Z hybridized to only a single pair of macrochromosomes in the Turkey vulture, except for paint GGA4, which hybridized to two distinct pairs, a medium submetacentric (pair 4) and a small metacentric (pair 9) one (Figure 1a–d). Pairs 6 and 8 of C. aura were hybridized by paints GGA6 and GGA8, respectively, but only in the proximal region of the long arm. The large terminal heterochromatic blocks found in these two pairs were not painted by any probe. Chromosome paints LAL3, 6, 7, 15 and 18 hybridized to CAU1; LAL 2, 4 and 20 to CAU2, and LAL 9, 13, 17 and 26 to GGA 3 (Figure 1 e–f). The homology map is shown in Figure 2.
Figure 1.
Representative FISH experiments with chromosome painting probes of Gallus gallus (GGA) (A–D) and Leucopternis albicollis (LAL) (E–F) hybridized onto Cathartes aura (CAU) metaphases. The chromosome probes used are indicated on the bottom left, in green (fluoroscein labelled) or red (biontin-cy3 labelled).
Figure 2.
Homology map between Cathartes aura and Gallus gallus (right) and Leucopternis albicollis (left).
Retention of a karyotype similar to the putative ancestor in many different groups of birds has been confirmed through the use of chromosome painting with Gallus probes. Some authors inferred that the relatively unchanged nature of the diploid chromosome number among the majority of avian species further implies that such a karyotype was and still is a highly successful means of genome organization (Griffin et al., 2007). Although some lineages, such as Falconiformes, Strigiformes and Charadriiformes, among others, show very derived karyotypes, most bird species present a 2n close to 80 and, apart from GGA4, which usually paints two different pairs, most syntenic groups of Gallus are conserved as one pair each in these karyotypes. Moreover, even though avian homologues to chicken microchromosomes remain to be defined, the similar morphology and number of microchromosomes in other birds is evidence of their retention as syntenic groups. On the other hand, in lineages with derived karyotypes, it has been shown that these small chromosomes are involved in a number of rearrangements, especially fusions (de Oliveira et al., 2005; Nishida et al., 2008; Nie et al., 2009).
Falconiformes (Accipitridae and Falconidae) have derived karyotypes and it is usually assumed that they have low diploid numbers, as the American Kestrel (Falco sparberius) with 2n = 50 (Shields, 1982). Nevertheless, high diploid numbers were found in Aquila adalberti, with 2n = 82 (Padilla et al., 1999) and caracaras (Caracara plancus) and other closely related species with 2n = 92 (Tagliarini et al., 2007). However, most species in these families show diploid numbers close to 2n = 66 or 68 (Amaral & Jorge, 2003).
Similarly, Ciconiiformes have karyotypes with lower diploid numbers, Ciconia nigra with 2n = 52, and Jabiru myctea with 2n = 56 (Belterman and de Boer, 1990). The distinction between macro- and microchromosomes is much clearer than in Falconiformes, and the lower number of dot-like chromosomes indicates possible fusion events specially involving these small pairs. Unfortunately, no species of Ciconiiformes has been analyzed by chromosome painting to assess this hypothesis.
In contrast to Falconiformes and Ciconiiformes, cathartid species have retained plesiomorphic characteristics in the karyotypes of all the species. The use of region-specific probes derived from whole-chromosome paints of L. albicollis reinforced the observation of the same general avian genomic organization in Cathartes aura, where these probes hybridized to similar regions in CAU1, 2 and 3, as was previously found in Gallus gallus (de Oliveira et al., 2010). These results show that not only the syntenic groups, but also the gene order seem to be conserved among different lineages of birds. This observation led us to conclude that New World vultures share cytotaxonomic symplesiomorphies with most lineages of birds, making it difficult to determine their correct phylogenetic position in relation to Falconiformes or Ciconiiformes. Nevertheless, the retention of this basic karyotype formula implies a basal position in relation to these two groups because both of them have species with karyotypes of various levels of derivation, although some diploid numbers are still close to 2n = 80.
Acknowledgments
The authors would like to thank Criadouro Gavião Real (Capitão Poço, Pará, Brazil) for biological samples, and Dr. Margarida Lima, Mr. Jorge Rissino and Mr. Claudio Ferreira for logistic support. This work was partially supported by CNPq (grant 472544/2006-3) and a grant to MAFS from the Wellcome Trust in support of the Cambridge Resource Centre for Comparative Genomics.
Footnotes
Associate Editor: Yatiyo Yonenaga-Yassuda
References
- Amadon D. Notes on the taxonomy of vultures. Condor. 1977;79:413–416. [Google Scholar]
- Amaral KF, Jorge W. The chromosomes of the order Falconiformes: A review. Ararajuba. 2003;11:65–73. [Google Scholar]
- Avise JC, Nelson WS, Sibley CG. DNA sequence support for a close phylogenetic relationship between some storks and New World vultures. Proc Natl Acad Sci USA. 1994;91:5173–5177. doi: 10.1073/pnas.91.11.5173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belterman RHR, De Boer LEM. A karyological study of 55 species of birds, including karyotypes of 39 species new to cytology. Genetica. 1984;93:17–29. [Google Scholar]
- de Boer LEM. Karyological heterogeneity in the Falconiformes (aves) Experientia. 1975;31:1138–1139. doi: 10.1007/BF02326755. [DOI] [PubMed] [Google Scholar]
- de Boer LEM. The somatic chromosomes of 16 species of Falconiformes (Aves) and the karyological relationships of the order. Genetica. 1976:4677–113. [Google Scholar]
- de Oliveira EHC, Habermann F, Lacerda O, Sbalqueiro IJ, Wienberg J, Müller S. Chromosome reshuffling in birds of prey: The karyotypes of the world's largest eagle (Harpy eagle, Harpia harpyja) compared to that of the chicken (Gallus gallus) Chromosoma. 2005;114:338–343. doi: 10.1007/s00412-005-0009-5. [DOI] [PubMed] [Google Scholar]
- de Oliveira EHC, Tagliarini MM, Rissino J, Nagamachi CY, Pieczarka JC, O'Brien P, Ferguson-Smith M. Reciprocal chromosome painting between white halk (Leucopternis albicollis) and chicken reveals extensions fusions and fissions during karyotype evolution of Accipitridae (Aves, Falconiformes) Chromosome Res. 2010;18:349–355. doi: 10.1007/s10577-010-9117-z. [DOI] [PubMed] [Google Scholar]
- Griffin DK, Robertson LBW, Tempest HG, Skinneret BM. The evolution of the avian genome as revealed by comparative molecular cytogenetics. Cytogenet Genome Res. 2007;117:64–77. doi: 10.1159/000103166. [DOI] [PubMed] [Google Scholar]
- Nie W, O'Brien PC, Ng BL, Fu B, Volobouev V, Carter NP, Ferguson-Smith MA, Yang F. Avian comparative genomics: Reciprocal chromosome painting between domestic chicken (Gallus gallus) and the stone curlew (Burhinus oedicnemus, Charadriiformes) - An atypical species with low diploid number. Chromosome Res. 2009;17:99–113. doi: 10.1007/s10577-009-9021-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishida C, Ishijima J, Kosaka A, Tanabe H, Habermann FA, Griffin DK, Matsuda Y. Characterization of chromosome structures of Falconinae (Falconidae, Falconiformes, Aves) by chromosome painting and delineation of chromosome rearrangements during their differentiation. Chromosome Res. 2008;16:171–181. doi: 10.1007/s10577-007-1210-6. [DOI] [PubMed] [Google Scholar]
- Padilla JA, Martinez-Trancon M, Rabasco A, Fernandez-Garcia JL. The karyotype of the Iberian imperial eagle (Aquila adalberti) analyzed by classical and DNA replication banding. Cytogenet Cell Genet. 1999;84:61–66. doi: 10.1159/000015216. [DOI] [PubMed] [Google Scholar]
- Raudsepp T, Houck ML, O'Brien PC, Ferguson-Smith MA, Ryder OA, Chowdhary BP. Cytogenetic analysis of California Condor (Gymnogyps californianus) chromosomes: Comparison with chicken (Gallus gallus) macrochromosomes. Cytogenet Genome Res. 2002;98:54–60. doi: 10.1159/000068532. [DOI] [PubMed] [Google Scholar]
- Sasaki M, Ikeuchi T, Maino S. A feather pulp culture for avian chromosomes with notes on the chromosomes of the peafowl and the ostrich. Experientia. 1968;24:1923–1929. doi: 10.1007/BF02146680. [DOI] [PubMed] [Google Scholar]
- Seibold I, Helbig AJ. Evolutionary history of New and Old World vultures inferred from nucleotide sequences of the mitochondrial cytochrome b gene. Phil Trans R Soc Lond B Biol Sci. 1995;350:163–178. doi: 10.1098/rstb.1995.0150. [DOI] [PubMed] [Google Scholar]
- Shields GF. Comparative avian genetics: A review. Condor. 1982;84:45–58. [Google Scholar]
- Sibley CG, Ahlquist JE, Monroe JA., Jr A classification of the living birds of the world based on DNA-DNA hybridization studies. Auk. 1988;105:409–423. [Google Scholar]
- Tagliarini MM, Nagamachi CY, Pieczarka JC, de Oliveira EHC. Description of two new karyotypes and cytotaxonomic considerations on Falconiformes. Rev Bras Ornit. 2007;15:261–266. [Google Scholar]
- Tagliarini MM, Nagamachi CY, Pieczarka JC, Rissino J, de Oliveira EHC. Chromosomal analysis in Cathartidae: Distribution of heterochromatic blocks and rDNA, and phylogenetic considerations. Genetica. 2009;135:299–304. doi: 10.1007/s10709-008-9278-2. [DOI] [PubMed] [Google Scholar]
- Takagi N, Sazaki A. A phylogenetic study of birds karyotypes. Chromosoma. 1974;46:91–120. doi: 10.1007/BF00332341. [DOI] [PubMed] [Google Scholar]