Abstract
This is an open access article. Unrestricted non-commercial use is permitted provided the original work is properly cited.
The reproducibility of mass spectrometry (MS) data collected using surface enhanced laser desorption/ionization-time of flight (SELDI-TOF) has been questioned. This investigation was designed to test the reproducibility of SELDI data collected over time by multiple users and instruments. Five laboratories prepared arrays once every week for six weeks. Spectra were collected on separate instruments in the individual laboratories. Additionally, all of the arrays produced each week were rescanned on a single instrument in one laboratory. Lab-to-lab and array-to-array variability in alignment parameters were larger than the variability attributable to running samples during different weeks. The coefficient of variance (CV) in spectrum intensity ranged from 25% at baseline, to 80% in the matrix noise region, to about 50% during the exponential drop from the maximum matrix noise. Before normalization, the median CV of the peak heights was 72% and reduced to about 20% after normalization. Additionally, for the spectra from a common instrument, the CV ranged from 5% at baseline, to 50% in the matrix noise region, to 20% during the drop from the maximum matrix noise. Normalization reduced the variability in peak heights to about 18%. With proper processing methods, SELDI instruments produce spectra containing large numbers of reproducibly located peaks, with consistent heights.
Keywords: mass spectrometry/wavelet, analysis of variance/reproducibility/SELDI
Introduction
Since its introduction onto the biotech market in 2000, researchers have used and continue to use the SELDI-TOF MS-based ProteinChip System for discovery of cancer-related biomarkers in addition to markers of other disease states such as peripheral arterial disease, thrombotic thrombocytopenic purpura, sickle cell disease, cystic fibrosis, hereditary thrombophilia, Peyronie disease and Alzheimer disease.1–7 Numerous examples of potential biomarkers for cancer detection,8–19 prognosis,20–22 response to therapy23–25 or survival26 identified using the SELDI platform continue to appear in the literature. Questions about the reproducibility of MS data collected using a SELDI system have, however, led to uncertainty about the overall usefulness of this technology in cancer proteomics research. One of the most thorough evaluations of SELDI- TOF-discovered biomarkers, the EDRN prostate cancer biomarker study,27,28 was launched following demonstration of acceptable reproducibility across laboratories.29 In the end, SELDI-discovered biomarkers appeared to lack diagnostic value for prostate cancer after a second stage of validation failed to separate cancer from normal controls. The authors pointed to possible flaws in study design such as the use of unfractionated serum. In support of such reservations, other investigators have suggested that some form of fractionation of serum or plasma will be required to detect consistently the more interesting, low abundance proteins.30–32
In addition to the use of patient blood, other cancer biomarker studies have been generated on the SELDI platform using such samples as urine, saliva, nipple aspirate fluid (NAF), pancreatic juice, tissue lysates and cell culture.33–39 Whatever the sample source or extent of preparation prior to analysis, it is important to remember that in designing experiments using biological samples, both pre-analytical and analytical factors contribute to the variability in measurements made using any assay method, including SELDI-TOF-MS. These factors include inter-individual biological differences, differences in sample handling, differences in protocol design and execution, and differences in instrument performance.
Studies designed specifically to assess SELDI peak reproducibility have consistently reported coefficient of variation (CV’s) in the 30% range, both on an intra-laboratory and inter-laboratory basis.29,40–42 A number of studies have attempted to ascertain factors that impact variability. Liggett et al determined that variance was greater between separate arrays (biochips) than between spots on a single array and suggested this variability might be decreased by improving the array manufacturing and handling processes.43 Some of these studies have explored pre-analytical factors. For example, Timms et al explored various types of tubes for blood collection, clotting time, and storage time on peak profiles.41 The authors concluded that the most stringent and the least stringent conditions had the least variability, presumably because the most stringent condition controls proteolysis and the least stringent condition allows for proteolysis to reach completion. Understanding the relative contribution of these factors to overall variability should permit researchers to better design and execute clinical proteomics studies.
This investigation, conducted in five individual laboratories at the University of Texas M.D. Anderson Cancer Center, was designed to test the reproducibility of SELDI data collected over time by multiple users and instruments. Our focus was to test the effect of consistent sample handling, preparation, and in particular, data acquisition among participating laboratories on the variability of data obtained over time. Arrays were manually prepared with a fresh serum sample weekly for six consecutive weeks in each laboratory. All laboratories used aliquots from the same batch of serum. Mass spectra were collected on separate instruments in the individual laboratories. In addition, all arrays were read on a common instrument in the same laboratory every week. In this report, we describe the analysis of the spectra collected over time on individual instruments as well as on the common instrument. The experimental design and the methods for assessing reproducibility are applicable to other forms of MS used in protein profiling, not just SELDI.
Materials and Methods
Instruments
All five laboratories followed a standard operating procedure (SOP). Briefly, five ProteinChip System instruments (Ciphergen Biosystems, Inc.) located in separate laboratories and operated by the respective laboratory personnel were used. Four were the PBSII series; one was a PBSIIc. Before the study, each instrument was checked by a service engineer and adjusted to perform at individual optimum levels. Instruments were not engineered to be ‘matched’ for identical data production. Heat Inactivated Fetal Bovine Serum (FBS) (Invitrogen-Life Technologies) was used as the quality control sample for all laboratories. Two commercially available (Ciphergen Biosystems, Inc.) cationic exchange arrays (CM10) were prepared manually with samples once every week for six weeks. CM10 arrays were pre-wetted twice with binding buffer and 50 μl of a denatured serum sample was added to each spot of the array in a bioprocessor. The arrays were incubated with sample for 30 min with shaking. The model of shaker varied across laboratories. After incubation, each spot was washed three times with binding buffer and then quickly two times with water. After air-drying, 1 μl of sinapinic acid (Ciphergen Biosystems) was applied twice. Before reading the prepared arrays, a new calibration curve was created from 12230, 16951 and 29023 Daltons each week from All in One Protein II mass calibrants (Ciphergen Biosystems), prepared by each user according to the manufacturer’s instructions. The spot protocol for calibration curve data collection was set identically on each instrument for parameters of lag time, warmings hots, mass optimization window, and number of total shots averaged (Supplementary SOP file). The PBSIIc allows for setting of a deflector mass and was set at 1000 Daltons. However, the sample-containing arrays were read using settings for low mass data collection that varied in laser intensity and detector sensitivity as was optimal for each instrument.
Study design
Each week for six weeks, investigators in five laboratories manually prepared protein samples from aliquots of the same batch of serum. Samples were applied to all spots of two CM10 ProteinChip arrays (16 spots per lab per week). Spectra were collected on the instruments in the individual laboratories from each spot using low intensity laser settings. In addition, we collected another set of spectra by rescanning all spots at low intensity on a common (PBCIIc) instrument in the same laboratory every week. Because of equipment failure, one laboratory produced no data during two weeks of the study. Because of a clerical error, data from one chip, along with three additional spots, from another laboratory were not scanned on the common instrument. As a result, we collected 445 spectra from individual instruments and 440 spectra on the common instrument.
Processing spectra
Spectra were collected with a digitizer rate of 250 MHz, which was dictated by the hardware; each intensity represents the number of ions hitting the detector during a period of τ = 4 nanoseconds. Raw spectra were aligned by the transformation t′ = Multiplier* t + Offset. The average of all raw spectra was the base spectrum to which all others were aligned. We computed 4 different shifts that maximized the cross-correlation between baseline-corrected spectra within 4 separate small time windows that cover 4 dominant peaks in the mean spectrum (Fig. S4A of Supporting Information). To perform baseline removal, we applied an undecimated discrete wavelet transform (UDWT) with a threshold of 500 to each spectrum (2000 for the average raw spectrum), subtracted the smooth part, and used the residuals as the baseline-corrected spectrum.44 By least squares approximation, we computed the values of Multiplier and Offset for which the transformation above yields the best approximation of the four shifts.
The spectra scanned on the common instrument were collected at 500 MHz (τ = 2 nanoseconds), which was again dictated by the hardware. Raw spectra were aligned by shifting by a small integer multiple of τ; the “multiplier” term, which represents physical differences between instruments, was unnecessary for data scanned on a single instrument. An arbitrary spectrum produced during week 1 from the first laboratory was the base spectrum. We baseline-corrected each spectrum (as above) and computed the shift that maximized the cross-correlation between corrected spectra, using MATLAB version 7.0.1 (The Mathworks, Natick MA).
We used the UDWT with a threshold of 20 to denoise the mean spectrum, using the Cromwell software package;44,45 local maxima in the denoised mean spectrum were identified as peaks. Intervals containing the peaks and the location of the maxima within intervals were recorded. Peak heights were quantified as the difference between the maxima and minima in the corresponding interval.
Linear models were fit in R version 2.7.1, using the functions lm and lme.46,47
Results
Alignment variability across instruments
All spectra were aligned as described above. For all but 13 of the 445 spectra, the offsets ranged between −10τ and +20τ. The 13 unusual spectra had offsets between 115τ and 120τ; they were produced in the same lab during the same week. These 13 spectra had poor alignment for t < 30 μs (Figs. S6 and S7 of Supporting Information); they were excluded from further analysis. For the spectra scanned on the common instrument, the offsets ranged between −10τ and +10τ for all but 3 of the 440 spectra. These 3 spectra had offsets of −42τ, −28τ, and +45τ; they were low quality by visual inspection and excluded.
We hypothesized that spot position on the array might affect the offset (Figs. S2 and S14 of Supporting Information), for two reasons. First, Peaks software (Ciphergen Biosystems) version 3.1.1 included a “spot correction factor” to adjust for the different positions of different spots relative to the laser and drift tube inside the instrument. Second, the measured pressure falls steadily from spot A to H (Figs. S3 and S15 of Supporting Information), since the vacuum pump continues to run while the instrument scans each spot. We tested linear models incorporating spot position (categorical) and pressure (continuous) to predict the offset. When used separately, both pressure (P = 0.1236) and spot position (P = 0.998) failed to predict the offset; and failed to predict the multiplier.
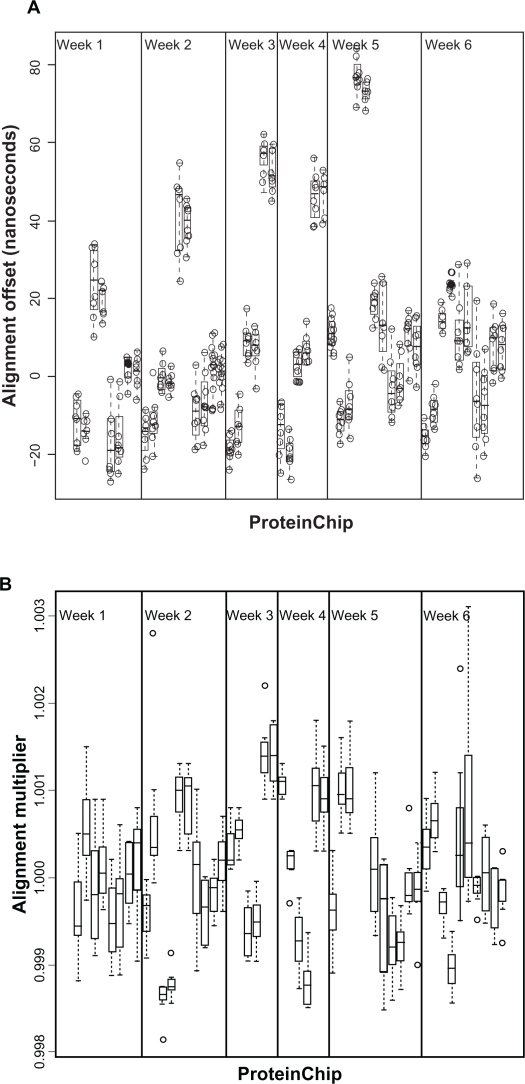
There were significant differences in offset between arrays (Fig. 1), which might be attributable to differences in arrays, in the instruments from week to week, or in the sample preparation protocol and scanning from laboratory to laboratory. We used a linear mixed-effects model to estimate these variances.47
| (1.1) |
Figure 1.
A) Alignment offsets by ProteinChip. The arrays on the x-axis are ordered by week and by laboratory within week. Pairs of arrays from the same laboratory in the same week are adjacent. B) Alignment multiplier by ProteinChip. The arrays on the x-axis are ordered by week and by laboratory within week. Pairs of arrays from the same laboratory in the same week are adjacent.
We write Yijkl for the alignment offset, or multiplier. Here i = 1, …, 8 ranges over the spots, j = 1, …, 5 ranges over laboratories, k = 1, …, 6 ranges over weeks, and l = 1,2 ranges over the duplicate arrays used each week in each laboratory. The term μ represents the overall mean of Y; other terms represent random effects. We let , , and represent the laboratory, week, array effects and residual error (unexplained spot-to-spot variability within a array), respectively. All random effects were assumed to be independent. All the confidence intervals for the estimates overlap (Table 1); all of the variance sources are of comparable importance. The best point estimates suggest that lab-to-lab and array-to-array variability in the alignment offset are likely to be larger than week-to-week variability. The same applies to the Multiplier.
Table 1.
Components of variance explaining the alignment offset and the multiplier.
| Random effect |
Offset |
Multiplier |
||||
|---|---|---|---|---|---|---|
| Standard deviation (nanoseconds) | 95% confidence interval | % variance explained | Standard deviation | 95% confidence interval | % variance explained | |
| Lab | σL = 16.69 | (8.55, 32.60) | 50.07 | σL = 3.10 × 10−4 | (1.21 × 10−4, 8.13 × 10−4) | 12.51 |
| Week | σW = 2.74 | (0.42, 17.73) | 1.35 | σW = 5.77 × 10−8 | NA | 0.00 |
| Array | σC = 15.28 | (12.32, 18.96) | 41.96 | σC = 7.20 × 10−4 | (5.80 × 10−4, 8.89 × 10−4) | 65.55 |
| Spot (residual) | σE = 6.07 | (5.66, 6.52) | 6.62 | σE = 4.20 × 10−4 | (3.97 × 10−4, 4.58 × 10−4) | 22.30 |
Alignment variability on a single instrument
For spectra scanned on the common instrument, both pressure (P = 6.033 × 10−6) and spot position (P = 2.196 × 10−8) were significant predictors of the offset. Spot position remained significant if pressure was already included in the model (P = 0.00015), but pressure became insignificant if added to a model that already included spot position (P = 0.6413). We only include a spot factor to explain the offset.
Similarly, there were significant differences in offset between arrays for the spectra on the common instrument (Fig. S8 of Supporting Information). Each term was significant by fitting the mixed-effect models. The best point estimates (Table S1 of Supporting Information) suggest that spot-to-spot and array-to-array variability are larger than the variability attributable to running samples prepared in different laboratories during different weeks.
Reproducibility of intensities in aligned spectra across instruments
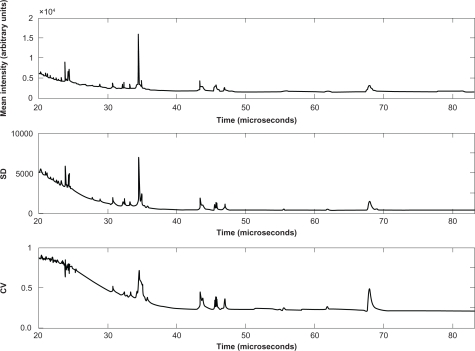
We computed the mean spectrum by averaging the intensities at each time point.45 Peaks of the mean spectra were higher and narrower after alignment (Figs. S4 and S16 of Supporting Information). We computed the standard deviation (SD) and coefficient of variation (CV) at each time point (Fig. 2 and Fig. S9 of Supporting Information). SD appeared to be roughly proportional to the mean spectral intensity. However, SD was relatively greater at the locations of the most prominent peaks; this added variability is due in part to additional variation in the peak locations that is not fully accounted for by the offsets used to align the spectra globally. Away from the peaks, the CV follows a simple pattern. The first 20 μs (about 2860 Daltons/charge) were discarded, since there was substantial matrix noise and saturation for most spectra. From 20 to 25 μs (about 4500 Daltons/charge), it is still the noise region that tends to be highly variable, with CVs reaching 90%. The CV then declines until about 40 μs (11500 Daltons/charge), where it stabilizes at around 25%.
Figure 2.
Reproducibility of spectrum intensity. These graphs contain plots of the mean intensity across 432 aligned spectra (top), the point-wise standard deviation (SD; center), and the point-wise coefficient of variation (CV; bottom) as functions of the time of flight.
Reproducibility of intensities in aligned spectra on a single instrument
For spectra on the common instrument, during the initial period of about 5 μs (214 Daltons/charge, the approximate time when the ionized matrix molecules reach the detector), the spectra are highly reproducible, with CV under 5%. From 5 to 15 μs (about 1545 Daltons/charge), there is a large matrix noise region that tends to be highly variable, with CVs reaching 50%. From here until 30 μs (6150 Daltons/charge), the base CV is near 20%. The CV then declines until about 50 μs (17160 Daltons/charge), where it stabilizes below 5%. From these results, the main variability sources in the spectra are the huge differences in baseline caused by the matrix noise, with additional spikes of variability from large peaks that are not perfectly aligned.
Reliability of peak localization across instruments
The occurrence of a maximum at the end of an interval suggests the interval may not contain a true peak. So, for each peak, we modeled the distribution of the relative locations across spectra as a beta distribution, Beta (α, β); parameters α and β are fit using the method of moments. A peak was “questionable” if both α < 1 and β < 1, since this ensured the distribution was concentrated at the ends of the interval. Otherwise, the peak was “reliable”.
We found 53 questionable peaks and 237 reliable peaks in the spectra collected from different instruments. The questionable peaks tended to be narrower and lower. The median width of intervals containing questionable or reliable peaks was 8τ or 17τ, respectively. The median height of questionable or reliable peaks was 157 or 295, resp. The heights of the questionable peaks usually had slightly higher CVs than other peaks occurring in similar portions of the spectrum. All questionable peaks occurred during the first 40 μs, which corresponds to the exponentially decaying part of the spectra, plus the first two clusters of dominant peaks. We retained the reliable peaks for further analysis.
Reliability of peak localization on a single instrument
On the common instrument, we found 106 questionable peaks and 477 reliable peaks; the increase in the number of peaks is likely attributable to the higher resolution of the common instrument. The median width of questionable or reliable peaks was 9τ or 37τ, resp., and the median height was 282 or 411, resp. A majority of the questionable peaks occurred: i) between 10 and 12 μs, ii) between 19 and 20 μs, and iii) between 26 and 30 μs. The first portion occurs at the beginning of the sharp rise in the matrix noise region; the other two occur as the matrix noise region is coming to an end, on either side of a cluster of prominent peaks.
Reproducibility of peak heights across instruments
After quantifying the heights of reliable peaks, we computed the CV for each peak (Figs. S1 and S13 of Supporting Information). The median CV was 72%, with inter-quartile range IQR = 10.41%. The CV was similar for most peaks. However, there is a slight drop in CV around t = 40 μs. Having a nearly constant CV suggests that the SD is roughly proportional to the mean. After log-two transformation, the median CV = 1.25% with IQR = 1.6%.
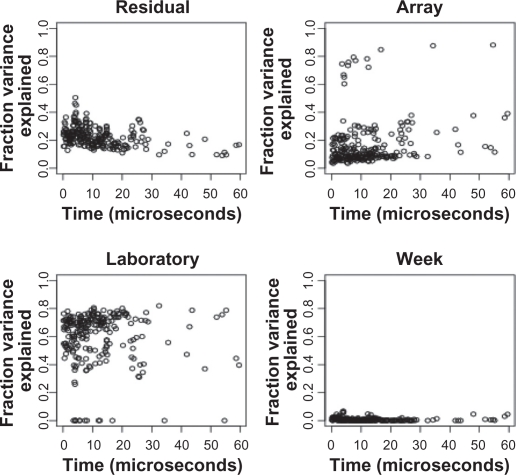
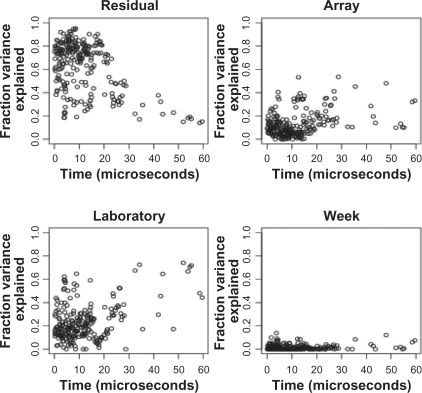
After log transformation, we used Equation (1.1) to construct a separate random-effects model for each peak. The random effects have the same interpretation, but Yijkl denote the logarithmic peak height. At most peaks, the lab-to-lab differences make the largest contribution to the total variance, between 60% and 80%. Week-to-week and array-to-array variation contribute about 10%, and residual contribute about 20% (Fig. 3). Overall, lab and array variation dominate the total variance.
Figure 3.
Results of a peak-by-peak decomposition of variance components before normalization. Each panel shows the percentage of variance of the log-transformed peak heights, as a function of the time-of-flight, for one of the factors (top left: residual; top right: array; lower left: laboratory; lower right: week).
Reproducibility of peak heights on a single instrument
For spectra scanned on the common instrument, the median CV was 33%, with IQR = 8.46%. The CV was similar for most peaks after reaching the maximum in the matrix noise region at around 10 μs. After log-two transformation, the median CV = 6.1% with IQR = 1.7%. At most peaks, the residual or spot-to-spot variability made the largest contribution, between 40% and 60%. Week-to-week and array-to-array differences contribute about 20%; lab-to-lab differences contribute less than 10% (Fig. S10 of Supporting Information). At both ends of the spectrum, however, the array-to-array variance dominates the spot-to-spot variance. Overall, spot and array effects are at least as large as week and laboratory effects.
The impact of normalization on reproducibility of peak heights across instruments
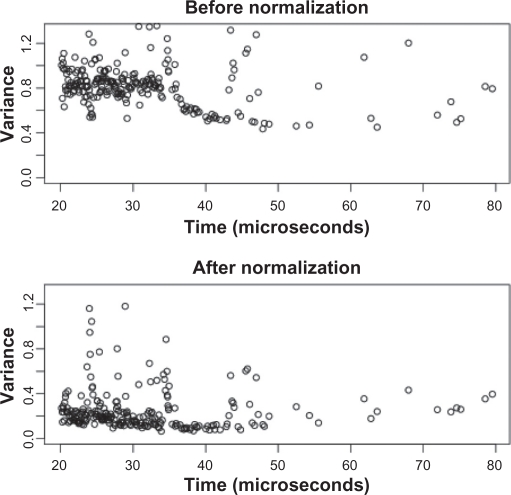
We globally normalized the log-transformed data within each spectrum by subtracting the mean log peak height for that spectrum. This procedure substantially reduced the variance in peak heights, from a median variance of 0.8177 before to 0.195 after normalization (Fig. 4). After normalization, we repeated the peak-by-peak variance analysis described previously (Fig. 5 and Fig. S12 of Supporting Information). The contribution of week-to-week differences is nearly zero. In general, the contribution of lab-to-lab differences was decreased substantially, while array-to-array stayed about the same. For most peaks, nearly all the remaining variability arises from the spot-to-spot differences.
Figure 4.
Variance of the log-transformed peak heights before and after normalization.
Figure 5.
Results of a peak-by-peak decomposition of variance components after normalization. Each panel shows the percentage of variance of the log-transformed peak heights, as a function of the time-of-flight, for one of the factors (top left: residual; top right: array; lower left: laboratory; lower right: week).
The impact of normalization on reproducibility of peak heights on a single instrument
On the common instrument, the same procedure reduced the peak height variance from 0.243 to 0.117 (Fig. S11 of Supporting Information). The week-to-week differences contribution is also nearly zero. In general, the percent of variance attributable to array differences was decreased, while lab-to-lab stayed about the same. For most peaks, nearly all the remaining variability arises from the residual spot-to-spot differences.
Discussion
In 2005, the Early Detection Research Network (EDRN) conducted the first study to assess the reproducibility of SELDI spectra.29 They reported that frequent and accurate calibrations were essential for reproducibility. While we agree with the EDRN’s assessment, we find that alignment of spectra (without calibration) is adequate for reproducibility. In the present study, we found that the offsets required to align the spectra correctly were on the order of −10τ to +20τ, where τ = 4 nanoseconds was the time resolution of the instrument. An analysis of the variability in the offset showed that the lab-to-lab and array-to-array variability was at least as large as the variability attributable to spot differences or different weeks when the experiments were performed. When the spectra were prepared in different laboratories but scanned on the same instrument, the offsets required to align the spectra correctly were on the order of ±10τ, where τ = 2 nanoseconds was the time resolution of the instrument. Moreover, part of this offset could be explained by a fixed spot effect arising from the different spot positions. An analysis of the remaining variability in the offset showed that the spot-to-spot and array-to-array variability was at least as large as lab-to-lab or week-to-week variability. When spectra are scanned on different instruments, those instrument differences are larger than differences over time. When spectra are merely prepared by different laboratories but scanned on a common instrument, the differences in alignment due to preparation using a standard protocol appear to be relatively small.
The EDRN based their assessment of the reproducibility on only three peaks, the largest and most prominent peaks in their samples, and concluded that the largest peaks had the best chances of being reproducible.28 Our study investigated the variability in all 237 peaks that we were able to detect in our spectra. Our approach yields a much more detailed picture of the reproducibility of the spectra as a whole, which we believe will be important when trying to use low intensity peaks as markers to distinguish between different disease states. In addition, this study included an analysis of spectra collected on the same physical instrument, whereas the EDRN study only used data from different instruments. By restricting our attention to spectra collected on a single instrument, we were able to assess the variability inherent in the sample preparation protocol, without confounding that variability with differences between instruments.
The EDRN found that the CV of the peak height (or intensity) ranged between 9% and 41%. This result is comparable to our findings on a common instrument, where the median CV of peaks in unnormalized spectra was 33%. Johnson and colleagues48 reported that the median CV for “ion abundance” of peaks was also around 30% on an extremely high-resolution Fourier-transform mass spectrometer. It is conceivable that a CV of around 30% is inherent in the technology that is being used to extract ions from samples, regardless of how the abundance of those ions is later measured in a mass spectrometer. Our study also assessed the differences between instruments. We found slightly higher CVs point-wise for all time points in the spectrum, as well as for the heights of individual peaks, where the median CV in unnormalized spectra was 72%. Normalization reduced the CV of peak heights to 18%–20% both on a common instrument and across different instruments. Studies using various quality control methodologies have reported CV’s in this range. For example, Hong used a correlation matrix tool to exclude outlier spectra, resulting in CV’s ∼20%.49 Normalization using total ion current resulted in CV’s of 15%–20%.50 Forshed et al used a combination of PCA to exclude outlier spectra as well as peak normalization to obtain CV’s between 10%–20%.51
We extended the EDRN analyses by decomposing the variance at each peak. Our main goal was to determine which variability source gave the greatest contribution: 1) differences in the array preparation by user and spectra generation on different instruments, 2) differences in the individual instruments from week-to-week, 3) differences between ProteinChip arrays, or 4) differences between individual spots on the ProteinChip arrays. Before normalization, differences between labs accounted for the largest portion of variability, and differences between spots were second. After normalization, the contribution of lab-to-lab differences was substantially reduced. Similarly, the EDRN prostate cancer reproducibility study demonstrated that intralaboratory variability was comparable to interlaboratory variability (range of intralaboratory CV’s of the three peaks was 16%–43%, 9%–23%, and 11%–20%, respectively where as the range of interlaboratory CV’s of the three peaks was 36, 17, and 15%, respectively).29 In that study, the normalization process within the ProteinChip software was used. In our study, normalization also reduced the total variance in peak heights by about 75%. Interestingly, the percentage of variance explained by array-to-array variation stayed about the same. This suggests that normalization alone is adequate to remove the variability arising from scanning the arrays prepared by different researchers on different instruments.
For the analysis of arrays scanned on a single instrument, our main goal was to determine which of the four sources of variability contributes most: 1) differences in the sample preparation protocol in different laboratories; all other three are same as above. Both before and after normalization, differences between spots accounted for the largest portion of variability, and differences between arrays were second. This suggests that most of the variability observed in this study is inherent in the surface chemistry on the array spots that first binds proteins and then releases ions. Normalization also reduced the total variance in peak heights by about half. Before normalization, differences in the instrument settings from week-to-week were about the same size as array differences. After normalization, these week-to-week differences virtually disappeared. This finding provides strong evidence that a simple global normalization can successfully correct for almost all the differences due to changes in the instrument, at least over a period of six weeks. The percentage of variation caused by array differences also declined after normalization. Interestingly, the percentage of variance explained by different laboratories stayed about the same, in contrast to the effect of normalization on spectra collected on different instruments. However, the amount of variation due to laboratory preparation in the setting of a common instrument started out smaller than the amount due to the combination of laboratory preparation and different instruments in the multi-instrument part of the analysis. Moreover, the final percentage of variance explained by laboratories differences was about the same in the common instrument and multi-instrument situations after normalization. This finding, together with the fact that the absolute amount of variance after normalization is similar in the two studies suggests that our normalization procedure can correct for differences between instruments, but that there are some intrinsic differences in individual sample preparation that cannot be removed simply by normalization. This further suggests that some improvement in reproducibility might be obtained by switching from manual to robotic preparation.
Lastly, the use of wavelets in analyzing mass spectra has been shown to improve peak detection and classification. For example, Du et al demonstrate that their wavelet procedure identifies more peaks (both large and small) while maintaining an acceptable false discovery rate.52 An advantage of the wavelet method is that no baseline subtraction is required, eliminating a step that can induce variable results depending on the parameters used.
Experiments that are intended to discover biomarkers that distinguish between disease states must use designs that avoid confounding the contrast of interest with experimental factors such as run order or laboratory. However, with proper experimental design and data processing, we have demonstrated that the SELDI ProteinChip system is a reproducible technology that can be used reliably for discovery of biomarkers both over time and between laboratories.
Supplimentary data: Protocol for FBS Serum Profiles for QC
Materials provided by Ciphergen:
Single Use Serum Aliquots: Fetal Bovine Serum, Heat Inactivated, Cat # 16140-063, Invitrogen-Life Technologies.
200 μl U9 aliquots (use 2 times, always refreeze at −20).
CM10 arrays-prepare 2 arrays each week (at same time).
CM10 binding/wash buffer-store at 4 °C: aliquot into 10 ml test tube for each use.
2 bioprocessor reservoirs (You will use two columns for each prep, save the reservoir and use different columns each time).
NP20 array (use for calibrants).
- SPA—prepare a fresh tube of SPA each week: 200 μl Acetonitrile (100%) + 200 μl of 1% TFA (also made fresh weekly!), vortex well for several minutes by taping the tube onto a vortex and let it mix continuously. Spin at 10000 rpm/2 min. Remove 200 μl (without touching bottom) into a fresh tube. Store in the dark until use.
- ○ 1% TFA: 100 ul + 10 mls of ddH20: (add TFA in fume hood).
All in One Protein Calibrants. Prepare according to insert directions. Store diluted aliquots at −20 °C. Collect a new spectrum each week. (Can use same array over again). Add 5 ul of SPA to the 5 ul aliquot of calibrants and spot 1 ul onto each spot of a NP20 chip. Keep prepared chip in the dark.
Thaw one serum and one U9 aliquot. Make sure U9 is completely thawed. Vortex well.
Centrifuge serum at 10000 rpm/10 min/4 °C.
Add 30 μl of U9 to a fresh 1.5 ml tube.
Add 20 μl of the centrifuged serum to the U9 = total of 50 μl.
Incubate with gentle shaking at 4 °C/20 minutes. Try to keep this time as consistent as possible.
While serum is incubating at 4 °C, set up a bioprocessor with 2 CM10 arrays. Make sure the gasket doesn’t cover the spot.
Pre-wet spots with 150 μl buffer/spot. Incubate with shaking/5 min/RT.
Remove and repeat buffer pre-wetting.
Add 1450 μl of the binding buffer to the serum/U9 mixture (final 1:75 dilution of the serum). Invert to mix well.
Remove final buffer wash from arrays and add 50 ul of diluted serum/well. Watch for airbubbles!
Cover wells (can use 96 well plate sealer). Shake at RT for 30 minutes. (If using a robot mixer, use Form = 48, amp = 9)
Remove samples.
Add 150 μl of buffer/well. Shake 5 min/RT. Remove.
Repeat 2 X for a total of 3 washes with mixing. 15. Add 150 μl of ddH20. Remove (no incubation). Repeat.
Quickly remove reservoir and wick off excess water from spots.
Let dry 5 min. Keep this time consistent! 18. Add 1 μl SPA/spot. Let dry. Repeat. 19. Read within next hour.
Quick cool centrifuge to 4 °C if necessary.
Take serum and U9 aliquot from freezer.
Thaw serum in your hand quickly, mix gently and begin centrifugation.
Hold U9 in hand until it is thawed. Mix well.
Set up bioprocessor with the 2 arrays.
During the U9/serum incubation, begin the buffer prewashes of the arrays.
During sample incubation on the arrays, prepare SPA.
Always save spectra into the database specified as the “QC project”. The user should be set to ‘QC’. Keep all calibration spectra in a ‘Calibration’ project folder. Create a new project file each week to for saving the weekly files.
Average 100 spectra from a spot with the All in One Protein II calibrants, manual or with a spot protocol. Be sure the intensity and sensitivity values that you specify don’t cause any of the peaks to go off scale. Use the same intensity/detector sensitivity settings each time you create a calibration spectrum for the most consistency. Most calibrant -containing spots will last several weeks and can be read over and over. If you need to increase the laser intensity alot to acquire the same calibrants peaks, then it is time to make a new chip.
Internally calibrate using the masses: 12230, 16951 and 29023. Save spectra. Set as default instrument calibration equation. Collection protocol: high mass = 50,000; optimize from 12,000–30,000 daltons; pulse settings—center; if you have a IIc, set the deflector to 5000 daltons.
Go to Options, choose “Configure dynamic spec tags”: make sure box is checked for adding to spectra as they are acquired. Add to tag: 1) Spot #, 2) Sample Name, 3) Sample Group.
When you enter a barcode, go to File, Open chips, and choose ‘new’. Add barcode. Then type in the information below in the correct spaces.
Sample Name = QC_1 (for first week, then change it to 2, etc).
Sample Group = the PI’s name (ie, Bast).
Sample Type = FBS (not included in the spectral tag).
LMW spot: high mass to acquire; 50,000 daltons; optimization range = 1,000 to 30,000 daltons; time lag focus- set to 700 ns; IIc deflector- set to 1000 daltons; set warming shots to collect 1 at the same intensity that you choose for all data collection, but don’t use warming shots; collect from 20 to 80 with 10 shots/area, delta 5.
HMW spot: high mass to acquire; 200,000 daltons; optimization range = 30,000–150,000 daltons; time lag focus- center; IIc deflector- set to 10,000 daltons; set warming shots to collect 1 at the same intensity that you choose for all data collection, but don’t use warming shots; collect from 23 to 78 with 10 shots/area, delta 5.
Calibration equation: For this study, always use a curve based on 12230, 16951 and 29023 daltons (for both LMW and HMW spectra).
Laser Intensity:
Detector Sensitivity:
- Time lag focus:
- ○ LMW: use 700 ns
- ○ HMW: center of optimization range between 30,000 and 150,000 daltons.
Read the spots with both a low mass acquisition spot protocol and a high mass acquisition spot protocol. The above listed variables should be adjusted so that you collect a QC spectrum that gives a peak pattern as closely resembling the ‘QC standard’ (spectrum saved on your computer) as possible.
First, read spots A–C and compare spectrum to the ‘standard’ stored on your instrument. If you compare a spectrum to one previously acquired in the weeks before and it doesn’t not appear comparable, you can adjust the laser intensity and detector sensitivity in the spot protocol and read again. Then, read spots D–H of that chip and all 8 spots of the second chip you prepare each week (using a chip protocol for both LMW and HMW acquire).
All chips prepared by the different labs will be read on one instrument to assess any preparation issues.
Voltage Detector: check setting and record in your notes: to access this setting, go to Instrument, configuration, in bottom section see Detector = PBSII-(1800 to 2200 V); IIc-(2700 to 3000 V). If you start to lose low intensity peaks from the spectrum, the detector voltage may need to be increased (use 50 unit increases).
Laser Focus (service engineer will change this if needed): If any of the peak shapes change (ie, poor resolution), focusing the laser can help optimize resolution.
Compare the peak shape and intensity in your acquired spectra to the examples stored in the QC database. Adjust setting as required to maintain spectra integrity from week to week.
Approx masses: 3984 da, 8490 da, 13.5, 15.2 and 15.8 kD, 33.3 kD.
Approx masses: 33.3 kD, 66 kD, and 133 kD.
Table S1.
Components of variance explaining the alignment offset for the setting scanned on a common instrument.
| Random effect | Standard deviation (nanoseconds) | 95% confidence interval | % variance explained |
|---|---|---|---|
| Lab | σL = 1.6164 | (0.5949, 4.3916) | 9.3 |
| Week | σW = 2.0608 | (0.9679, 4.3876) | 15.2 |
| Array | σC = 3.5024 | (2.7909, 4.3952) | 43.8 |
| Spot (residual) | σE = 2.9763 | (2.7725, 3.1951) | 31.7 |
Financial relationship with Company 1:
Nature of Relevant Financial Relationship:
□ Full-time/Part-time Employee
□ Grant/Research Support Recipient
□ Advisor/Board Member
□ Consultant/Independent Contractor
□ Stock Shareholder (self managed)
□ Speaker’s Bureau
□ Honorarium Recipient
□ Other (please describe the Other Relationship)
Financial relationship with Company 2:
Nature of Relevant Financial Relationship:
□ Full-time/Part-time Employee
□ Grant/Research Support Recipient
□ Advisor/Board Member
□ Consultant/Independent Contractor
□ Stock Shareholder (self managed)
□ Speaker’s Bureau
□ Honorarium Recipient
□ Other (please describe the Other Relationship)
Financial relationship with Company 3:
Nature of Relevant Financial Relationship:
□ Full-time/Part-time Employee
□ Grant/Research Support Recipient
□ Advisor/Board Member
□ Consultant/Independent Contractor
□ Stock Shareholder (self managed)
□ Speaker’s Bureau
□ Honorarium Recipient
□ Other (please describe the Other Relationship)
Financial relationship with Company 4:
Nature of Relevant Financial Relationship:
□ Full-time/Part-time Employee
□ Grant/Research Support Recipient
□ Advisor/Board Member
□ Consultant/Independent Contractor
□ Stock Shareholder (self managed)
□ Speaker’s Bureau
□ Honorarium Recipient
□ Other (please describe the Other Relationship)
Financial relationship with Company 5:
Nature of Relevant Financial Relationship:
□ Full-time/Part-time Employee
□ Grant/Research Support Recipient
□ Advisor/Board Member
□ Consultant/Independent Contractor
□ Stock Shareholder (self managed)
□ Speaker’s Bureau
□ Honorarium Recipient
□ Other (please describe the Other Relationship)
Financial relationship with Company 6:
Nature of Relevant Financial Relationship:
□ Full-time/Part-time Employee
□ Grant/Research Support Recipient
□ Advisor/Board Member
□ Consultant/Independent Contractor
□ Stock Shareholder (self managed)
□ Speaker’s Bureau
□ Honorarium Recipient
□ Other (please describe the Other Relationship)
□ No, I have no relevant financial relationship(s).
Acknowledgments
This research was funded in part by NIH/NCI grants P30 CA016672, P01 CA106451-01, R01 CA107304-02, P50 CA070907-05 and the M.D. Anderson Ovarian SPORE P50 CA083639-10.
Dr. Bast serves on the Scientific Advisory Board of Vermillion Diagnostics.
Abbreviations
- EDRN
Early Detection Research Network
- FBS
Fetal Bovine Serum
- IQR
inter-quartile range
- SOP
standard operating procedure
- UDWT
undecimated discrete wavelet transform
Footnotes
Disclosure
This manuscript has been read and approved by all authors. This paper is unique and is not under consideration by any other publication and has not been published elsewhere. The authors and peer reviewers of this paper report no conflicts of interest. The authors confirm that they have permission to reproduce any copyrighted material.
References
- 1.Tumblin A, Tailor A, Hoehn, et al. Apolipoprotein A-I and serum amyloid a plasma levels are biomarkers of acute painful episode in patients with sickle cell disease. Haematologica. 2010 Apr 7; doi: 10.3324/haematol.2009.018044. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.MacGregor G, Gray RD, Hilliard TN, et al. Biomarkers for cystic fibrosis lung disease: application of SELDI-TOF mass spectrometry to BAL fluid. J Cyst Fibros. 2008 Sep 7;(5):352–8. doi: 10.1016/j.jcf.2007.12.005. [DOI] [PubMed] [Google Scholar]
- 3.Yang S, Xu L, Wu HM. Rapid multiplexed genotyping for hereditary thrombophilia by SELDI- TOF mass spectrometry. Diagn Mol Pathol. 2010 Mar;19(1):54–61. doi: 10.1097/PDM.0b013e3181a814bf. [DOI] [PubMed] [Google Scholar]
- 4.De Young LX, Bella AJ, O’Gorman DB, Gan BS, Lim KB, Brock GB. Protein biomarker analysis of primary Peyronie’s disease cells. J Sex Med. 2010 Jan;7(1 Pt 1):99–106. doi: 10.1111/j.1743-6109.2009.01556.x. [DOI] [PubMed] [Google Scholar]
- 5.Simonsen AH, McGuire J, Podust VN, et al. Identification of a novel panel of cerebrospinal fluid biomarkers of Alzeheimer’s disease. Neurobiol Aging. 2008 Jul;29(7):961–8. doi: 10.1016/j.neurobiolaging.2007.01.011. [DOI] [PubMed] [Google Scholar]
- 6.Jin M, Cataland S, Bissell M, Wu HM. A rapid test for the diagnosis of thrombotic thrombocytopenic purpura uing surface enhanced laser desorption/ionization time-of-flight (SELDI-TOF)-mass spectrometry. J Thromb Haemost. 2006 Feb;4(2):333–8. doi: 10.1111/j.1538-7836.2006.01758.x. [DOI] [PubMed] [Google Scholar]
- 7.Wilson AM, Kimura E, Harada RK, et al. β2-Microglobulin as a biomarker in peripheral arterial disease: proteomic profiling and clinical studies. Circulation. 2007 Sep 18;116(12):1396–403. doi: 10.1161/CIRCULATIONAHA.106.683722. [DOI] [PubMed] [Google Scholar]
- 8.Fan Y, Wang J, Yang Y, et al. Detection and identification of potential biomarkers of breast cancer. J Cancer Res Clin Oncol. 2010 Mar 17; doi: 10.1007/s00432-010-0775-1. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shi L, Zhang J, Wu P, et al. Discovery and identification of potential biomarkers of pediatric acute lymphoblastic leukemia. Proteome Sci. 2009 Mar 16;7:7. doi: 10.1186/1477-5956-7-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Caron J, Mange A, Guillot B, Solassol J. Highly sensitive detection of melanoma based on serum proteomic profiling. J Cancer Res Clin Oncol. 2009 Sep;135(9):1257–64. doi: 10.1007/s00432-009-0567-7. [DOI] [PubMed] [Google Scholar]
- 11.Wei YS, Zheng YH, Liang WB, et al. Identification of serum biomarkers for nasopharyngeal carcinoma by proteomic analysis. Cancer. 2008 Feb 1;112(3):544–51. doi: 10.1002/cncr.23204. [DOI] [PubMed] [Google Scholar]
- 12.Yang SY, Xiao XY, Zhang WG, et al. Application of serum SELDI proteomic patterns in diagnosis of lung cancer. BMC Cancer. 2005 Jul 20;5:83. doi: 10.1186/1471-2407-5-83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lu HB, Zhou JH, Ma YY, et al. Five serum proteins identified using SELDI- TOF-MS as potential biomarkers of gastric cancer. Jpn J Clin Oncol. 2010 Apr;40(4):336–42. doi: 10.1093/jjco/hyp175. [DOI] [PubMed] [Google Scholar]
- 14.Zhang Z, Bast RC, Jr, Yu Y, et al. Three biomarkers identified from serum proteomic analysis for the detection of early stage ovarian cancer. Cancer Res. 2004 Aug 15;64(16):5882–90. doi: 10.1158/0008-5472.CAN-04-0746. [DOI] [PubMed] [Google Scholar]
- 15.Fan Y, Shi L, Liu Q, et al. Discovery and identification of potential biomarkers of papillary thyroid carcinoma. Mol Cancer. 2009 Sep 28;:8–79. doi: 10.1186/1476-4598-8-79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liu M, Li CF, Chen HS, et al. Differential expression of proteomics models of colorectal cancer, colorectal benign disease and healthy controls. Proteome Sci. 2010 Mar 25;8(1):16. doi: 10.1186/1477-5956-8-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Saito Y, Oba N, Nishinadagawa S, et al. Identification of beta2-microgloblin as a candidate for early diagnosis of imaging- invisible hepatocellular carcinoma in patient with liver cirrhosis. Oncol Rep. 2010 May;23(5):1325–30. doi: 10.3892/or_00000767. [DOI] [PubMed] [Google Scholar]
- 18.Kakehashi A, Kato A, Inoue M, et al. Cytokeratin 8/18 as a new marker of mouse liver preneoplastic lesions. Toxicol Appl Pharmacol. 2010 Jan 1;242(1):47–55. doi: 10.1016/j.taap.2009.09.013. [DOI] [PubMed] [Google Scholar]
- 19.Guo J, Wang W, Liao P, et al. Identification of serum biomarkers for pancreatic adenocarcinoma by proteomic analysis. Cancer Sci. 2009 Dec;100(12):2292–301. doi: 10.1111/j.1349-7006.2009.01324.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Vermaat JS, Vander Tweel I, Mehra N, et al. Two-protein signature of novel serological markers apolipoprotein-A1 and serum amyloid alpha predicts prognosis in patients with metastatic renal cell cancer and improves the currently used prognostic survival models. [DOI] [PubMed]
- 21.Bouamrani A, Ramus C, Gay E, et al. Increased phosphorylation of vimentin in noninfiltrative meningiomas. PLoS One. 2010 Feb 16;5(2):e9238. doi: 10.1371/journal.pone.0009238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Al-Ruwaili JA, Larkin SE, Zeidan BA, et al. Discovery of serum protein biomarkers for prostate cancer progression by proteomic analysis. Cancer Genomics Proteomics. 2010 Mar-Apr;7(2):93–103. [PubMed] [Google Scholar]
- 23.Han M, Liu Z, Yu J, Zheng S. Identification of candidate molecular markers predicting chemotherapy resistance in non-small cell lung cancer. Clin Chem Lab Med. 2010 Mar 26; doi: 10.1515/CCLM.2010.169. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- 24.He J, Shen D, Chung DU, et al. Tumor proteomic profiling predicts the susceptibility of breast cancer to chemotherapy. Int J Oncol. 2009 Oct;35(4):683–92. doi: 10.3892/ijo_00000380. [DOI] [PubMed] [Google Scholar]
- 25.Dalenc F, Doisneau-Sixou SF, Allal BC, et al. Tipifarnib plus tamoxifen in tamoxifen-resistant metastatic breast cancer: a negative phase II and screening of potential therapeutic markers by proteomic analysis. Clin Cancer Res. 2010 Feb 15;16(4):1264–71. doi: 10.1158/1078-0432.CCR-09-1192. [DOI] [PubMed] [Google Scholar]
- 26.Petrik V, Saadoun S, Loosemore A, et al. Serum alpha 2-HS glycoprotein predicts survival in patients with glioblastoma. Clin Chem. 2008 Apr;54(4):713–22. doi: 10.1373/clinchem.2007.096792. [DOI] [PubMed] [Google Scholar]
- 27.Grizzle WE, Semmes OJ, Basler J, et al. The early detection research network surface-enhanced laser desorption and ionization prostate cancer detection study: A study in biomarker validation in genitourinary oncology. [DOI] [PubMed]
- 28.McLerran D, Grizzle WE, Feng Z, et al. SELDI-TOF whole serum proteomic profiling with IMAC surface does not reliably detect prostate cancer. Clin Chem. 2008 Jan;54(1):53–60. doi: 10.1373/clinchem.2007.091496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Semmes OJ, Feng Z, Adam BL, et al. Evaluation of serum protein profiling by surface-enhanced laser desorption/ionization time-of-flight mass spectrometry for the detection of prostate cancer: I. Assessment of platform reproducibility. Clin Chem. 2005 Jan;51(1):102–12. doi: 10.1373/clinchem.2004.038950. [DOI] [PubMed] [Google Scholar]
- 30.Linke T, Doraiswamy S, Harrison EH. Rat plasma proteomics: effects of abundant protein depletion on proteomic analysis. J Chromatogr B Analyt Technol Biomed Life Sci. 2007 Apr 15;849(1–2):273–81. doi: 10.1016/j.jchromb.2006.11.051. [DOI] [PubMed] [Google Scholar]
- 31.Andersen JD, Boylan KL, Xue FS, et al. Identification of candidate biomarkers in ovarian cancer serum by depletion of highly abundant proteins and differential in-gel electrophoresis. Electrophoresis. 2010 Jan;31(4):599–610. doi: 10.1002/elps.200900441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Orvisky E, Drake SK, Martin BM, et al. Enrichment of low molecular weight fration of serum for MS analysis of peptides associated with hepatocellular carcinoma. Proteomics. 2006 May;6(9):2895–902. doi: 10.1002/pmic.200500443. [DOI] [PubMed] [Google Scholar]
- 33.Zhou J, Trock B, Tsangaris RN, et al. A unique proteolytic fragment of alpha1-antitrypsin is elevated in ductal fluid of breast cancer patient. Breast Cancer Res Treat. 2009 Nov 10; doi: 10.1007/s10549-009-0625-5. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- 34.Jansen C, Hebeda KM, Linkels M, et al. Protein profiling of B-cell lymphomas using tissue biopsies: A potential tool for small samples in pathology. Cell Oncol. 2008;30(1):27–38. doi: 10.1155/2008/898356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ward DG, Nyangoma S, Joy H, et al. Proteomic profiling of urine for the detection of colon cancer. Proteome Sci. 2008 Jun 16;6:19. doi: 10.1186/1477-5956-6-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shirai Y, Sogawa K, Yamaguchi T, et al. Protein profiling in pancreatic juice for detection of intraductal papillary mucinous neoplasm of the pancreas. Hepatogastroenterology. 2008 Sep-Oct;55(86–87):1824–9. [PubMed] [Google Scholar]
- 37.Melle C, Ernst G, Winkler R, et al. Proteomic analysis of human papillomavirus-related orgal squamous cell carcinoma: identification of thioredoxin and epidermal-fatty acid binding protein as upregulated protein markers in microdissected tumor tissue. Proteomics. 2009 Apr;9(8):2193–201. doi: 10.1002/pmic.200800882. [DOI] [PubMed] [Google Scholar]
- 38.Sun W, Ye Z, Mi Z, Shi T, Han C, Guo S. A comparative study on proteomics between LNCap and DU145 cells by quantitative detection and SELDI analysis. J Hauzhong Univ Sci Technolog Med Sci. 2008 Apr;28(2):174–8. doi: 10.1007/s11596-008-0215-5. [DOI] [PubMed] [Google Scholar]
- 39.Shintani S, Hamakawa H, Ueyama Y, Hatori M, Toyoshima T. Identification of a truncated cystatin SA-I as a saliva biomarker for oral squamous cell carcinoma using the SELDI ProteinChip platform. Int J Oral Maxillofac Surg. 2010 Jan;39(91):68–74. doi: 10.1016/j.ijom.2009.10.001. [DOI] [PubMed] [Google Scholar]
- 40.Albrethsen J, Bøgebo R, Olsen J, Raskov H, Gammeltoft S. Preanalytical and analytical variation of surface-enhanced laser desorption-ionization time-of-flight mass spectrometry of human serum. Clin Chem Lab Med. 2006;44(10):1243–52. doi: 10.1515/CCLM.2006.228. [DOI] [PubMed] [Google Scholar]
- 41.Timms JF, Arslan-Low E, Gentry-Maharaj A, et al. Preanalytic influence of sample handling on SELDI-TOF serum protein profiles. Clin Chem. 2007 Apr;53(4):645–56. doi: 10.1373/clinchem.2006.080101. [Epub Feb 15 2007]. [DOI] [PubMed] [Google Scholar]
- 42.Rai AJ, Stemmer PM, Zhang Z, et al. Analysis of Human Proteome Organization Plasma Proteome Project (HUPO PPP) reference specimens using surface enhanced laser desorption/ionization-time of flight (SELDI-TOF) mass spectrometry: multi-institution correlation of spectra and identification of biomarkers. Proteomics. 2005 Aug;5(13):3467–74. doi: 10.1002/pmic.200401320. [DOI] [PubMed] [Google Scholar]
- 43.Liggett WS, Barker PE, Semmes OJ, Cazares LH. Measurement reproducibility in the early stages of biomarker development. Disease Markers. 2004;20:295–307. doi: 10.1155/2004/726517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Coombes KR, Tsavachidis S, Morris JS, Baggerly KA, Hung MC, Kuerer HM. Improved peak detection and quantification of mass spectrometry data acquired from surface-enhanced laser desorption and ionization by denoising spectra with the undecimated discrete wavelet transform. Proteomics. 2005;5:4107–17. doi: 10.1002/pmic.200401261. [DOI] [PubMed] [Google Scholar]
- 45.Morris JS, Coombes KR, Koomen JM, Baggerly KA, Kobayashi R. Feature extraction and quantification for mass spectrometry in biomedical applications using the mean spectrum. Bioinformatics. 2005;21:1764–75. doi: 10.1093/bioinformatics/bti254. [DOI] [PubMed] [Google Scholar]
- 46.R Development Core Team R . 2004. A language and environment for statistical computing Vienna, R Foundation for Statistical Computing. ISBN 3-900051-07-0. [Google Scholar]
- 47.Pinheiro JC, Bates DM. New York: Springer-Verlag; 2000. Mixed-Effects Models in S and S-Plus. ISBN 0-387-98957-0. [Google Scholar]
- 48.Johnson KL, Mason CJ, Muddiman DC, Eckel JE. Analysis of the low molecular weight fraction of serum by LC-dual ESI-FT-ICR mass spectrometry: precision of retention time, mass, and ion abundance. Anal Chem. 2004;76:5097–103. doi: 10.1021/ac0497003. [DOI] [PubMed] [Google Scholar]
- 49.Hong H, Dragan Y, Epstein J, et al. Quality control and quality assessment of data from surface enhanced laser desorption/ionization (SELDI) time of flight (TOF) mass spectrometry. BMC Bioinformatics. 2005;6(Suppl 2):S5. doi: 10.1186/1471-2105-6-S2-S5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Semmes OJ, Cazares LH, Ward MD, et al. Discrete serum protein signatures discriminate between human retrovirus associated hematologic and neurologic disease. Leukemia. 2005;19:1229–38. doi: 10.1038/sj.leu.2403781. [DOI] [PubMed] [Google Scholar]
- 51.Forshed J, Pernemalm M, Seng Tan C, et al. Proteomic data analysis workflow for discovery of candidate biomarker peaks predictive of clinical outcome for patients with acute myeloid leukemia. J Proteome Res. 2008;7(6):2332–41. doi: 10.1021/pr070482e. [DOI] [PubMed] [Google Scholar]
- 52.Du P, Kibbe W, Lin S. Improved peak detection in mass spectrum by incorporating continuous wavelet transform-based pattern matching. Bioinformatics. 2006;22(17):2059–65. doi: 10.1093/bioinformatics/btl355. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Materials provided by Ciphergen:
Single Use Serum Aliquots: Fetal Bovine Serum, Heat Inactivated, Cat # 16140-063, Invitrogen-Life Technologies.
200 μl U9 aliquots (use 2 times, always refreeze at −20).
CM10 arrays-prepare 2 arrays each week (at same time).
CM10 binding/wash buffer-store at 4 °C: aliquot into 10 ml test tube for each use.
2 bioprocessor reservoirs (You will use two columns for each prep, save the reservoir and use different columns each time).
NP20 array (use for calibrants).
- SPA—prepare a fresh tube of SPA each week: 200 μl Acetonitrile (100%) + 200 μl of 1% TFA (also made fresh weekly!), vortex well for several minutes by taping the tube onto a vortex and let it mix continuously. Spin at 10000 rpm/2 min. Remove 200 μl (without touching bottom) into a fresh tube. Store in the dark until use.
- ○ 1% TFA: 100 ul + 10 mls of ddH20: (add TFA in fume hood).
All in One Protein Calibrants. Prepare according to insert directions. Store diluted aliquots at −20 °C. Collect a new spectrum each week. (Can use same array over again). Add 5 ul of SPA to the 5 ul aliquot of calibrants and spot 1 ul onto each spot of a NP20 chip. Keep prepared chip in the dark.
Thaw one serum and one U9 aliquot. Make sure U9 is completely thawed. Vortex well.
Centrifuge serum at 10000 rpm/10 min/4 °C.
Add 30 μl of U9 to a fresh 1.5 ml tube.
Add 20 μl of the centrifuged serum to the U9 = total of 50 μl.
Incubate with gentle shaking at 4 °C/20 minutes. Try to keep this time as consistent as possible.
While serum is incubating at 4 °C, set up a bioprocessor with 2 CM10 arrays. Make sure the gasket doesn’t cover the spot.
Pre-wet spots with 150 μl buffer/spot. Incubate with shaking/5 min/RT.
Remove and repeat buffer pre-wetting.
Add 1450 μl of the binding buffer to the serum/U9 mixture (final 1:75 dilution of the serum). Invert to mix well.
Remove final buffer wash from arrays and add 50 ul of diluted serum/well. Watch for airbubbles!
Cover wells (can use 96 well plate sealer). Shake at RT for 30 minutes. (If using a robot mixer, use Form = 48, amp = 9)
Remove samples.
Add 150 μl of buffer/well. Shake 5 min/RT. Remove.
Repeat 2 X for a total of 3 washes with mixing. 15. Add 150 μl of ddH20. Remove (no incubation). Repeat.
Quickly remove reservoir and wick off excess water from spots.
Let dry 5 min. Keep this time consistent! 18. Add 1 μl SPA/spot. Let dry. Repeat. 19. Read within next hour.
Quick cool centrifuge to 4 °C if necessary.
Take serum and U9 aliquot from freezer.
Thaw serum in your hand quickly, mix gently and begin centrifugation.
Hold U9 in hand until it is thawed. Mix well.
Set up bioprocessor with the 2 arrays.
During the U9/serum incubation, begin the buffer prewashes of the arrays.
During sample incubation on the arrays, prepare SPA.
Always save spectra into the database specified as the “QC project”. The user should be set to ‘QC’. Keep all calibration spectra in a ‘Calibration’ project folder. Create a new project file each week to for saving the weekly files.
Average 100 spectra from a spot with the All in One Protein II calibrants, manual or with a spot protocol. Be sure the intensity and sensitivity values that you specify don’t cause any of the peaks to go off scale. Use the same intensity/detector sensitivity settings each time you create a calibration spectrum for the most consistency. Most calibrant -containing spots will last several weeks and can be read over and over. If you need to increase the laser intensity alot to acquire the same calibrants peaks, then it is time to make a new chip.
Internally calibrate using the masses: 12230, 16951 and 29023. Save spectra. Set as default instrument calibration equation. Collection protocol: high mass = 50,000; optimize from 12,000–30,000 daltons; pulse settings—center; if you have a IIc, set the deflector to 5000 daltons.
Go to Options, choose “Configure dynamic spec tags”: make sure box is checked for adding to spectra as they are acquired. Add to tag: 1) Spot #, 2) Sample Name, 3) Sample Group.
When you enter a barcode, go to File, Open chips, and choose ‘new’. Add barcode. Then type in the information below in the correct spaces.
Sample Name = QC_1 (for first week, then change it to 2, etc).
Sample Group = the PI’s name (ie, Bast).
Sample Type = FBS (not included in the spectral tag).
LMW spot: high mass to acquire; 50,000 daltons; optimization range = 1,000 to 30,000 daltons; time lag focus- set to 700 ns; IIc deflector- set to 1000 daltons; set warming shots to collect 1 at the same intensity that you choose for all data collection, but don’t use warming shots; collect from 20 to 80 with 10 shots/area, delta 5.
HMW spot: high mass to acquire; 200,000 daltons; optimization range = 30,000–150,000 daltons; time lag focus- center; IIc deflector- set to 10,000 daltons; set warming shots to collect 1 at the same intensity that you choose for all data collection, but don’t use warming shots; collect from 23 to 78 with 10 shots/area, delta 5.
Calibration equation: For this study, always use a curve based on 12230, 16951 and 29023 daltons (for both LMW and HMW spectra).
Laser Intensity:
Detector Sensitivity:
- Time lag focus:
- ○ LMW: use 700 ns
- ○ HMW: center of optimization range between 30,000 and 150,000 daltons.
Read the spots with both a low mass acquisition spot protocol and a high mass acquisition spot protocol. The above listed variables should be adjusted so that you collect a QC spectrum that gives a peak pattern as closely resembling the ‘QC standard’ (spectrum saved on your computer) as possible.
First, read spots A–C and compare spectrum to the ‘standard’ stored on your instrument. If you compare a spectrum to one previously acquired in the weeks before and it doesn’t not appear comparable, you can adjust the laser intensity and detector sensitivity in the spot protocol and read again. Then, read spots D–H of that chip and all 8 spots of the second chip you prepare each week (using a chip protocol for both LMW and HMW acquire).
All chips prepared by the different labs will be read on one instrument to assess any preparation issues.
Voltage Detector: check setting and record in your notes: to access this setting, go to Instrument, configuration, in bottom section see Detector = PBSII-(1800 to 2200 V); IIc-(2700 to 3000 V). If you start to lose low intensity peaks from the spectrum, the detector voltage may need to be increased (use 50 unit increases).
Laser Focus (service engineer will change this if needed): If any of the peak shapes change (ie, poor resolution), focusing the laser can help optimize resolution.
Compare the peak shape and intensity in your acquired spectra to the examples stored in the QC database. Adjust setting as required to maintain spectra integrity from week to week.
Approx masses: 3984 da, 8490 da, 13.5, 15.2 and 15.8 kD, 33.3 kD.
Approx masses: 33.3 kD, 66 kD, and 133 kD.
Table S1.
Components of variance explaining the alignment offset for the setting scanned on a common instrument.
| Random effect | Standard deviation (nanoseconds) | 95% confidence interval | % variance explained |
|---|---|---|---|
| Lab | σL = 1.6164 | (0.5949, 4.3916) | 9.3 |
| Week | σW = 2.0608 | (0.9679, 4.3876) | 15.2 |
| Array | σC = 3.5024 | (2.7909, 4.3952) | 43.8 |
| Spot (residual) | σE = 2.9763 | (2.7725, 3.1951) | 31.7 |
Financial relationship with Company 1:
Nature of Relevant Financial Relationship:
□ Full-time/Part-time Employee
□ Grant/Research Support Recipient
□ Advisor/Board Member
□ Consultant/Independent Contractor
□ Stock Shareholder (self managed)
□ Speaker’s Bureau
□ Honorarium Recipient
□ Other (please describe the Other Relationship)
Financial relationship with Company 2:
Nature of Relevant Financial Relationship:
□ Full-time/Part-time Employee
□ Grant/Research Support Recipient
□ Advisor/Board Member
□ Consultant/Independent Contractor
□ Stock Shareholder (self managed)
□ Speaker’s Bureau
□ Honorarium Recipient
□ Other (please describe the Other Relationship)
Financial relationship with Company 3:
Nature of Relevant Financial Relationship:
□ Full-time/Part-time Employee
□ Grant/Research Support Recipient
□ Advisor/Board Member
□ Consultant/Independent Contractor
□ Stock Shareholder (self managed)
□ Speaker’s Bureau
□ Honorarium Recipient
□ Other (please describe the Other Relationship)
Financial relationship with Company 4:
Nature of Relevant Financial Relationship:
□ Full-time/Part-time Employee
□ Grant/Research Support Recipient
□ Advisor/Board Member
□ Consultant/Independent Contractor
□ Stock Shareholder (self managed)
□ Speaker’s Bureau
□ Honorarium Recipient
□ Other (please describe the Other Relationship)
Financial relationship with Company 5:
Nature of Relevant Financial Relationship:
□ Full-time/Part-time Employee
□ Grant/Research Support Recipient
□ Advisor/Board Member
□ Consultant/Independent Contractor
□ Stock Shareholder (self managed)
□ Speaker’s Bureau
□ Honorarium Recipient
□ Other (please describe the Other Relationship)
Financial relationship with Company 6:
Nature of Relevant Financial Relationship:
□ Full-time/Part-time Employee
□ Grant/Research Support Recipient
□ Advisor/Board Member
□ Consultant/Independent Contractor
□ Stock Shareholder (self managed)
□ Speaker’s Bureau
□ Honorarium Recipient
□ Other (please describe the Other Relationship)
□ No, I have no relevant financial relationship(s).