Abstract
During progression of breast cancer, CCN6 protein exerts tumor inhibitory functions. CCN6 is a secreted protein that modulates the insulin-like growth factor-1 (IGF-1) signaling pathway. Knockdown of CCN6 in benign mammary epithelial cells triggers an epithelial to mesenchymal transition (EMT), with upregulation of the transcription factor ZEB1/δEF1. How CCN6 regulates ZEB1 expression is unknown. We hypothesized that CCN6 might regulate ZEB1, EMT and breast cancer invasion by modulating IGF-1 signaling. Exogenously added human recombinant CCN6 protein was sufficient to downregulate ZEB1 mRNA and protein levels in CCN6-deficient (CCN6 KD) HME cells and MDA-MB-231 breast cancer cells. Recombinant CCN6 protein decreased invasion of CCN6 KD cells compared with controls. We discovered that knockdown of CCN6 induced IGF-1 secretion in HME cells cultivated in serum-free medium to higher concentrations than found in MDA-MB-231 cells. Treatment with recombinant CCN6 protein was sufficient to decrease IGF-1 protein and mRNA to control levels, rescuing the effect of CCN6 knockdown. Specific inhibition of IGF-1 receptors using the pharmacological inhibitor NVP-AE541 or short hairpin shRNAs revealed that ZEB1 upregulation due to knockdown of CCN6 requires activation of IGF-1 receptor signaling. Recombinant CCN6 blunted IGF-1-induced ZEB1 upregulation in MDA-MB-231 cells. Our data define a pathway in which CCN6 attenuates IGF-1 signaling to decrease ZEB1 expression and invasion in breast cancer. These results suggest that CCN6 could be a target to prevent or halt breast cancer invasion.
Key words: CCN6, WISP3, IGF, Insulin-like growth factor, Epithelial to mesenchymal transition, ZEB1, Breast cancer
Introduction
Invasive breast carcinoma is a group of malignant epithelial tumors characterized by invasion of adjacent tissues and propensity to metastasize to distant sites. The vast majority of breast cancers are adenocarcinomas that are embedded in a desmoplastic stroma containing fibroblasts, myofibroblasts, immune system cells, structural proteins, and vessels. It has become clear in recent years that this rich microenvironment exerts a powerful influence on the behavior of epithelial cells and that epithelial and stromal crosstalk maintains tissue homeostasis. Matricellular proteins are extracellular proteins that connect signaling pathways and regulate their influence on epithelial cells rather than exerting a structural role (Bornstein and Sage, 2002; Perbal, 2008). The matricellular protein CCN6 (also known as WNT1-inducible-signaling pathway protein 3 or WISP3) was found to have tumor growth and invasion inhibitory functions in inflammatory breast cancer and aggressive non-inflammatory breast cancer (Huang et al., 2010; Huang et al., 2008; Kleer et al., 2002). Our lab has demonstrated that CCN6 is secreted from epithelial cells in the breast and decreases activation of the IGF-1 signaling pathway in breast cancer (Kleer, 2004; Zhang et al., 2005). Downregulation of CCN6 in benign breast cells triggers an epithelial to mesenchymal transition (EMT) through upregulation of the transcriptional repressor ZEB1 (Huang et al., 2008). The mechanism by which CCN6 regulates ZEB1-mediated EMT and invasion is unknown.
The transcriptional factor ZEB1/δEF1 has emerged as an important regulator of EMT and invasion in breast cancer and other neoplasms (Eger et al., 2005; Graham et al., 2008). ZEB1 is a member of the ZFH family of transcription factors with multiple zinc fingers and one or more homeodomains. ZEB1 mainly acts as a transcriptional repressor (Cabanillas et al., 2001; Vandewalle et al., 2009) that is capable of transcriptional auto-regulation (Manavella et al., 2007). ZEB1 has an important role in cell differentiation pathways such as skeletal patterning and chondrogenesis (Takagi et al., 1998), T-cell lymphopoiesis (Higashi et al., 1997), myogenesis and neurogenesis (Darling et al., 2003; Postigo and Dean, 1999). Recent studies show that ZEB1 is a central regulator of EMT and invasion in solid tumors through transcriptional repression of the gene encoding E-cadherin (Eger et al., 2005) and genes responsible for maintaining cell junctions and cell polarity (Aigner et al., 2007). In addition, ZEB1 inhibits the expression of the miR-200 family, which are inducers of epithelial differentiation in solid tumors (Gregory et al., 2008; Park et al., 2008; Wellner et al., 2009). Very limited data exist on how ZEB1 is regulated during tumorigenesis, but IGF-1 has been reported to have a role (Graham et al., 2008).
In this study, we hypothesize that CCN6 regulates EMT and breast cancer invasion through IGF-1-mediated regulation of ZEB1. We show that human recombinant CCN6 protein reduces activation of the IGF-1 pathway and leads to a reduction of ZEB1 expression. Functionally, exogenous CCN6 protein induces an epithelial phenotype and blocks invasion in CCN6 knockdown human mammary epithelial (HME) cells compared with controls. Mechanistic studies with gain-of-function and loss-of-function approaches demonstrate that the effect of CCN6 on ZEB1 is dependent on the integrity of the IGF-1 pathway.
Results
Human recombinant CCN6 protein downregulates mRNA and protein levels of ZEB1
The mechanism by which CCN6 regulates ZEB1 is unknown. We first assessed the endogenous expression of ZEB1 in a panel of breast cell lines including breast cancer cells, benign human mammary epithelial cells with stable knockdown of CCN6 and controls. The expression of CCN6 in these cells has been previously reported by our group (Huang et al., 2010; Huang et al., 2008). CCN6-expressing cell lines (benign HME cells, MCF10A cells and MCF7 well-differentiated invasive but not metastasizing breast cancer cells) had low ZEB1 expression and high E-cadherin protein. By contrast, CCN6-deficient metastasizing breast cancer cells MDA-MB-231 exhibited high expression of ZEB1 and undetectable levels of E-cadherin (Fig. 1A, left). A mesenchymal-like cell line (HME-shCCN6) derived from HME with short hairpin inhibition of CCN6 in a lentivirus vector (CCN6 KD) (Huang et al., 2010) showed upregulation of ZEB1 and decreased E-cadherin compared with scrambled shRNA controls (Fig. 1A, right).
Fig. 1.
CCN6 regulates ZEB1 expression and the epithelial-mesenchymal phenotype. (A) Inmunoblot of a set of human mammary cell lines lysates showing endogenous expression of ZEB1 and E-cadherin proteins. (B) Western blot of ZEB1 and vimentin after incubation with the indicated amounts of rCCN6 for 4 hours. UT, untreated HME CCN6 KD cells. (C) Immunofluorescence of indicated cells labeled with Alexa Fluor 488 phalloidin (green) and Alexa Fluor 568 (red) for F-actin and vimentin filaments, respectively. Nuclei were stained with DAPI (blue). Arrows indicate the presence of F-actin stress fibers in untreated cells and changes in vimentin subcellular localization (‘cap’-like shape) in treated cells. Results are representative of 24 hours of incubation. (D) RT-qPCR showing the effect of rCCN6 treatment on ZEB1 mRNA levels. The results indicate mean ± s.e.m. of two experiments performed in triplicate. ***P<0.001.
Because CCN6 is a secreted protein, we hypothesized that extracellular CCN6 protein might reduce ZEB1 expression and promote phenotypic changes from mesenchymal-like to epithelial in HME CCN6 KD cells. We investigated the dose and time-dependent effect of exogenous human recombinant CCN6 (rCCN6) on the expression of ZEB1 and vimentin on CCN6 KD cells. Whereas untreated CCN6 KD cells showed high ZEB1 expression, addition of rCCN6 for 4 hours induced a dose-dependent inhibition of ZEB1 and vimentin protein levels, with 500 ng/ml rCCN6 having the maximal inhibitory effect (Fig. 1B). Longer incubations showed similar results on ZEB1 and vimentin expression (not shown).
Immunofluorescence analyses further strengthened these data. Although control HME cells contained organized F-actin fibers located mainly at the cell–cell junction, CCN6 KD cells demonstrated formation of stress fibers and lamellipodia (Fig. 1C, top). After 24 hours, 500 ng/ml rCCN6 reduced stress fibers and lamellipodia to control levels. We noted that knockdown of CCN6 upregulated vimentin expression in HME cells. Addition of rCCN6 resulted in reorganization of vimentin filaments to form perinuclear aggregates (Fig. 1C, bottom). Interestingly, this pattern of vimentin reorganization is sufficient to induce mesenchymal cells to adopt epithelial shapes and decrease motility (Mendez et al., 2010).
To investigate the mechanism by which exogenous rCCN6 reduces ZEB1 expression we performed real-time RT-PCR analysis. After 3 hours of incubation, a single dose of 500 ng/ml rCCN6 significantly reduced ZEB1 mRNA levels, partially rescuing the effect of CCN6 knockdown (Fig. 1D). Taken together, these data demonstrate that exogenous CCN6 protein can regulate mRNA and protein levels of ZEB1 and is able to partially induce cytoplasmic filament reorganization and expression towards an epithelial phenotype.
Exogenous CCN6 protein attenuates the IGF-1 signaling pathway
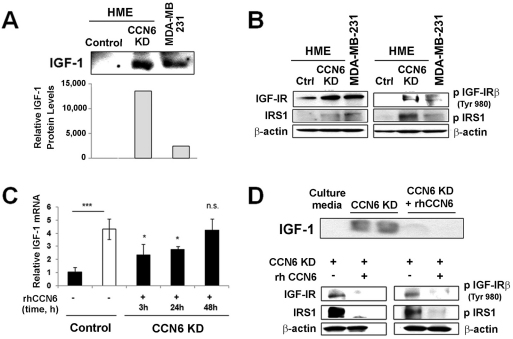
CCN6 has been reported to interfere with IGF-1 signaling in breast cells and knockdown of CCN6 in HME cells increased phosphorylation of the IGF-1 receptor (IGF-1R) and IRS1 (insulin receptor substrate-1) compared with levels in the control (Kleer et al., 2004; Zhang et al., 2005). However, the mechanism by which CCN6 influences IGF-1 signaling is unknown. We found that CCN6 KD cells cultured under serum deprivation conditions had increased IGF-1 protein levels in the medium (Fig. 2A). IGF-1 concentration in the conditioned medium of CCN6 KD cells was higher than that in the medium of MDA-MB-231 breast cancer cells (Fig. 2A). This result was not surprising based on our previous work (Kleer et al., 2004). Consistently, CCN6 knockdown in HME cells increased the phosphorylation of IGF-1R and one of its main downstream effectors, IRS1, to higher levels than those detected in MDA-MB-231 cells (Fig. 2B).
Fig. 2.
CCN6 regulates IGF-1 expression and pathway activation. (A) IGF-1 secretion in the culture medium of MDA-MB-231 and HME CCN6 KD cells under serum starvation conditions by immunoblot (upper panel) and its quantification (lower panel) in relation to the total amount of protein in the medium. Both cell lines were treated at a confluency of 70%. (B) Western blotting was performed for IGF-1R, phosphorylated IGF-1R, IRS1 and phosphorylated IRS1 in MDA-MB-231 and HME CCN6 KD cells. (C) HME control and HME CCN6 KD cells were incubated with placebo or rCCN6, respectively. IGF1 mRNA levels were determined by RT-qPCR. Bars show mean ± s.e.m. of two experiments done in triplicate. *P<0.05; ***P<0.001, n.s., not significant compared with untreated control. (D) Western blotting of IGF-1 in the medium (top) and IGF-1R, phosphorylated IGF-1R, IRS1 and phosphorylated IRS1 (bottom) after incubation with 500 ng/ml rCCN6 for 4 hours.
Real-time RT-PCR revealed that CCN6 knockdown triggered a fourfold increase in the level of IGF1 mRNA compared with control levels (Fig. 2C). We next determined the effect of increasing concentrations of rCCN6 at different incubation times on levels of IGF-1 mRNA and protein. rCCN6 attenuated the effect of CCN6 knockdown by significantly decreasing IGF1 mRNA levels after 3 hours and 24 hours of treatment (Fig. 2C). This model also shows a dependence on exogenous CCN6 for regulation of IGF1 mRNA, because IGF1 mRNA levels returned to the control value 48 hours after the last treatment with rCCN6, by which time rCCN6 has probably been degraded.
Even though exogenous rCCN6 protein partially rescued the effect of CCN6 knockdown on IGF1 mRNA levels, addition of rCCN6 protein reduced the levels of IGF-1 protein detected in the conditioned medium (Fig. 2D). Consistent with these results, rCCN6 protein rescued the increased IGF-1R and IRS1 phosphorylation triggered by CCN6 knockdown in HME cells (Fig. 2D). Collectively, these data show that CCN6 knockdown increases IGF1 mRNA and results in elevated IGF-1 protein levels in the extracellular medium. Mechanistically, rCCN6 functions in an autocrine or paracrine manner to decrease the concentration of IGF-1 in the medium and to attenuate the IGF-1 signaling pathway, which might have therapeutic implications.
ZEB1 upregulation due to CCN6 knockdown requires activation of IGF-1 receptor signaling
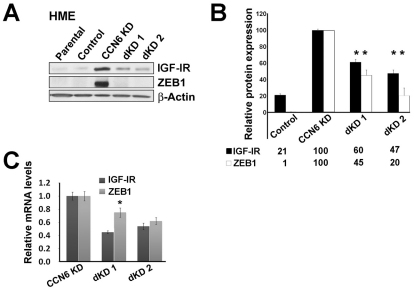
Based on our results that CCN6 knockdown enhances the IGF-1 signaling pathway, and recently published data showing that IGF-1 can upregulate ZEB1 in human prostate cancer cells (Graham et al., 2008), we hypothesized that CCN6 regulates ZEB1 through modulation of the IGF-1 signaling pathway. To test this hypothesis, we first used shRNA knockdown of IGF-1R in CCN6 KD HME cells and controls. shRNA inhibition of IGF-1R and CCN6 (IGF-1R/CCN6 double KD, dKD) decreased mRNA and protein levels of IGF-1R and ZEB1 (Fig. 3A-C). We obtained two stable clones of dKD cells derived from different shRNAs sequences targeting IGF-1R, which was validated to have no cross matches with the genes of interest for these studies (Fig. 3A). Both dKD cell lines showed a significant decrease in IGF1R and ZEB1 mRNA measured by real-time RT PCR (Fig. 3C). IGF-1R and CCN6 double knockdown led to downregulation of IGF-1R protein (dKD1 60±5%, dKD2 47±5%) and reduced ZEB1 protein (dKD1 45±7%, dKD2 20±10%) compared with controls (Fig. 3B).
Fig. 3.
Knockdown of IGF-1R restores ZEB1 expression in CCN6 KD cells. (A) Western blot of ZEB1 and IGF-1R on HME cells with CCN6 KD, double KD of CCN6 and IGF-1R (dKD clones 1 and 2), and scrambled controls. (B) Quantification of average ZEB1 and IGF-1R protein expression using densitometry relative to β-actin. (C) Quantification of levels of ZEB1 and IGF1R mRNA. For B and C, values are expressed as mean ± s.d. (n=2). *P<0.05 vs HME-CCN6 KD.
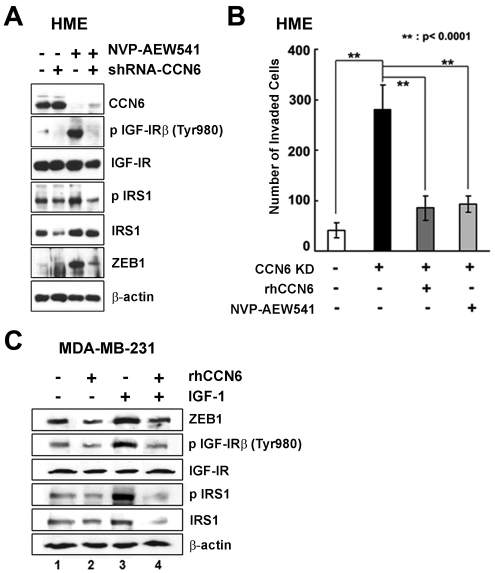
To further test whether CCN6 regulates ZEB1 by modulation of the IGF-1R signaling pathway, we used NVP-AEW541, which is a potent and highly selective inhibitor of IGF-1R kinase. This inhibitor has been previously validated (Garcia-Echeverria et al., 2004; Gariboldi et al., 2010). Western blots confirmed the inhibition of IGF-1R and IRS1 phosphorylation in CCN6 KD cells by NVP-AEW541 (Fig. 4A). NVP-AEW541 reversed the ZEB1 upregulation induced by CCN6 knockdown. This was accompanied by a significant decrease of the CCN6 KD invasive phenotype (Fig. 4B). rCCN6 had a similar invasion inhibitory effect on CCN6 KD HME cells as NVP-AEW541 (Fig. 4B).
Fig. 4.
CCN6 knockdown requires activation of IGF-1 receptor signaling to induce ZEB1 expression and invasion. (A) Western blot of CCN6, phosphorylated IGF-1R, IGF-1R, phosphorylated IRS1, IRS1 and ZEB1 in HME-CCN6 KD cells (shRNA-CCN6) and controls untreated or treated with 10 μM NVP-AEW541. (B) Invasion ability of HME controls and HME CCN6 KD cells untreated or treated with rCCN6 (500 ng/ml) or NVP-AEW541 (10 μM). (C) Western blot of ZEB1, IGF-1R, phosphorylated IGF-1R, IRS1 and phosphorylated IRS1 in MDA-MB-231 cells and controls untreated or treated with 500 ng/ml rhCCN6 and/or 100 ng/ml IGF-1.
The relevance of this pathway was next investigated in MDA-MB-231 breast cancer cells. Single treatment with rCCN6 decreased ZEB1 expression levels (Fig. 4C, lane 2). Single treatment with IGF-1 protein upregulated ZEB1 expression (Fig. 4C, lane 3). Of note, rCCN6 protein decreased the activation of IGF-1 signaling and reduced IGF-1-induced ZEB1 upregulation (Fig. 4, lane 4 compare with lane 3). Taken together, these data provide direct evidence that CCN6 decreases ZEB1 protein levels by attenuation of IGF-1 signaling in breast cancer, and that the invasive ZEB1-overexpressing phenotype conferred by CCN6 knockdown in HME cells requires activation of the IGF-1 signaling pathway (Fig. 5).
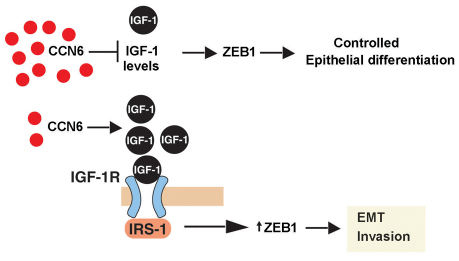
Fig. 5.
Working model for the role of CCN6 in breast cancer invasion. Our data support the hypothesis that CCN6 regulates the levels and activity of IGF-1R signaling pathway in mammary epithelial cells. Low levels of CCN6, as found in aggressive invasive carcinomas of the breast, result in activation of IGF-1R signaling and increased ZEB1 levels and function.
Discussion
The link between stimulus of EMT by the tumor microenvironment and invasion in cancer has been established in recent years. One group of microenvironmental proteins is the CCN family, which has six members (CCN1/Cyr61, CCN2/CTGF, CCN3/Nov, CCN4/WISP1, CCN5/WISP2, CCN6/WISP3) (Brigstock et al., 2003; Lau and Lam, 1999; Leask and Abraham, 2006). CCN proteins function as matricellular proteins that connect signaling pathways and mediate epithelial and stromal crosstalk in multiple organ systems. In the breast, CCN6 mRNA levels are decreased in inflammatory breast cancer, the most lethal form of locally advanced breast cancer, compared with stage-matched, non-inflammatory tumors (van Golen et al., 1999). Our laboratory has demonstrated that CCN6 protein is expressed in normal breast tissues, but it is reduced or lost in 60% of invasive carcinomas of the breast (Huang et al., 2008). CCN6 is secreted from breast epithelial cells and, consistent with its reduced expression in a subset of invasive carcinomas, has been shown to act as a tumor suppressor in breast cancer (Huang et al., 2010; Huang et al., 2008; Kleer et al., 2002; Kleer et al., 2004). The mechanistic details of how CCN6 regulates EMT and invasion need further investigation. In the present study, we delineate a pathway in which CCN6 attenuates the IGF-1 signaling pathway to decrease ZEB1 expression, thereby decreasing invasion.
IGFs have a central role in breast cancer development (Pollak, 2008). Compelling epidemiological and clinical data show that high concentrations of IGF-1 in serum are associated with increased mammographic density (one of the strongest predictors of breast cancer risk), and also reliably predict increased breast cancer risk specifically in premenopausal women (Pollak, 2008; Samani et al., 2007). In vitro and in vivo studies have shown that IGFs promote the proliferation, survival and metastatic ability of breast cancer cells (Law et al., 2008; Pollak, 2008; Samani et al., 2007; Witsch et al., 2010). IGF-1R promotes breast cancer growth and metastasis, and its hyperactivation has been linked with increased radioresistance and breast cancer recurrence (Law et al., 2008; Samani et al., 2007). Although we found that knockdown of CCN6 in breast cells activates the IGF-1 signaling pathway, the underlying mechanism remained unknown (Zhang et al., 2005). The present study demonstrates that this activation is due to induction of IGF-1 levels in the extracellular medium. We found that downregulation of CCN6 in benign mammary epithelial cells under serum starvation increases the levels of IGF1 mRNA as well as the concentration of IGF-1 in the conditioned medium compared with control levels. The levels of IGF-1 protein in the conditioned medium and the induction of IGF-1R and IRS1 phosphorylation by CCN6 knockdown are higher than those exhibited by the highly aggressive breast cancer cell line MDA-MB-231.
Strengthening our data and providing further mechanistic insights, exogenous human recombinant CCN6 protein decreased IGF1 mRNA and protein levels in the conditioned medium. As expected, the IGF-1R signaling pathway was attenuated by recombinant CCN6 protein, as demonstrated by the absence of activated forms of the receptor (IGF-1Rβ-P) and its main downstream mediator IRS1 (IRS1-P). Our data provide evidence that exogenous CCN6 regulates the levels of IGF-1 and the activation of IGF-1R signaling pathway in breast cells. Our data also suggest that extracellular CCN6 is able to regulate IGF-1 at the transcriptional level, either directly or indirectly, which warrants further investigation.
ZEB1 is a master regulator of EMT, not only for its repressor function on E-cadherin promoter, but also through direct downregulation of a variety of genes involved in maintenance of the epithelial phenotype and its ability to induce cell invasion (Aigner et al., 2007; Spaderna et al., 2008; Vandewalle et al., 2009). Limited data exist on the mechanisms responsible for ZEB1 regulation. Transforming growth factor β (TGFβ), a potent inducer of EMT has been shown to regulate expression of ZEB1 and ZEB2 in mouse mammary NmuMG epithelial cells and to induce EMT (Shirakihara et al., 2007). It has been demonstrated that the miR-200 family, in conjunction with miR-205, inhibits expression of ZEB1 and ZEB2 in epithelial cells and has a major role in preventing these factors from triggering EMT (Gregory et al., 2008; Park et al., 2008). Bracken and colleagues reported the existence of a double-negative feedback regulatory loop between ZEB1 and the miR-200 family (Bracken et al., 2008). A recent study provided evidence that IGF-1 is implicated in ZEB1 regulation in prostate cells (Graham et al., 2008). Enhanced IGF-1 signaling was sufficient to increase ZEB1 expression and migration of prostate cancer cells in vitro (Graham et al., 2008). Given our previous results showing that CCN6 knockdown in breast epithelial cells triggers EMT through ZEB1-dependent repression of E-cadherin (Huang et al., 2010), we hypothesized that extracellular CCN6 regulates ZEB1 by blocking IGF-1 signaling. We first tested the effect of CCN6 on ZEB1 mRNA and protein levels using gain- and loss-of-function approaches. CCN6 recombinant protein in a dose- and time-dependent manner (not shown) decreased ZEB1 mRNA and protein levels. This was accompanied by a reduction in cell invasion in vitro, and phenotypic changes from mesenchymal-like to epithelial. Our study shows that the invasive CCN6 KD phenotype requires activation of the IGF-1 signaling pathway through the IGF-1R. Pharmacological and shRNA inhibition of IGF-1R activation in CCN6 KD cells blunted the increase in ZEB1 compared with levels in the control. Functionally, inhibition of IGF-1R activation abrogated the invasive abilities of CCN6 KD cells compared with controls. The effect of CCN6 was also evident on invasive breast cancer MDA-MB-231 cells. Recombinant human CCN6 protein inhibits the IGF-1 signaling pathway and blocks IGF-1-mediated ZEB1 upregulation in breast cancer cells.
In summary, our study provides evidence that the matricellular protein CCN6 regulates the levels of IGF-1 in the extracellular medium of cultured breast cells. We define a pathway by which CCN6 attenuates IGF-1R signaling to decrease ZEB1 levels, thereby decreasing invasion. Our results also suggest that CCN6 should be explored as a target for future therapeutic action to prevent or halt breast cancer invasion.
Materials and Methods
Cell culture
The HME cell line was developed and provided by Stephen P. Ethier (Karmanos Cancer Institute, Detroit, MI). Stable CCN6 KD HME cell lines and scrambled controls have been previously developed and reported by our laboratory (Huang et al., 2010) and maintained as described previously (Huang et al., 2010). MCF-7 cells were obtained from AATC and cultured in complete medium consisting of Eagle's Minimum Essential Medium (Gibco) supplemented with 10% fetal bovine serum (Gibco) and 10 μg/ml insulin. MDA-MB-231 cells (ATCC) were maintained in Dulbecco's modified Eagle's medium (Gibco) supplemented with 10% FBS. MCF-7 and MDA-MB-231 cells were grown at 37°C under 5% CO2. Streptomycin (100 μg/ml) and Penicillin G (100 U/ml) were added to the respective media. For IGF-1 secretion and pathway activation assays, we used serum deprivation medium which consisted of F-12 or D-MEM supplemented with 100 U/ml penicillin and 100 μg/ml streptomycin. This medium was deprived of exogenous factors and serum.
Human recombinant CCN6 protein treatment
HME CCN6 KD and HME control cells were cultured in serum deprivation medium for 16 hours and incubated with 500 ng/ml (unless stated) of recombinant human CCN6 (PeproTech, Rocky Hill, NJ) for 4 hours for western blotting; 3, 24 and 48 hours for RT-qPCR analysis and 24 hours treatment for immunofluorescence assays. MDA-MB-231 cells were incubated with 500 ng/ml of rCCN6 for 24 hours for western blot analysis.
Human recombinant IGF-1 protein treatment
MDA-MB-231 cells were cultured in serum deprivation medium for 16 hours and incubated with were treated with vehicle or physiologic concentrations of IGF-1 (100 ng/ml) of recombinant human IGF-1 (Chemicon-Millipore, Billerica, MA) for 8 hours. Cell lysates were analyzed by western blotting.
NVP-AEW541 treatment
HME Control and CCN6 KD cells were cultured in serum deprivation medium. After starvation for 24 hours, cells were treated with the IGF-1R kinase inhibitor NVP-AEW541 (gift from Novartis, Basel, Switzerland) at a concentration 10 μM for 24 hours.
Lentiviral transduction
To generate stable HME Cell Lines with double knockdown of CCN6 and IGF-1R (CCN6/IGF-1R dKD), we used short hairpin RNAs (shRNAs). Sequence I (clone ID TRCN00005116) (for dKD1) and II (clone ID TRCN0000010361) (for dKD2) targeting human IGF-1R (NM_000875) (Open Biosystems, Huntsville, AL; Cat No. RHS4533-NM_000875) were cloned into pLKO.1-puro vector (Open Biosystems). Both shRNA-containing plasmids were packaged into lentiviral particles at the Vector Core (University of Michigan, Ann Arbor, MI) and transduced in HME CCN6 KD cells and controls (scrambled shRNA). The method for generation of shRNA CCN6 HME cells was previously reported (Huang et al., 2010).
Western blot analysis
Cell lysates were collected using modified RIPA lysis buffer. Lysates and conditioned media were added with a mixture of protease and/or phosphatase inhibitors (Roche, Indianapolis, IN) and used for western blot assays as previously reported (Huang et al., 2010). Primary antibodies including anti-ZEB1 (H-102), anti-IGF-1 (H-70), anti-IGF-1R (7G11) (Santa Cruz Biotechnology, Santa Cruz, CA); anti-phospho-IGF-1Rβ (Tyr980) (14A11) (Cell Signaling, Boston, MA); anti-IRS1, anti-phospho-IRS1 (Upstate Biotechnology, Billerica, MA); anti-E-cadherin (BD Biosciences, CA); anti-vimentin (SP20), anti-cytokeratin-18 (AB32118) (Abcam), anti-β-actin (Sigma) were used at the manufacturer's recommended dilutions and conditions. After washing in Tris-buffered saline containing Tween 20, the blot was incubated with horseradish-peroxidase-conjugated secondary antibodies at 1:2000 (Amersham Bioscience, Piscataway, NJ), and the immunoblot signals were visualized by a chemiluminescence system (Thermo Scientific, Rockford, IL). Digital images were processed by ImageJ (NIH).
RNA preparation and RT-qPCR
Total cellular RNA was isolated from HME Control, HME CCN6 KD, HME dKD1 and HME dKD2 cells using the RNeasy Mini kit (Qiagen, Valencia, CA) and quantified by UV absorption; complementary DNA (cDNA) was synthesized using Superscript III kit (Invitrogen, Carlsbad, CA), with random hexamers as primers. To amplify cDNA encoding ZEB1, IGF-1 and IGF-1R, we used TaqMan Universal PCR Master Mix (Applied Biosystems, Foster City, CA) according to the manufacturer's instruction and inventoried TaqMan Gene Expression Assays (Applied Biosystems): Hs00232783_m1 for ZEB1, Hs01547656_m1 for IGF1 and Hs00609566_m1 for IGF1R. GAPDH RNA was used as the endogenous control. The reaction product was quantified with an ABI PRISM 7900HT (Applied Biosystems).
Inmunofluorescence and confocal microscopy
2.5×105 HME Control and HME CCN6 KD cells were grown on chamber slides (Nalgen Nunc International, Naperville, IL), for 8 hours, starved and treated with rCCN6 (PeproTech) as stated above. Cells were fixed for 40 minutes with 4% paraformaldehyde in PBS at room temperature, permeabilized and blocked with 5% non-fat dry milk, 1% BSA, 0.5% Triton X-100 and incubated overnight with rabbit monoclonal anti-vimentin (Abcam, Cambridge, UK) primary antibody (1:300) or Alexa Fluor 488 Phalloidin Conjugate (Molecular Probes, Eugene, OR) (1:10,000) to stain F-actin filaments, in blocking solution at 4°C in the dark. After washing the slides with anti-vimentin antibody, rabbit secondary anti-Alexa Fluor 568 antibody (Molecular Probes) (1:2000) was added in blocking solution for 1hour. 4′6-diamidino-2-phenylindole was applied to stain nuclei. Confocal images were taken with an Olympus FluoView 500 Laser Scanning confocal microscope (Microscopy & Image Analysis Laboratory, University of Michigan) or a Zeiss LSM 5 Pascal microscope (Laboratorio de Microscopía Óptica, CIBICI-CIQUIBIC, Universidad Nacional de Córdoba, Argentina).
Matrigel invasion chamber assay
The invasive ability of HME Control and CCN6 KD cells was tested by using a Matrigel invasion chamber (BD Biocoat Cellware, Jose, CA). Cells were seeded at a density of 5×104 per insert in serum-free medium onto the extracellular matrix layer. Cells were treated with 500 ng/ml rCCN6 (PeproTech) or 10 μM NVP-AEW541 (Novartis). Triplicate wells were used for each treatment. After incubation for 24 hours, the non-invading cells from the interior of the insert were removed gently with a cotton swab. The invasive cells on the lower surface of membrane were fixed and stained using Diff-Quik staining kit (Allegiance), air dried, and photographed. Cells were counted under the microscope.
Statistical analysis
Statistical analysis was performed using 95% confidence intervals for the estimates of the means. Means were compared by one-way ANOVA. Student–Newman–Keuls (SNK) tests or paired t-tests were performed to test the significance of differences among means. Data represent means ± s.e.m or s.e. P<0.05 was considered statistically significant and statistical probability is expressed as *P<0.05, **P<0.01 and ***P<0.001 unless is indicated.
Acknowledgments
We are thankful to Pilar Crespo from Laboratorio de Microscopía Óptica, CIBICI-CIQUIBIC, Universidad Nacional de Córdoba for her invaluable help with confocal microscopy and image processing. We thank Maria E. Gonzalez from the Kleer lab for insightful discussions and technical advice. This work was supported by NIH grants 2R01CA107469, R01CA125577 and U01CA154224 (to C.G.K.), and by the National Institutes of Health through the University of Michigan's Cancer Center Support Grant (5 P30 CA46592), support from the Fashion Footwear Charitable Foundation of New York/QVC Presents Shoes on Sale™ and by International Collaboration Grant from National Research Council (CONICET, Argentina) (to A.M.C.). Travel expenses for G.L. were supported by a Journal of Cell Science Travelling Fellowship from the Company of Biologists; A.M.C. was a 2009 Fulbright-CONICET Visitor Scholar. Deposited in PMC for release after 12 months.
References
- Aigner K., Dampier B., Descovich L., Mikula M., Sultan A., Schreiber M., Mikulits W., Brabletz T., Strand D., Obrist P., et al. (2007). The transcription factor ZEB1 (deltaEF1) promotes tumour cell dedifferentiation by repressing master regulators of epithelial polarity. Oncogene 26, 6979-6988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bornstein P., Sage E. H. (2002). Matricellular proteins: extracellular modulators of cell function. Curr. Opin. Cell Biol. 14, 608-616 [DOI] [PubMed] [Google Scholar]
- Bracken C. P., Gregory P. A., Kolesnikoff N., Bert A. G., Wang J., Shannon M. F., Goodall G. J. (2008). A double-negative feedback loop between ZEB1-SIP1 and the microRNA-200 family regulates epithelial-mesenchymal transition. Cancer Res. 68, 7846-7854 [DOI] [PubMed] [Google Scholar]
- Brigstock D. R., Goldschmeding R., Katsube K. I., Lam S. C., Lau L. F., Lyons K., Naus C., Perbal B., Riser B., Takigawa M., et al. (2003). Proposal for a unified CCN nomenclature. Mol. Pathol. 56, 127-128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cabanillas A. M., Smith G. E., Darling D. S. (2001). T3-activation of the rat growth hormone gene is inhibited by a zinc finger/homeodomain protein. Mol. Cell. Endocrinol. 181, 131-137 [DOI] [PubMed] [Google Scholar]
- Darling D. S., Stearman R. P., Qi Y., Qiu M. S., Feller J. P. (2003). Expression of Zfhep/deltaEF1 protein in palate, neural progenitors, and differentiated neurons. Gene Expr. Patterns 3, 709-717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eger A., Aigner K., Sonderegger S., Dampier B., Oehler S., Schreiber M., Berx G., Cano A., Beug H., Foisner R. (2005). DeltaEF1 is a transcriptional repressor of E-cadherin and regulates epithelial plasticity in breast cancer cells. Oncogene 24, 2375-2385 [DOI] [PubMed] [Google Scholar]
- Garcia-Echeverria C., Pearson M. A., Marti A., Meyer T., Mestan J., Zimmermann J., Gao J., Brueggen J., Capraro H. G., Cozens R., et al. (2004). In vivo antitumor activity of NVP-AEW541-A novel, potent, and selective inhibitor of the IGF-1R kinase. Cancer Cell 5, 231-239 [DOI] [PubMed] [Google Scholar]
- Gariboldi M. B., Ravizza R., Monti E. (2010). The IGFR1 inhibitor NVP-AEW541 disrupts a pro-survival and pro-angiogenic IGF-STAT3-HIF1 pathway in human glioblastoma cells. Biochem. Pharmacol. 80, 455-462 [DOI] [PubMed] [Google Scholar]
- Graham T. R., Zhau H. E., Odero-Marah V. A., Osunkoya A. O., Kimbro K. S., Tighiouart M., Liu T., Simons J. W., O'Regan R. M. (2008). Insulin-like growth factor-I-dependent up-regulation of ZEB1 drives epithelial-to-mesenchymal transition in human prostate cancer cells. Cancer Res. 68, 2479-2488 [DOI] [PubMed] [Google Scholar]
- Gregory P. A., Bert A. G., Paterson E. L., Barry S. C., Tsykin A., Farshid G., Vadas M. A., Khew-Goodall Y., Goodall G. J. (2008). The miR-200 family and miR-205 regulate epithelial to mesenchymal transition by targeting ZEB1 and SIP1. Nat. Cell Biol. 10, 593-601 [DOI] [PubMed] [Google Scholar]
- Higashi Y., Moribe H., Takagi T., Sekido R., Kawakami K., Kikutani H., Kondoh H. (1997). Impairment of T cell development in deltaEF1 mutant mice. J. Exp. Med. 185, 1467-1479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang W., Zhang Y., Varambally S., Chinnaiyan A. M., Banerjee M., Merajver S. D., Kleer C. G. (2008). Inhibition of CCN6 (Wnt-1-induced signaling protein 3) down-regulates E-cadherin in the breast epithelium through induction of snail and ZEB1. Am. J. Pathol. 172, 893-904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang W., Gonzalez M. E., Toy K. A., Banerjee M., Kleer C. G. (2010). Blockade of CCN6 (WISP3) activates growth factor-independent survival and resistance to anoikis in human mammary epithelial cells. Cancer Res. 70, 3340-3350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleer C. G., Zhang Y., Pan Q., van Golen K. L., Wu Z. F., Livant D., Merajver S. D. (2002). WISP3 is a novel tumor suppressor gene of inflammatory breast cancer. Oncogene 21, 3172-3180 [DOI] [PubMed] [Google Scholar]
- Kleer C. G., Zhang Y., Pan Q., Merajver S. D. (2004). WISP3 (CCN6) is a secreted tumor-suppressor protein that modulates IGF signaling in inflammatory breast cancer. Neoplasia 6, 179-185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau L. F., Lam S. C. (1999). The CCN family of angiogenic regulators: the integrin connection. Exp. Cell Res. 248, 44-57 [DOI] [PubMed] [Google Scholar]
- Law J. H., Habibi G., Hu K., Masoudi H., Wang M. Y., Stratford A. L., Park E., Gee J. M., Finlay P., Jones H. E., et al. (2008). Phosphorylated insulin-like growth factor-i/insulin receptor is present in all breast cancer subtypes and is related to poor survival. Cancer Res. 68, 10238-10246 [DOI] [PubMed] [Google Scholar]
- Leask A., Abraham D. J. (2006). All in the CCN family: essential matricellular signaling modulators emerge from the bunker. J. Cell Sci. 119, 4803-4810 [DOI] [PubMed] [Google Scholar]
- Manavella P. A., Roqueiro G., Darling D. S., Cabanillas A. M. (2007). The ZFHX1A gene is differentially autoregulated by its isoforms. Biochem. Biophys. Res. Commun. 360, 621-626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendez M. G., Kojima S., Goldman R. D. (2010). Vimentin induces changes in cell shape, motility, and adhesion during the epithelial to mesenchymal transition. FASEB J. 24, 1838-1851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park S. M., Gaur A. B., Lengyel E., Peter M. E. (2008). The miR-200 family determines the epithelial phenotype of cancer cells by targeting the E-cadherin repressors ZEB1 and ZEB2. Genes Dev. 22, 894-907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perbal B. (2008). Matricellular CCN proteins. J. Cell Commun. Signal. 2, 57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollak M. (2008). Insulin and insulin-like growth factor signalling in neoplasia. Nat. Rev. Cancer 8, 915-928 [DOI] [PubMed] [Google Scholar]
- Postigo A. A., Dean D. C. (1999). ZEB represses transcription through interaction with the corepressor CtBP. Proc. Natl. Acad. Sci. USA 96, 6683-6688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samani A. A., Yakar S., LeRoith D., Brodt P. (2007). The role of the IGF system in cancer growth and metastasis: overview and recent insights. Endocr. Rev. 28, 20-47 [DOI] [PubMed] [Google Scholar]
- Shirakihara T., Saitoh M., Miyazono K. (2007). Differential regulation of epithelial and mesenchymal markers by deltaEF1 proteins in epithelial mesenchymal transition induced by TGF-beta. Mol. Biol. Cell 18, 3533-3544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spaderna S., Schmalhofer O., Wahlbuhl M., Dimmler A., Bauer K., Sultan A., Hlubek F., Jung A., Strand D., Eger A., et al. (2008). The transcriptional repressor ZEB1 promotes metastasis and loss of cell polarity in cancer. Cancer Res. 68, 537-544 [DOI] [PubMed] [Google Scholar]
- Takagi T., Moribe H., Kondoh H., Higashi Y. (1998). DeltaEF1, a zinc finger and homeodomain transcription factor, is required for skeleton patterning in multiple lineages. Development 125, 21-31 [DOI] [PubMed] [Google Scholar]
- van Golen K. L., Davies S., Wu Z. F., Wang Y., Bucana C. D., Root H., Chandrasekharappa S., Strawderman M., Ethier S. P., Merajver S. D. (1999). A novel putative low-affinity insulin-like growth factor-binding protein, LIBC (lost in inflammatory breast cancer), and RhoC GTPase correlate with the inflammatory breast cancer phenotype. Clin. Cancer Res. 5, 2511-2519 [PubMed] [Google Scholar]
- Vandewalle C., Van Roy F., Berx G. (2009). The role of the ZEB family of transcription factors in development and disease. Cell. Mol. Life Sci. 66, 773-787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wellner U., Schubert J., Burk U. C., Schmalhofer O., Zhu F., Sonntag A., Waldvogel B., Vannier C., Darling D., zur Hausen A., et al. (2009). The EMT-activator ZEB1 promotes tumorigenicity by repressing stemness-inhibiting microRNAs. Nat. Cell Biol. 11, 1487-1495 [DOI] [PubMed] [Google Scholar]
- Witsch E., Sela M., Yarden Y. (2010). Roles for growth factors in cancer progression. Physiology (Bethesda) 25, 85-101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Pan Q., Zhong H., Merajver S. D., Kleer C. G. (2005). Inhibition of CCN6 (WISP3) expression promotes neoplastic progression and enhances the effects of insulin-like growth factor-1 on breast epithelial cells. Breast Cancer Res. 7, R1080-R1089 [DOI] [PMC free article] [PubMed] [Google Scholar]