Abstract
Chemotherapy resistance is a key contributor to the dismal prognoses for lung cancer patients. While the majority of studies have focused on sequence mutations and expression changes in protein-coding genes, recent reports have suggested that microRNA (miRNA) expression changes also play an influential role in chemotherapy response. However, the role of genetic alterations at miRNA loci in the context of chemotherapy response has yet to be investigated. In this study, we demonstrate the application of an integrative, multidimensional approach in order to identify miRNAs that are associated with chemotherapeutic resistance and sensitivity utilizing publicly available drug response, miRNA loci copy number, miRNA expression, and mRNA expression data from independent resources. By instigating a logical stepwise strategy, we have identified specific miRNAs that are associated with resistance to several chemotherapeutic agents and provide a proof of principle demonstration of how these various databases may be exploited to derive relevant pharmacogenomic results.
1. Introduction
Lung cancer is the most common cause of cancer-related deaths worldwide, with a five-year survival rate of less than 15% [1]. The high incidence of late-stage diagnosis and a lack of efficient therapeutic strategies remain key contributors to the dismal survival statistics. Thus, to improve lung cancer patient outcome, improvement in early detection and a better understanding of the underlying tumor biology that governs response to therapy are necessary. Response to systemic therapy has been shown to be strongly associated with a variety of clinical and molecular features. For example, the chemotherapeutics Avastin and Permetrexed have shown differential response or adverse effects in different histological subtypes of lung cancer [2, 3]. Tyrosine kinase inhibitors (TKIs) targeting the epidermal growth factor receptor (EGFR) have shown preferential efficacy in Asian females who typically harbor sequence mutations in EGFR as well as those individuals who harbored EGFR amplifications, EGFR mutations, and the absence of KRAS mutations [4–6]. Very recently, inhibitors to ALK rearrangement also showed significant response in patients who harbor this genetic alteration [7].
In addition to molecular features that can predict sensitivity, there are also examples of features that can predict resistance. In ovarian cancer, resistance to therapy was observed in those individuals who carried amplifications of genes such as P-glycoprotein as well as specific regions in the genome such as 19q12 and 20q11.22-q13.12 [8, 9]. With respect to lung cancer, while there are individuals who do respond to TKIs, a large proportion will develop resistance to these therapies by acquiring an additional EGFR mutation (T790M), amplification of the c-MET oncogene, or hypermethylation of the PTEN locus [10–12]. High levels of ERCC1 mRNA and protein, a key player in nucleotide excision repair, have been associated with resistance to platinum-based chemotherapy [13]. Similarly, low levels of RRM1/2 mRNA and protein were associated with favorable gemcitabine response in NSCLC patients [14].
Although alterations in protein-coding genes remain a main focus to elucidate sensitivity or resistance to chemotherapy, deregulation of microRNAs (miRNAs) has recently been shown to play a role in chemotherapy response [15–17]. miRNAs are small noncoding RNAs approximately 18–25 nucleotides in length that negatively regulate gene expression posttranscriptionally [18, 19]. miRNA biogenesis begins with a long, double-stranded RNA known as a pri-miRNA, typically hundreds to thousands of nucleotides in length, which is processed into sequentially shorter double-stranded RNA sequences by the endonucleases Drosha and Dicer that are of 70 and 22 nucleotides in size, respectively [20, 21]. Dissociation of the duplex and incorporation of the mature strand into the RNA-induced silencing complex (RISC) guides RISC to the target mRNA, where the miRNA exhibits its effect [22]. miRNAs bind target transcripts based on sequence similarity—typically in the 3′UTR of the transcript and sometimes in the 5′UTR and the coding region—resulting in inhibition of translation or transcript degradation [18, 19, 23].
The relevance of miRNA deregulation to cancer biology arises because increased expression of certain miRNAs can result in downregulation of tumor suppressor genes, while decreased expression of other miRNAs can lead to increased expression of oncogenes [20, 21]. Often located at chromosomal breakpoint regions, fragile sites, and minimal regions of loss of heterozygosity or amplification, miRNA loci are highly susceptible to genomic alterations and subsequently, deregulated expression [24–27]. Aberrant miRNA expression is a common feature of both dysplasia and cancer, and miRNA expression profiles have been associated with prognosis, disease progression, survival, and outcome prediction [28, 29]. Further, miRNA expression profiles have been found to be superior to global mRNA expression profiles for the accurate definition of cancer types [30, 31]. Lung cancer drug response has been associated with the deregulation of several miRNAs. For example, sensitivity of nonsmall cell lung cancer (NSCLC) to cisplatin treatment was linked to upregulation of miR-181a, while resistance was conferred by upregulation of miR-630 [32]. Sensitivity to another chemotherapeutic agent, Gefitinib, was correlated with loss of miR-128b [33]. Several studies have shown that the overexpression of specific miRNAs, such as miR-134 and let-7a, can increase drug sensitivity, demonstrating the therapeutic potential of miRNAs [34, 35].
In this study, we sought to determine the role of DNA copy number alterations at miRNA loci in chemotherapy response. As a proof of principle, making use of datasets generated by multiple institutions, encompassing we performed an integrative and comparative DNA dosage and expression alteration analysis of miRNA loci in highly sensitive and resistant lung cancer cell lines for 18 different chemotherapeutics. Using a rigorous, stepwise analysis strategy, we identified four miRNAs which were frequently gained and overexpressed in lung cancer cell lines resistant to one or two of five different chemotherapeutic agents. Subsequent gene expression and gene network analyses for each set of mRNA targets of a given miRNA revealed functions such as DNA replication and repair and cellular assembly and maintenance that were overrepresented in all four sets. These findings demonstrate the feasibility and the value of integrative analysis of multidimensional publicly accessible databases as a strategy for pharmacogenomics discovery.
2. Material and Methods
2.1. Drug Response Profiles of Cancer Cell Lines
Drug response IC50 data for 18 different chemotherapeutics across 350 cancer cell lines (See Supplementary Material available online at doi: 10.1155/2011/474632 Supplemental Table 1) was generated as part of the Wellcome Trust Sanger Institute and Massachusetts General Hospital's (MGH) joint Genomics of Drug Sensitivity in Cancer Project. Data was downloaded from the following website: (http://www.sanger.ac.uk/genetics/CGP/translation/compound_sens_data.shtml). Briefly, IC50 is the required concentration of a particular drug to cause in vitro growth to be inhibited by 50%, and thus, a measure of drug effectiveness. A low IC50 indicates that a drug is very effective at inhibiting growth while a high IC50 indicates that a drug is less effective and thus requires a higher dosage to function. Of the 350 cancer cell lines, 73 cell lines were of lung origin.
2.2. Generation of DNA Copy Number Profiles for Cancer Cell Lines
Affymetrix SNP 6.0 data for the cancer cell lines were obtained from the Wellcome Trust Sanger Institute CGP Data Archive (http://www.sanger.ac.uk/genetics/CGP/Archive/). Of the 73 lung cancer cell lines with drug response data, 67 of them also had matching SNP array hybridization data (Supplemental Table 2). SNP array data were normalized using default parameters in Partek Genomics Suite (PGS, Partek Inc, St. Louis, MI). Whole genome copy number profiles were visualized using SIGMA2 software [36].
2.3. miRNA and mRNA Expression Data for Cancer Cell Lines
The current annotation of autosomal miRNAs and their genomic coordinates were obtained from the UCSC Genome Browser (http://www.genome.ucsc.edu/) using the NCBI36/hg18 mapping [37]. miRNA and mRNA expression profiles for lung cancer cell lines were downloaded from the Broad Institute (http://www.broadinstitute.org/cgi-bin/cancer/datasets.cgi) under the “Sanger Cell Line Project.” Affymetrix HG-U133A mRNA expression data were RMA-normalized using the “affy” package in Bioconductor in R [38–40]. Mapping of probes to genes was performed using the Affymetrix NetAffx annotation file (version NA31). Of the 73 lung cancer cell lines with drug response data, 64 had matching miRNA expression while 68 had matching mRNA expression data (Supplemental Table 2).
2.4. Determination of Predicted miRNA Targets
TargetSpy (version 1.0) and TargetScan (version 5.1) miRNA target prediction software were used to identify mRNA targets for further analyses [41–44]. For TargetSpy, the “no seed requirement, high sensitivity” set of targets were used, while for TargetScan, the nonconserved miRNA-mRNA targets were used. For the miRNAs that were further assessed for target analysis, only miRNA-mRNA target pairs that were present in both databases were assessed for gene expression differences.
2.5. Statistical Analysis
For DNA alteration analysis, copy number profiles of the cancer cell lines were determined against a pooled reference comprised of 72 cytogenetically normal individuals in the HapMap collection. SNP 6.0 data for the HapMap individuals were obtained through Affymetrix. Subsequently, to determine copy number gains and losses, copy number profiles were subjected to segmentation analysis using the “Genomic Segmentation” algorithm in PGS with the following parameters: minimum genomic markers = 20, P-value threshold for adjacent regions having significantly different means = 1 × 10−6, and P-value threshold for deviation from normal (diploid) copy number = 1 × 10−6. In addition to meeting P-value thresholds, a region was deemed gained if the cell line had >2.3 copies while a region was deemed lost if the cell line has <1.7 copies. For each cell line, the copy number status for individual miRNA loci were determined by mapping the genomic coordinates of the miRNA loci to the identified regions of alteration.
To determine miRNA loci in differentially altered regions of copy number between highly resistant and sensitive cell lines, for each chemotherapeutic, cell lines were ranked based on their IC50 value. The frequency of copy number gain, loss, and retention were compared between the top 1/3 and bottom 1/3 of cancer cell lines using a 3 by 2 Fisher's exact test. A miRNA was deemed significant if the P value from the Fisher's exact test was ≤.05.
For miRNA and mRNA expression analysis, similar to the differential copy number analysis, cell lines were ranked based on IC50 for each drug. Subsequently, for each miRNA, the expression in the top and bottom tertiles of cell lines was compared using a nonparametric Mann Whitney U test. A miRNA was deemed significant if the P value from the Mann Whitney U test was ≤.05.
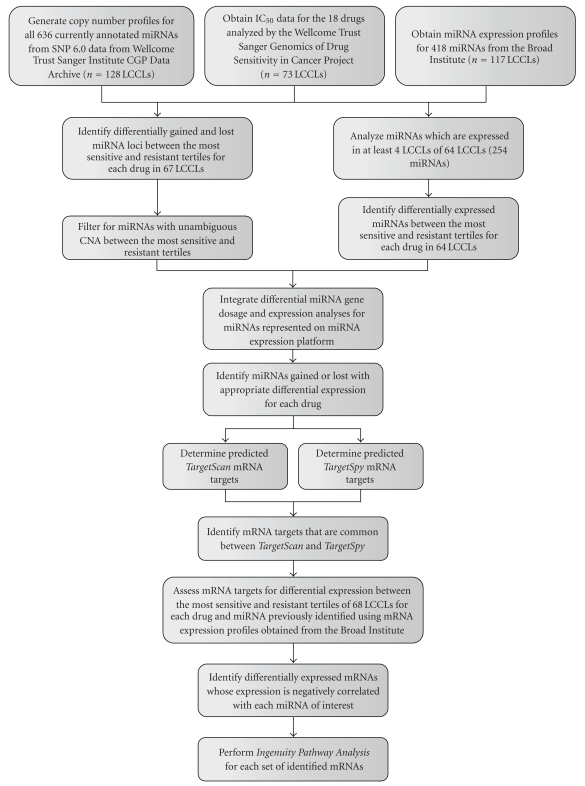
Upon identifying which lung cancer cell lines (LCCLs) contained matching DNA copy number and drug response profiles, for each chemotherapeutic, we compared the patterns of copy number alteration between the most sensitive and resistant LCCLs for 636 miRNA loci. Of the resulting differentially altered miRNA identified using the above statistical criteria, we filtered out those miRNAs which were both preferentially gained and lost in either highly resistant or highly sensitive LCCLs. We defined these variably altered miRNAs as those whose differential alteration frequency (DAF) of gain, frequency of gain in highly resistant minus the frequency of gain in highly sensitive, was within 10% of the DAF of loss, which is the frequency of loss in highly resistant minus the frequency of loss in highly sensitive. In parallel, upon identifying LCCLs with both miRNA expression and drug response profiles, we compared the miRNA expression profiles between the most sensitive and resistant LCCLs for 254 miRNAs using the above-mentioned statistical methods. Although 418 unique miRNAs are represented on the microarray platform, we restricted this analysis to the 254 miRNAs that were expressed in at least 4 LCCLs. Subsequently, for each drug, we identified the miRNAs which were both significantly different at the DNA copy number and expression levels that matched in the same direction that is, if a miRNA had higher copy number in the highly resistant LCCLs as compared to the highly sensitive LCCLs, then the expression would also have to be higher, and vice versa. Next, for each significant miRNA, bioinformatic analysis was performed to identify target mRNAs, and mRNA expression profiles for these genes were compared in a similar manner to that performed in the differential DNA copy number and miRNA expression analyses (using TargetSpy and TargetScan; see above). Restricting to those targets whose mRNA expression profiles negatively correlate with miRNA expression profiles, we performed gene network and function analysis using Ingenuity Pathway Analysis to identify significantly overrepresented functions that were common to all sets of differentially expressed miRNA targets. A flow chart illustrating this strategy is shown in Figure 1.
Figure 1.
The search for drug response-related miRNAs began with data acquisition from several independent databases. Drug response data for lung cancer cell lines (LCCLs) was integrated independently with copy number and expression data, and unique filtering criteria were applied. The integration of all three dimensions applied further filtration criteria, and the remaining miRNAs underwent predicted target analysis. The resulting mRNA target expression was anticorrelated with miRNA expression, and cellular functions of the final mRNA target list were derived by Ingenuity Pathway Analysis.
2.6. Gene Network and Pathway Analysis
For each miRNA, the set of differentially expressed target genes were analyzed using Ingenuity Pathway Analysis (Ingenuity Systems, Redwood City, CA) to determine statistically overrepresented networks and pathways. Briefly, a right tailed Fisher's exact test was employed to calculate a P-value for the probability that enrichment of functions within the gene list of interest and the entire list of genes in the human genome is due to chance alone. Only the Molecular and Cellular Functions within the Biological Functions analysis were assessed.
3. Results
3.1. Copy Number Alterations of miRNA Loci Correlate with Drug Response in Lung Cancer Cell Lines
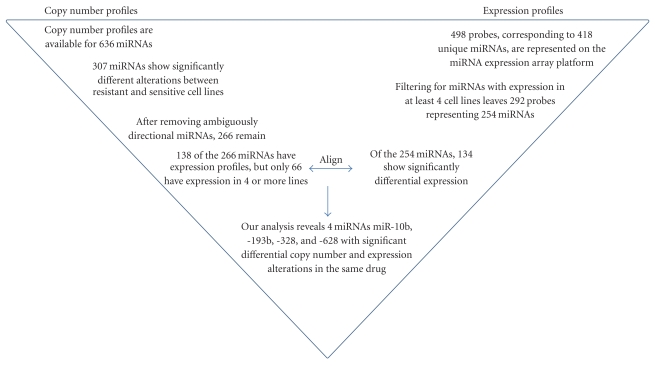
Sixty seven lung cancer cell lines with available IC50 data were used to analyze miRNA copy number alterations. For each drug, cell lines were sorted based on IC50 values and the frequencies of DNA copy number gain, loss, and retention were compared between the highest (most resistant, n = 22) and the lowest (most sensitive, n = 22) tertile of cell lines. Of the 636 miRNAs assessed, 307 miRNAs (48.3%) were significantly different between high and low IC50 for at least one drug, and 20 miRNAs (3.1%) were different for at least four drugs (P ≤ .05 Fisher's exact test, Table 1, Supplemental Table 3). In addition, among the 307 miRNAs, 58.4% were either more frequently gained in high IC50 or more frequently lost in low IC50 while 23.6% were either more frequently lost in high IC50 or more frequently gained in Iow IC50 lung cancer cell lines. The remaining 41 miRNAs (17.9%), although significantly different, had less than a 10% DAF difference between resistant and sensitive lines and were, therefore, deemed variably altered (see methods) and subsequently removed. This brought the total number of miRNAs with significant differences in copy number to 266 (Figure 2). In terms of the drug with the most striking pattern of differential alteration between high and low IC50 cell lines, TAE684, a small molecule ALK fusion kinase inhibitor, had 66 miRNAs that were significantly different between the most resistant and most sensitive cell lines (Figure 3). Conversely, miR-662 was the most frequently differentially-altered miRNA across all of the drugs, appearing significant in 6 of 18 drugs.
Table 1.
List of miRNA with most frequent differential copy number alterations.
| miRNA | Significant drugs |
|---|---|
| hsa-mir-662 | 6 (AZ, Erl, Gel, Gö, HKI, MK) |
| hsa-mir-124-2 | 5 (Erl, HKI, Ra, Sor, Sun) |
| hsa-mir-1285-2 | 5 (MG, PF, PH, Ra, Sun) |
| hsa-mir-548h-2 | 5 (Gel, HKI, MK, PD, Sor) |
| hsa-mir-1208 | 4 (Cy, Erl, HKI, Sun) |
| hsa-mir-1225 | 4 (AZ, Gö, MK, Sor) |
| hsa-mir-1228 | 4 (Gel, MK, PF, TAE) |
| hsa-mir-1299 | 4 (AZD, Erl, MK, PH) |
| hsa-mir-147 | 4 (Erl, HKI, MK, TAE) |
| hsa-mir-181a-2 | 4 (Erl, HKI, MK, TAE) |
| hsa-mir-181b-2 | 4 (Erl, HKI, MK, TAE) |
| hsa-mir-1827 | 4 (AZ, Gel, Pac, PF) |
| hsa-mir-1972 | 4 (AZ, Gö, MK, Sor) |
| hsa-mir-492 | 4 (Gel, Im, Pac, PF) |
| hsa-mir-548c | 4 (Gel, MK, PF, TAE) |
| hsa-mir-548d-1 | 4 (Cy, Gö, HKI, Sun) |
| hsa-mir-548f-2 | 4 (Gö, MG, PF, TAE) |
| hsa-mir-600 | 4 (Erl, HKI, MK, TAE) |
| hsa-mir-601 | 4 (Erl, HKI, MK, TAE) |
| hsa-mir-940 | 4 (AZ, Gö, MK, Sor) |
Figure 2.
Flowchart summarizing the process for the identification of the four miRNAs which correlated significantly with drug response.
Figure 3.
Comparison of the frequency of alteration of 636 miRNA loci between highly sensitive and highly resistant lung cancer cell lines (LCCLs) to agent TAE684. Highly sensitive LCCLs were represented by the lowest tertile of IC50 while the highly resistant were represented by the highest tertile of IC50. miRNA genomic position information was obtained from the UCSC Genome Browser database [37], and miRNAs on chromosomes X and Y were excluded. Copy number alterations frequencies were plotted using SIGMA2 software [36]. Vertical lines denote the frequency of alteration, where 1 or −1 signifies the alteration that occurs in 100% of samples. Horizontal bars depict miRNAs, with the frequency of copy gains and losses of each miRNA displayed to the right and left of 0, respectively. miRNAs disrupted in resistant lines are displayed in red, those occurring in sensitive lines are displayed in green, and regions of overlapping frequencies are shown in black.
3.2. miRNA Expression Levels Correlate with Drug Response in Lung Cancer Cell Lines
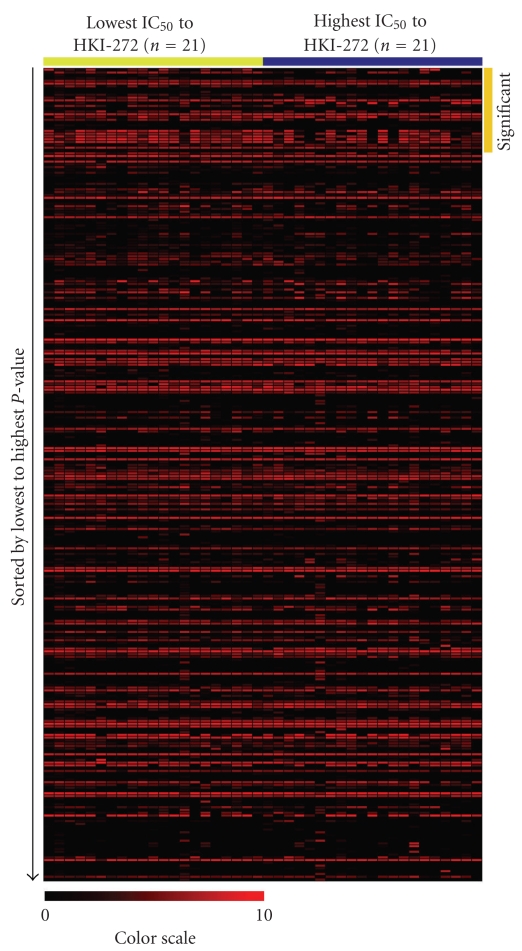
miRNA expression was assessed in 64 lung cancer cell lines using a similar method to that applied for identifying copy number alteration differences, comparison of the highest and lowest tertile of cell lines (n = 21) based on IC50 values for each drug. miRNA expression profiles were available for 498 probes measuring 418 unique miRNAs. However, a number of miRNAs have little to no expression. To account for these cases, miRNAs with expression in less than four cell lines were removed, leaving 292 probes which corresponded to 254 unique miRNAs (Figure 2). One hundred thirty four miRNAs (represented by 146 probes) of the 254 (52.8%) miRNAs with available expression data were significant in at least one drug (P ≤ .05, Mann Whitney U test) (Supplemental Table 4), with 18 miRNAs significant in at least four drugs (Table 2). Of the 134 differentially expressed miRNAs, 40% had higher expression in high IC50, while 60% had higher expression in low IC50 lung cancer cell lines. HKI-272 had the most miRNAs [30] that were significantly different at the expression level (Figure 4), and miR-625 was the most frequently differentially expressed miRNA, appearing significant in 7 of 18 drugs.
Table 2.
Most frequently different miRNAs at the expression level.
| miRNA | Significant drugs |
|---|---|
| hsa-mir-625 | 7 (Erl, Gö, HKI, MG, Pa, PHA, Ra) |
| hsa-mir-130a | 6 (Erl, Gö, HKI, MG, Pa, Sun) |
| hsa-mir-148a | 6 (AZ, Erl, Gel, Gö, HKI, Pa) |
| hsa-mir-215 | 5 (Gel, Gö, MG, MK, Pa) |
| hsa-mir-518b | 5 (Cy, PF, PHA, Ra, Sun) |
| hsa-mir-100 | 5 (Erl, Gö, HKI, Sun, TAE) |
| hsa-mir-192 | 5 (Gel, Gö, MG, MK, Pa) |
| hsa-mir-375 | 5 (Erl, Gel, Gö, HKI, Pa) |
| hsa-mir-503 | 5 (Cy, Erl, MK, PHA, Ra) |
| hsa-mir-193b | 4 (AZ, MK, PHA, Ra) |
| hsa-mir-521 | 4 (Cy, Erl, Im, PHA) |
| hsa-mir-95 | 4 (Cy, Erl, MG, Sun) |
| hsa-mir-194 | 4 (Gel, Gö, MG, Pa) |
| hsa-mir-205 | 4 (Erl, Gel, Gö, HKI) |
| hsa-mir-222 | 4 (AZ, Erl, Gel, HKI) |
| hsa-mir-27a | 4 (AZD, Erl, Gel, Gö) |
| hsa-mir-377 | 4 (MG, Pa, Sor, Sun) |
| hsa-mir-382 | 4 (AZD, MG, Pa, Sun) |
Figure 4.
Heatmap visualization of the miRNA expression of the 254 miRNAs (represented by 292 unique probes) that passed expression filtering criteria for the 21 most sensitive (yellow bar, low IC50) and 21 most resistant (blue bar, high IC50) to drug HKI-272. In total, 30 miRNAs were found to be significantly differentially expressed between the most sensitive and resistant lung cancer cell lines (LCCLs, orange bar). For this visualization, since a value of 4 represented no expression, all expression values were subtracted by 4 such that baseline expression would be shown as 0 (black).
3.3. Integrative Analysis of miRNA Gene Dosage and Expression Levels in Lung Cancer Cell Lines
To determine if miRNA dosage modulates expression, we compared the 266 miRNAs differentially altered at the copy number level and the 134 miRNAs differentially expressed in at least one of the drugs analyzed. Considering only those miRNAs that were significant at both the copy number and miRNA expression level for the same drug, the intersection of these two lists yielded five miRNAs, miR-10b, -191,-193b, -328, and -628 (Figure 2). Of these five, only expression of four miRNAs, mir-10b, -193b, -328, and -628 matched the direction of their respective copy number alterations. For example, miR-628 is more frequently gained in high IC50 lines compared to low IC50 lines treated with agent PF-2341066 and also shows higher expression in high IC50 lines compared to low IC50 lines (Table 3), whereas miR-191 is frequently gained in low IC50 lines when treated with TAE684, but shows higher expression in high IC50 lines.
Table 3.
List of miRNAs with significant copy number and expression alterations in the same drug.
| miRNA | Copy number alteration | Expression alteration | Drug in which significant |
|---|---|---|---|
| hsa-mir-10b | Gained, High IC50 | Overexpressed, High IC50 | MG-132 |
| hsa -mir-193b | Gained, High IC50 | Overexpressed, High IC50 | AZ628, MK-0457 |
| hsa-mir-328 | Gained, High IC50 | Overexpressed, High IC50 | Geldanamycin |
| hsa-mire-628 | Gained, High IC50 | Overexpressed, High IC50 | PF-2341066 |
3.4. Gene Expression Analysis of mRNA Targets of miR-10b, miR-193b, miR-328, and miR-628
The target prediction software TargetScan and TargetSpy were used to identify putative mRNA targets of miRNAs found to be significantly different at the copy number and miRNA expression levels between high-IC50 and low-IC50 cell lines. For the four miRNAs (miR-10b, miR-328, miR-193b, and miR-628) identified by integrative analyses, only miRNA-mRNA targets present in both databases were used for further analysis.
miR-10b was identified as having a significant association with response to the proteosome inhibitor MG-132. In total, target prediction analysis found 636 genes that were deemed as putative targets of miR-10b (Supplemental Table 5). Comparison of the gene expression profiles between lung cancer cell lines with high and low IC50 for MG-132 revealed 48 of these target genes to be differentially expressed (P ≤ .05, Mann Whitney U test), with 32 of them showing the expected direction of differential expression (i.e., anticorrelated mRNA expression to miRNA expression) (Table 4).
Table 4.
Differentially expressed mRNA targets for the four identified miRNAs.
| miRNA | 10b | 328 | 193b | 193b | 628 |
|---|---|---|---|---|---|
| Drug | MG-132 (26S Proteasome inhibitor) | Geldanamycin (HSP90 inhibitor) | AZ628 (RAF inhibitor) | MK-0457 (Aurora kinase inhibitor) | PF-2341066 (MET, ALK inhibitor) |
| Targets | SMARCC1 | SLC16A1 | PHF15 | EIF2S1 | DCTD |
| MKRN2 | ANGEL1 | RPP30 | ARIH2 | NUP188 | |
| CMTM6 | UBR5 | RRP1B | TMEM231 | ELAC1 | |
| CSDE1 | SEMA3C | EIF4B | GABBR1 | DACT1 | |
| MAP4 | MEIS2 | WDR48 | IKZF1 | CYP7B1 | |
| MYO10 | TRIM32 | PTPN21 | FKTN | RPA4 | |
| NKTR | GM2A | CLEC2D | GPATCH8 | GAR1 | |
| RYBP | EIF2S1 | MRPS16 | EZH1 | UCKL1 | |
| ZNF532 | RAC2 | NUDT15 | OLFML2A | IL7 | |
| SENP5 | OLFML2A | NECAP2 | PSME3 | ABCE1 | |
| CBL | HIP1 | AGTPBP1 | CASP3 | ||
| WHSC1 | PRKD3 | NMT2 | WBP4 | ||
| FOXJ2 | VDR | ADARB1 | KLF9 | ||
| APOLD1 | YME1L1 | GREB1 | ACTN2 | ||
| PEA15 | TP63 | SMC5 | GPM6A | ||
| RAD1 | TFAP2C | CCDC28A | LSM12 | ||
| CDC6 | CHP2 | RNMT | PRKCA | ||
| UBE2K | STK24 | C15orf29 | ADAT1 | ||
| BNIP2 | GPR126 | RRP1B | PBRM1 | ||
| PARD6B | SERTAD2 | AGPAT4 | SP2 | ||
| FGD6 | DSC2 | UBA52 | TRAIP | ||
| MMP14 | PTGIS | GOSR1 | MAGOHB | ||
| OLFML2A | RAB22A | PRKD3 | |||
| GNL3L | FBXW2 | SYF2 | |||
| CTDSPL | HS2ST1 | SSX2IP | |||
| MME | RPP30 | ANKH | |||
| PPFIBP1 | SLC16A3 | ZBTB39 | |||
| M6PR | MAN1A2 | KBTBD11 | |||
| KPNA6 | TGIF2 | SEC16A | |||
| RMI1 | CLEC2D | PHC3 | |||
| RBM15B | ZNF510 | ||||
| SS18L2 | RTF1 | ||||
| CYLD | |||||
| WDR48 | |||||
| BTRC | |||||
| CCNT2 | |||||
| RAB36 | |||||
Interestingly, miR-193b alteration was significantly associated with response to two therapeutics: AZ628 and MK0457 (RAF and aurora kinase inhibitors, resp.). When a similar analysis to hsa-miR-10b was performed for miR-193b, 518 genes were identified as putative targets of miR-193b (Supplemental Table 5). For the analysis of gene expression between highly sensitive and resistant AZ628 cells, 28 of these targets were differentially expressed, with ten of these genes matching the expected direction of differential expression. For MK-0457, 67 of these target genes were differentially expressed with over half (37) matching the expected direction (Table 4).
Alteration of miR-328 was significantly associated with the response to Hsp90 inhibitor Geldanamycin in lung cancer cell lines. Of the 437 genes targeted by miR-328, 49 of these genes were significantly differentially expressed between highly resistant and sensitive cell lines, with 31 of the genes matching the expected direction (Supplemental Tables 5 and 4). Finally, for miR-628, whose alteration was significantly associated with the MET inhibitor PF-2341066 response, 392 targets genes were identified with 49 of them being differentially expressed and 22 of those in the appropriate direction (Supplemental Tables 5 and 4).
4. Discussion
Chemotherapy response can be influenced by a number of clinicopathological and molecular factors. At the molecular level, while a large focus revolves around the role of activating and inactivating sequence mutations as well as copy number amplifications and deletions in protein coding genes, there has been an increasing emphasis on examining the role of miRNAs and response to chemotherapy. Recent studies have focused on differentially expressed miRNAs in conjunction to resistance and sensitivity to a variety of chemotherapeutics [32–35, 45, 46]. However, the influence of copy number alterations at miRNA loci (or gene dosage) in the context of drug response has not been thoroughly investigated. To this end, we have performed an integrative analysis of genome-wide miRNA copy number, miRNA expression, mRNA expression, and drug sensitivity data from 18 different chemotherapeutics on a panel of lung cancer cell lines to identify miRNAs that are significantly different at the copy number and expression levels between the most sensitive and resistant cell lines for a given drug.
Upon comparison of the 636 annotated miRNAs throughout the human genome, it was found that 266 of them revealed significant differences in copy number alteration pattern between sensitive and resistant cancer cell lines for at least one drug (Supplemental Table 3). Moreover, of the 266 miRNAs, there were more miRNAs with increased copy number for the highly resistant versus the highly sensitive lung cancer cell lines than vice versa. The miRNA that was found to have a differential pattern of copy number alteration between sensitive and resistant cancer cell lines for the most drugs was miR-662, and, conversely, the drug with most significantly different miRNAs was TAE684. miR-662 is located on chromosomal region 16p13.3 and was found to be more frequently gained in cell lines highly resistant to AZ628, Erlotinib, Geldanamycin, Gö-6976, HKI-272 (Neratinib), and MK-0457. All of these drugs, except for Geldanamycin, which is an antibody that targets HSP90, are kinase inhibitors [47]. While not much is known of miR-662, it was recently shown that it is transiently upregulated in response to high doses of X-ray radiation in human fibroblasts [48]. It should also be noted that miR-124-2, miR-1285-2, and miR-548h-2 were significantly altered for five drugs (Table 1). Similar to miR-662, little is known of miR-548h-2. However, miR-124 has been shown to play a tumor suppressive role in cervical cancer, hepatocellular carcinoma, and glioblastoma, while miR-1285 inhibits p53 and p21 expression by targeting the 3′UTR of p53 transcript [49–52].
To identify differentially expressed miRNAs in our dataset, we employed the same approach used to identify differential copy number alterations by assessing the expression of 254 miRNAs in 64 lung cancer cell lines. We identified 134 unique miRNAs significantly different at the expression level between resistant and sensitive lines in at least one drug. Of which, 40% overexpressed in highly resistant and 60% overexpressed in highly sensitive lung cancer cell lines. Of these 134 miRNAs, miR-625, about which little is known regarding function, was the most frequently differentially expressed. It was significantly differentially expressed in the analyses of agents Paclitaxel, HKI-272, Gö-6976, Erlotinib, Rapamycin, PHA665752, and MG-132. In terms of the drug comparisons with the highest number of differentially expressed miRNAs, the comparison between LCCLs highly sensitive and resistant to HKI-272, an irreversible tyrosine kinase inhibitor of HER2, revealed 30 differentially expressed miRNAs.
Previous studies of miRNA deregulation with respect to response of some of the drugs used in our analyses have identified a number of miRNAs whose expression correlates with drug sensitivity. For example, underexpression of miR-34a and overexpression of miR-125b, 2-21, -222 , and -923 confer Paclitaxel resistance in prostate cancer [53] and breast cancer [54], respectively, while for hepatocellular carcinomas expression of let-7c [55], miR-122 [56] and miR-193b [57] confer sensitivity to Sorafenib. Notably, Sorafenib is a multikinase inhibitor with highest potency for RAF; this is consistent with our findings that link mir-193b with resistance to the RAF inhibitor AZ628. In addition, underexpression of miR-130a and -126 was correlated with resistance to Paclitaxel [58] and Imatinib [59], respectively. From our analyses, we observed miR-130 and -126 to be overexpressed in lung cancer cell lines sensitive to Paclitaxel and Imatinib.
Cancer genomes are characterized by widespread genetic aberrations including high-level amplifications, deletions, DNA methylation, mutations, and chromosomal rearrangements. Within the hundreds of alterations in a cancers genome, only a small subset of these alterations drive tumor initiation and progression and DNA alterations with corresponding expression alterations are more likely to contribute to tumorigenesis [60, 61]. To identify miRNAs likely implicated in drug resistance, we integrated the 266 miRNAs that were significantly different at the copy number level and the 134 miRNAs that were significantly different at the expression level and subsequently filtered for those miRNAs that were differentially expressed and altered in the same drug. Our analysis identified four miRNAs, miR-10b, -193b, -328, and -628, that met these criteria. While the overlap of significant miRNAs in the same drug is minimal, stringent selection criteria such as P ≤ .05 for both copy number and expression alterations, and limited miRNA expression data, likely contributed to the small number of overlapping miRNAs. Importantly, many of the miRNAs most frequently differentially altered at the copy number level (128 of 266, 48.1%) were not represented on the microarray platform. Moreover, when we factored in our expression criteria of expression in at least four cell lines, the number of miRNAs with expression profiles and significantly different copy number alterations was reduced to 66. The copy number profiles of these miRNAs suggest they may play an important role in drug resistance, dictating the importance and need to assess these uninvestigated miRNAs at the expression level.
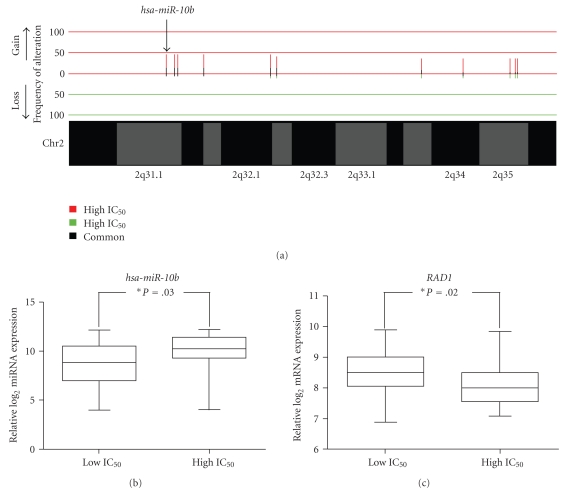
The observation that miR-10b is differentially gained and overexpressed in resistant cell lines treated with MG-132 is consistent with previous findings (Figures 5(a) and 5(b)). miR-10b is an oncomir whose overexpression has been identified in a variety of cancers [62–67]. Specifically, overexpression of miR-10b has been demonstrated to promote the development of metastatic disease in breast cancer and correlate with clinical breast cancer progression, poor overall survival in gastric cancer, and higher grades of malignant glioma. It was also found to be an effective therapeutic target by using antagomirs to reduce expression of HOXD10, subsequently suppressing breast cancer metastasis [62, 64, 66, 68]. Bioinformatic and gene expression analysis of mRNA targets of miR-10b revealed 32 of 636 target genes that were underexpressed in highly resistant cell lines, which have high expression of miR-10b. Amongst the identified genes was RAD1 (Figure 5(c)). RAD1 is part of a complex of proteins known as the 9-1-1 complex, which functions as a heterotrimeric cell cycle checkpoint [69]. The complex, which functions in DNA repair, is recruited to the site of DNA damage or incomplete replication where it recruits DNA polymerases and DNA repair enzymes. RAD1 has been shown to be important in preventing tumor development in response to DNA damage in mice, whereas deletion of RAD1 greatly increased the susceptibility for skin tumor development [70]. In addition, RAD1 is an important component of nucleotide excision repair (NER) which can have drastic effects on chemotherapy drug response. In drugs that instill double stranded DNA breaks, such as the platinum based treatment Cisplatin, upregulation of NER increases drug resistance while in certain non-DNA damage based chemotherapies, downregulation of NER has been shown to increase resistance [71, 72]. In NSCLC patients that have low expression of ERCC1, a gene also involved with NER, have decreased survival when compared to patients with high ERCC1 expression [73], and in both murine and human cells, low XPC expression, another gene involved in NER, correlated with resistance to the Doxorubicin derivative, Nemorubicin [72]. Intriguingly, one of the overrepresented functions identified by Ingenuity Pathway Analysis of the 32 differentially expressed target genes was DNA Replication, Recombination, and Repair (Figure 6).
Figure 5.
Example of a miRNA showing differentially copy number alteration, differential miRNA expression, and differential target gene expression. (a) Copy number alteration comparison between cell lines which are highly resistant and sensitive to agent MG-132 revealed that the hsa-miR-10b locus, on chromosomal region 2q31.1, is more frequently gained in the highly resistant cell lines (P < .05, Fisher's exact test). (b) miRNA expression analysis of miR-10b shows that expression is significantly higher in highly resistant cell lines as compared to sensitive cell lines to MG-132 (P = .03, Mann Whitney U test). (c) mRNA expression analysis of RAD1, a gene identified by bioinformatics prediction analysis as a putative target of miR-10b, shows anticorrelative expression to miR-10b expression. Specifically, decreased expression of RAD1 in highly resistant cell lines to MG-132 relative to highly sensitive lines is observed.
Figure 6.
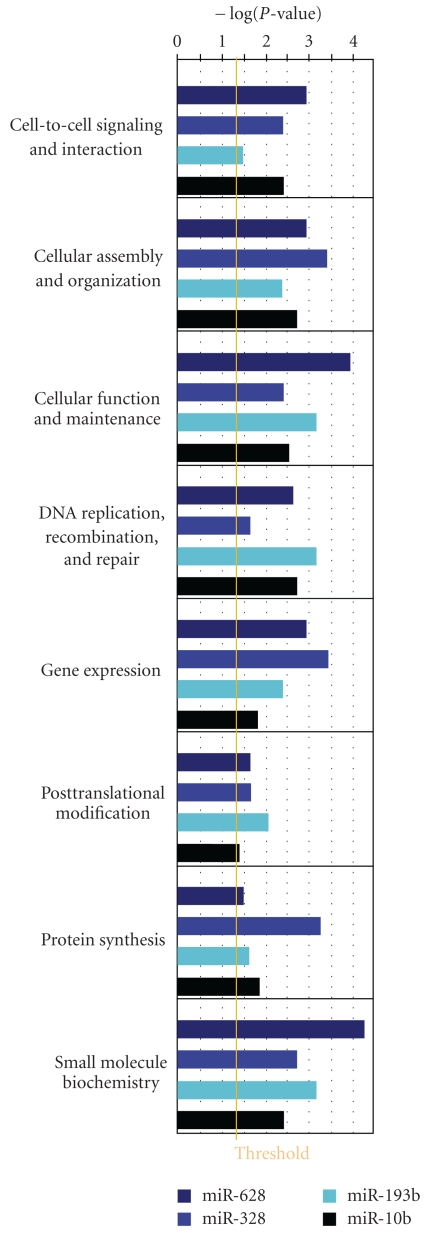
Overrepresented cellular and molecular functions that are common to all four sets of differentially expressed target genes. A “Core Analysis Comparison” was performed using Ingenuity Pathway Analysis and within the Biological Functions, only functions within Molecular and Cellular Functions were assessed. In total, eight of these functions were significant in all four sets. The orange threshold line corresponds to a P-value of .05.
Expression patterns of miR-193b in human cancers, unlike miR-10b, are largely variable. High expression of miR-193b is frequently observed in head and neck squamous cell carcinomas and is associated with a high risk of metastatic disease in uveal melanoma [74, 75]. Conversely, in other cancer types, overexpression of miR-193b has elicited increased tumor suppression as well as sensitivity to specific chemotherapeutics [57, 76, 77]. Moreover, conflicting results have also been observed within a given cancer type. In malignant cutaneous melanoma, overexpression of miR-193b predicts disease outcome and is associated with poor survival, while induced overexpression in cell lines repressed proliferation through the downregulation of Cyclin D1 [78, 79]. Subsequent gene expression analysis of target mRNAs of miR-193b revealed 10 genes that were underexpressed in cell lines highly resistant to AZ628and 37 genes that were underexpressed in cell lines highly resistant to MK-0457. One of the target genes that was also differentially expressed was IKAROS family zinc finger 1 (IKZF1). This transcription regulating gene functions through associations with complexes that are both histone deacetylase (HDAC)-dependent and HDAC-independent [80]. Previous studies have shown that nonhigh-risk ALL9 patients with IKZF1 deletions show a 12-fold higher rate of relapse compared to patients without IKZF1 deletions and IKZF1 deletion has also been implicated in tyrosine kinase inhibitor (TKI) resistance and disease progression in patients with chronic phase- (CP-) chronic myeloid leukemia (CML) [81, 82]. Overexpression of an Isoform of IKZF1 lacking a DNA binding domain, IK6, in acute lymphoblastic leukemia (ALL) patients with the Philadelphia chromosome has also been associated with TKI resistance [83]. Interestingly, MK-0457 is a small molecule inhibitor chemotherapy drug that targets aurora kinase. Underexpression of IKZF1 as a result of miR-193b targeted degradation may increase resistance to MK-0457 in a similar mechanism to TKI resistance.
Similar to miR-193b, evidence supporting the role of miR-328 in cancer is also unclear. In lung adenocarcinoma, miR-328 has been shown to be overexpressed in tumor tissue relative to matched nonmalignant tissue regardless of EGFR or KRAS mutation status [28]. However, in other cancer types, miR-328 underexpression, for example, enables drug resistance through the upregulation of ABCG2 and correlates with cancer progression [84–86]. Our analyses revealed miR-328 to be gained and overexpressed in lung cancer cell lines resistant to Geldanamycin, an antibody against HSP90. Target and gene expression analysis of miR-328 identified 31 genes underexpressed in cell lines highly resistant to Geldanamycin, with one of the targets being the Vitamin D receptor (VDR). VDR and its downstream components, have been previously shown to have antiproliferative effects in a wide variety of cancer types. The anticancer effects of VDR signaling are mostly mediated through its active metabolite, 1,25-dihydroxyvitamin D (calcitriol), which has been shown to exhibit anti-inflammatory effects as well as the suppression of tumor angiogenesis, invasion, and metastasis [87, 88]. Expression of VDR has also been shown to be associated with increased survival in breast, colorectal cancer, and cholangiocarcinoma. It has been recently shown that nuclear VDR status may be a prognostic marker of improved survival in patients with NSCLC [88]. Another intriguing finding for miR-328-associated mRNAs is the implication of both H(+)-monocarboxylate cotransporter (MCT) proteins 1 and 4 (SLC16A1/MCT1 and SLC16A3/MCT4). MCT1 and 4 are involved in lactate uptake and pH balance. Inhibition of MCT1 in tumors can shift aerobic cancer cells from oxidative phosphorylation (lactate metabolism) to glycolysis (glucose), resulting in the death of hypoxic tumor cells due to glucose deprivation [89].
Relatively little has been reported with regard to the role of miR-628. A recent study revealed that miR-628 was expressed in neuroblastomas with favorable prognosis, while those with unfavorable prognosis were devoid of expression, suggesting a tumor suppressive role in this type of cancer [90]. From our analyses, we identified miR-628 to be gained and overexpressed in resistant lung cancer cell lines treated with agent PF-2341066, a MET and ALK kinase inhibitor which has recently shown tremendous efficacy in a subset of lung cancer patients [7]. While the direction of expression contradicts the findings in neuroblastoma, miRNA tissue specificity may play a role in differential expression patterns. Regardless, further analysis of miR-628 is required to better elucidate its role in human cancers. Target and gene expression analysis of miR-628 revealed 22 genes which were underexpressed in cell lines highly resistant to PF-2341066, with one of these differentially expressed being caspase 3 (CASP3). CASP3 is a gene involved in the caspase apoptosis cascade by activating caspases 6, 7, and 9 through cleavage [91]. Moreover, it is also used as a general indicator of cell death and apoptosis. Notably, PF-2341066, which functions as a TKI inhibitor, was found to induce the caspase cell death cascade in vitro through increased levels of CASP3 [92]. Thus, CASP3 downregulation, as a result of miR-628 targeting, may play a significant role in resistance to PF-2341066.
While all involved in response to different drugs, the targets of these miRNAs share certain biological functions. Figure 6 illustrates the functions in which the targets of all four miRNAs participate at a statistically significant level. Broadly, if roles such as cellular maintenance and DNA repair were compromised, such cell populations could develop tolerance to the accumulation of mutations, some of which could dictate resistance. Participation in small molecule biochemistry has implications in the alteration of how these administered drugs are processed. Cellular organization and cell-to-cell signaling, if altered, could confer a more invasive phenotype, contributing to drug resistance.
5. Conclusions
In conclusion, we have demonstrated our method of integrative analysis of multiple dimensions of data including genome-wide miRNA copy number, miRNA expression, mRNA expression, and drug sensitivity data, all available in the public domain, can be a powerful tool to identify miRNAs and genes involved in drug sensitivity. Through these initial analyses, we have identified miRNAs that may have a role in conferring chemoresistance to a number of drugs. Further in vitro and in vivo analyses of the miRNAs and their respective mRNA targets will be necessary to confirm the findings from this study. In addition, given that nearly half of the miRNAs that were differentially altered were not even represented on the miRNA platform analyzed, evaluation of these miRNAs may prove fruitful when new data becomes available. It should also be noted that miRNA target prediction approaches and algorithms are constantly evolving and increasing number of miRNA-mRNA interactions being experimentally validated, potentially revealing important target genes that are not currently implicated. Finally, since the MGH/Sanger collaboration aims to generate drug response profiles for a large number of chemotherapeutics in over 1000 cancer cell lines, as more data becomes available, our approach could identify candidate miRNAs that are associated with multiple drugs which have similar mechanisms of action. Moreover, our strategy could also be repeated in a more specific and clinically relevant manner, which could ultimately lead to the identification of prognostic biomarkers and therapeutic indicators for better disease management and patient outcome.
Supplementary Material
Supplemental Table 1 - List of 18 drugs analyzed in this study.
Supplemental Table 2 - List of lung cancer cell lines with either SNP 6.0 copy number, miRNA expression, or IC50 response.
Supplemental Table 3 - Summary of copy number alteration comparisons for all miRNAs across all drugs.
Supplemental Table 4 - miRNAs significantly differentially expressed across 18 drugs.
Supplemental Table 5 - mRNA targets found by TargetScan and TargetSpy for the four identified miRNAs.
Acknowledgments
The authors wish to thank Kelsie Thu for assistance in data processing. This work was supported by funds from the Canadian Institutes for Health Research (MOP 86731, MOP 77903), Canadian Cancer Society (CCS20485), and the United States Department of Defense Congressionally Directed Medical Research Program - Lung Cancer Research Program (LC090634P2). C. E. Alvarez was supported by the United States National Institutes of Health (HG004663). Declaration required by the data source: SNP 6.0 cell line data were downloaded from the Wellcome Trust Sanger Institute CGP Data Archive (http://www.sanger.ac.uk/genetics/CGP/Archive/). Authors declare that those carried out the original analysis and collection of the data bear no responsibility for the further analysis or interpretation of it by the authors. Contributed equally to this work. K. S. S. Enfield, G. L. Stewart, and L. A. Pikor.
References
- 1.Jemal A, Siegel R, Xu J, Ward E. Cancer statistics, 2010. CA: Cancer Journal for Clinicians. 2010;60(5):277–300. doi: 10.3322/caac.20073. [DOI] [PubMed] [Google Scholar]
- 2.Wakelee H, Belani CP. Optimizing first-line treatment options for patients with advanced NSCLC. Oncologist. 2005;10(3):1–10. doi: 10.1634/theoncologist.10-90003-1. [DOI] [PubMed] [Google Scholar]
- 3.Scagliotti G, Hanna N, Fossella F, et al. The differential efficacy of pemetrexed according to NSCLC histology: a review of two phase III studies. Oncologist. 2009;14(3):253–263. doi: 10.1634/theoncologist.2008-0232. [DOI] [PubMed] [Google Scholar]
- 4.Fukuoka M, Yano S, Giaccone G, et al. Multi-institutional randomized phase II trial of gefitinib for previously treated patients with advanced non-small-cell lung cancer. Journal of Clinical Oncology. 2003;21(12):2237–2246. doi: 10.1200/JCO.2003.10.038. [DOI] [PubMed] [Google Scholar]
- 5.Mok TS, Wu YL, Thongprasert S, et al. Gefitinib or carboplatin-paclitaxel in pulmonary adenocarcinoma. The New England Journal of Medicine. 2009;361(10):947–957. doi: 10.1056/NEJMoa0810699. [DOI] [PubMed] [Google Scholar]
- 6.Sos ML, Michel K, Zander T, et al. Predicting drug susceptibility of non-small cell lung cancers based on genetic lesions. Journal of Clinical Investigation. 2009;119(6):1727–1740. doi: 10.1172/JCI37127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kwak EL, Bang Y-J, Camidge DR, et al. Anaplastic lymphoma kinase inhibition in non-small-cell lung cancer. The New England Journal of Medicine. 2010;363(18):1693–1703. doi: 10.1056/NEJMoa1006448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Buys TPH, Chari R, Lee EHL, et al. Genetic changes in the evolution of multidrug resistance for cultured human ovarian cancer cells. Genes Chromosomes and Cancer. 2007;46(12):1069–1079. doi: 10.1002/gcc.20492. [DOI] [PubMed] [Google Scholar]
- 9.Etemadmoghadam D, Defazio A, Beroukhim R, et al. Integrated genome-wide DMA copy number and expression analysis identifies distinct mechanisms of primary chemoresistance in ovarian carcinomas. Clinical Cancer Research. 2009;15(4):1417–1427. doi: 10.1158/1078-0432.CCR-08-1564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kobayashi S, Boggon TJ, Dayaram T, et al. EGFR mutation and resistance of non-small-cell lung cancer to gefitinib. The New England Journal of Medicine. 2005;352(8):786–792. doi: 10.1056/NEJMoa044238. [DOI] [PubMed] [Google Scholar]
- 11.Pao W, Miller VA, Politi KA, et al. Acquired resistance of lung adenocarcinomas to gefitinib or erlotinib is associated with a second mutation in the EGFR kinase domain. PLoS Medicine. 2005;2(3, article e73) doi: 10.1371/journal.pmed.0020073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bianco R, Shin I, Ritter CA, et al. Loss of PTEN/MMAC1/TEP in EGF receptor-expressing tumor cells counteracts the antitumor action of EGFR tyrosine kinase inhibitors. Oncogene. 2003;22(18):2812–2822. doi: 10.1038/sj.onc.1206388. [DOI] [PubMed] [Google Scholar]
- 13.Kirschner K, Melton DW. Multiple roles of the ERCC1-XPF endonuclease in DNA repair and resistance to anticancer drugs. Anticancer Research. 2010;30(9):3223–3232. [PubMed] [Google Scholar]
- 14.Rosell R, Felip E, Taron M, et al. Gene expression as a predictive marker of outcome in stage IIB-IIIA-IIIB non-small cell lung cancer after induction gemcitabine-based chemotherapy followed by resectional surgery. Clinical Cancer Research. 2004;10(12, part 2):4215s–4219s. doi: 10.1158/1078-0432.CCR-040006. [DOI] [PubMed] [Google Scholar]
- 15.Ferracin M, Veronese A, Negrini M. Micromarkers: MiRNAs in cancer diagnosis and prognosis. Expert Review of Molecular Diagnostics. 2010;10(3):297–308. doi: 10.1586/erm.10.11. [DOI] [PubMed] [Google Scholar]
- 16.Garofalo M, Condorelli G, Croce CM. MicroRNAs in diseases and drug response. Current Opinion in Pharmacology. 2008;8(5):661–667. doi: 10.1016/j.coph.2008.06.005. [DOI] [PubMed] [Google Scholar]
- 17.Hummel R, Hussey DJ, Haier J. MicroRNAs: predictors and modifiers of chemo- and radiotherapy in different tumour types. European Journal of Cancer. 2010;46(2):298–311. doi: 10.1016/j.ejca.2009.10.027. [DOI] [PubMed] [Google Scholar]
- 18.Anglicheau D, Muthukumar T, Suthanthiran M. MicroRNAs: small RNAs with big effects. Transplantation. 2010;90(2):105–112. doi: 10.1097/TP.0b013e3181e913c2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shyu AB, Wilkinson MF, van Hoof A. Messenger RNA regulation: to translate or to degrade. The EMBO Journal. 2008;27(3):471–481. doi: 10.1038/sj.emboj.7601977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Calin GA, Croce CM. MicroRNA signatures in human cancers. Nature Reviews Cancer. 2006;6(11):857–866. doi: 10.1038/nrc1997. [DOI] [PubMed] [Google Scholar]
- 21.Garzon R, Calin GA, Croce CM. MicroRNAs in cancer. Annual Review of Medicine. 2009;60:167–179. doi: 10.1146/annurev.med.59.053006.104707. [DOI] [PubMed] [Google Scholar]
- 22.Iorio MV, Croce CM. MicroRNAs in cancer: small molecules with a huge impact. Journal of Clinical Oncology. 2009;27(34):5848–5856. doi: 10.1200/JCO.2009.24.0317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mallick R, Patnaik S, Yendamuri S. MicroRNAs and lung cancer: biology and applications in diagnosis and prognosis. Journal of Carcinogenesis. 2010;9, article 8 doi: 10.4103/1477-3163.67074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang L, Huang J, Yang N, et al. microRNAs exhibit high frequency genomic alterations in human cancer. Proceedings of the National Academy of Sciences of the United States of America. 2006;103(24):9136–9141. doi: 10.1073/pnas.0508889103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Calin GA, Sevignani C, Dumitru CD, et al. Human microRNA genes are frequently located at fragile sites and genomic regions involved in cancers. Proceedings of the National Academy of Sciences of the United States of America. 2004;101(9):2999–3004. doi: 10.1073/pnas.0307323101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Starczynowski DT, Morin R, McPherson A, et al. Genome-wide identification of human microRNAs located in leukemia-associated genomic alterations. Blood. 2011;117(2):595–607. doi: 10.1182/blood-2010-03-277012. [DOI] [PubMed] [Google Scholar]
- 27.Starczynowski DT, Kuchenbauer F, Argiropoulos B, et al. Identification of miR-145 and miR-146a as mediators of the 5q-syndrome phenotype. Nature Medicine. 2010;16(1):49–58. doi: 10.1038/nm.2054. [DOI] [PubMed] [Google Scholar]
- 28.Dacic S, Kelly L, Shuai Y, Nikiforova MN. MiRNA expression profiling of lung adenocarcinomas: correlation with mutational status. Modern Pathology. 2010;23(12):1577–1582. doi: 10.1038/modpathol.2010.152. [DOI] [PubMed] [Google Scholar]
- 29.Wu XM, Xiao HS. Mirnas modulate the drug response of tumor cells. Science in China, Series C. 2009;52(9):797–801. doi: 10.1007/s11427-009-0114-4. [DOI] [PubMed] [Google Scholar]
- 30.Chuang JC, Jones PA. Epigenetics and microRNAs. Pediatric Research. 2007;61(5, part 2):24R–29R. doi: 10.1203/pdr.0b013e3180457684. [DOI] [PubMed] [Google Scholar]
- 31.Lu J, Getz G, Miska EA, et al. MicroRNA expression profiles classify human cancers. Nature. 2005;435(7043):834–838. doi: 10.1038/nature03702. [DOI] [PubMed] [Google Scholar]
- 32.Galluzzi L, Morselli E, Vitale I, et al. miR-181a and miR-630 regulate cisplatin-induced cancer cell death. Cancer Research. 2010;70(5):1793–1803. doi: 10.1158/0008-5472.CAN-09-3112. [DOI] [PubMed] [Google Scholar]
- 33.Weiss GJ, Bemis LT, Nakajima E, et al. EGFR regulation by microRNA in lung cancer: correlation with clinical response and survival to gefitinib and EGFR expression in cell lines. Annals of Oncology. 2008;19(6):1053–1059. doi: 10.1093/annonc/mdn006. [DOI] [PubMed] [Google Scholar]
- 34.Cho WC. MicroRNAs as therapeutic targets for lung cancer. Expert Opinion on Therapeutic Targets. 2010;14(10):1005–1008. doi: 10.1517/14728222.2010.522399. [DOI] [PubMed] [Google Scholar]
- 35.Oh JS, Kim JJ, Byun JY, Kim IA. Lin28-let7 modulates radiosensitivity of human cancer cells with activation of K-Ras. International Journal of Radiation Oncology Biology Physics. 2010;76(1):5–8. doi: 10.1016/j.ijrobp.2009.08.028. [DOI] [PubMed] [Google Scholar]
- 36.Chari R, Coe BP, Wedseltoft C, et al. SIGMA: a system for the integrative genomic multi-dimensional analysis of cancer genomes, epigenomes, and transcriptomes. BMC Bioinformatics. 2008;9, article 422 doi: 10.1186/1471-2105-9-422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rhead B, Karolchik D, Kuhn RM, et al. The UCSC genome browser database: update 2010. Nucleic Acids Research. 2009;38(1, database issue):D613–D619. doi: 10.1093/nar/gkp939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Irizarry RA, Hobbs B, Collin F, et al. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics. 2003;4(2):249–264. doi: 10.1093/biostatistics/4.2.249. [DOI] [PubMed] [Google Scholar]
- 39.Gautier L, Cope L, Bolstad BM, Irizarry RA. Affy—analysis of Affymetrix GeneChip data at the probe level. Bioinformatics. 2004;20(3):307–315. doi: 10.1093/bioinformatics/btg405. [DOI] [PubMed] [Google Scholar]
- 40.Gentleman RC, Carey VJ, Bates DM, et al. Bioconductor: open software development for computational biology and bioinformatics. Genome Biology. 2004;5(10):p. R80. doi: 10.1186/gb-2004-5-10-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sturm M, Hackenberg M, Langenberger D, Frishman D. TargetSpy: a supervised machine learning approach for microRNA target prediction. BMC Bioinformatics. 2010;11, article 292 doi: 10.1186/1471-2105-11-292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120(1):15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- 43.Grimson A, Farh KKH, Johnston WK, Garrett-Engele P, Lim LP, Bartel DP. MicroRNA targeting specificity in mammals: determinants beyond seed pairing. Molecular Cell. 2007;27(1):91–105. doi: 10.1016/j.molcel.2007.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Friedman RC, Farh KKH, Burge CB, Bartel DP. Most mammalian mRNAs are conserved targets of microRNAs. Genome Research. 2009;19(1):92–105. doi: 10.1101/gr.082701.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ma J, Dong C, Ji C. MicroRNA and drug resistance. Cancer Gene Therapy. 2010;17(8):523–531. doi: 10.1038/cgt.2010.18. [DOI] [PubMed] [Google Scholar]
- 46.Zheng T, Wang J, Chen X, Liu L. Role of microRNA in anticancer drug resistance. International Journal of Cancer. 2010;126(1):2–10. doi: 10.1002/ijc.24782. [DOI] [PubMed] [Google Scholar]
- 47.DeBoer C, Meulman PA, Wnuk RJ, Peterson DH. Geldanamycin, a new antibiotic. Journal of Antibiotics. 1970;23(9):442–447. doi: 10.7164/antibiotics.23.442. [DOI] [PubMed] [Google Scholar]
- 48.Maes OC, An J, Sarojini H, Wu H, Wang E. Changes in microRNA expression patterns in human fibroblasts after low-LET radiation. Journal of Cellular Biochemistry. 2008;105(3):824–834. doi: 10.1002/jcb.21878. [DOI] [PubMed] [Google Scholar]
- 49.Wilting SM, van Boerdonk RAA, Henken FE, et al. Methylation-mediated silencing and tumour suppressive function of hsa-miR-124 in cervical cancer. Molecular Cancer. 2010;9:p. 167. doi: 10.1186/1476-4598-9-167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Silber J, Lim DA, Petritsch C, et al. miR-124 and miR-137 inhibit proliferation of glioblastoma multiforme cells and induce differentiation of brain tumor stem cells. BMC Medicine. 2008;6, article 14 doi: 10.1186/1741-7015-6-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Furuta M, Kozaki KI, Tanaka S, Arii S, Imoto I, Inazawa J. miR-124 and miR-203 are epigenetically silenced tumor-suppressive microRNAs in hepatocellular carcinoma. Carcinogenesis. 2009;31(5):766–776. doi: 10.1093/carcin/bgp250. [DOI] [PubMed] [Google Scholar]
- 52.Tian S, Huang S, Wu S, Guo W, Li J, He X. MicroRNA-1285 inhibits the expression of p53 by directly targeting its 3′ untranslated region. Biochemical and Biophysical Research Communications. 2010;396(2):435–439. doi: 10.1016/j.bbrc.2010.04.112. [DOI] [PubMed] [Google Scholar]
- 53.Kojima K, Fujita Y, Nozawa Y, Deguchi T, Ito M. MiR-34a attenuates paclitaxel-resistance of hormone-refractory prostate cancer PC3 cells through direct and indirect mechanisms. Prostate. 2010;70(14):1501–1512. doi: 10.1002/pros.21185. [DOI] [PubMed] [Google Scholar]
- 54.Zhou M, Liu Z, Zhao Y, et al. MicroRNA-125b confers the resistance of breast cancer cells to paclitaxel through suppression of pro-apoptotic Bcl-2 antagonist killer 1 (Bak1) expression. Journal of Biological Chemistry. 2010;285(28):21496–21507. doi: 10.1074/jbc.M109.083337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Shimizu S, Takehara T, Hikita H, et al. The let-7 family of microRNAs inhibits Bcl-xL expression and potentiates sorafenib-induced apoptosis in human hepatocellular carcinoma. Journal of Hepatology. 2010;52(5):698–704. doi: 10.1016/j.jhep.2009.12.024. [DOI] [PubMed] [Google Scholar]
- 56.Bai S, Nasser MW, Wang B, et al. MicroRNA-122 inhibits tumorigenic properties of hepatocellular carcinoma cells and sensitizes these cells to sorafenib. Journal of Biological Chemistry. 2009;284(46):32015–32027. doi: 10.1074/jbc.M109.016774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Braconi C, Valeri N, Gasparini P, et al. Hepatitis C virus proteins modulate microRNA expression and chemosensitivity in malignant hepatocytes. Clinical Cancer Research. 2010;16(3):957–966. doi: 10.1158/1078-0432.CCR-09-2123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sorrentino A, Liu CG, Addario A, Peschle C, Scambia G, Ferlini C. Role of microRNAs in drug-resistant ovarian cancer cells. Gynecologic Oncology. 2008;111(3):478–486. doi: 10.1016/j.ygyno.2008.08.017. [DOI] [PubMed] [Google Scholar]
- 59.San José-Enériz E, Román-Gómez J, Jiménez-Velasco A, et al. MicroRNA expression profiling in Imatinib-resistant Chronic Myeloid Leukemia patients without clinically significant ABL1-mutations. Molecular Cancer. 2009;8, article 1476:p. 69. doi: 10.1186/1476-4598-8-69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Coe BP, Chari R, Lockwood WW, Lam WL. Evolving strategies for global gene expression analysis of cancer. Journal of Cellular Physiology. 2008;217(3):590–597. doi: 10.1002/jcp.21554. [DOI] [PubMed] [Google Scholar]
- 61.Luo B, Hiu WC, Subramanian A, et al. Highly parallel identification of essential genes in cancer cells. Proceedings of the National Academy of Sciences of the United States of America. 2008;105(51):20380–20385. doi: 10.1073/pnas.0810485105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ma L, Teruya-Feldstein J, Weinberg RA. Tumour invasion and metastasis initiated by microRNA-10b in breast cancer. Nature. 2007;449(7163):682–688. doi: 10.1038/nature06174. [DOI] [PubMed] [Google Scholar]
- 63.Jukic DM, Rao UNM, Kelly L, et al. Microrna profiling analysis of differences between the melanoma of young adults and older adults. Journal of Translational Medicine. 2010;8, article 27 doi: 10.1186/1479-5876-8-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Sasayama T, Nishihara M, Kondoh T, Hosoda K, Kohmura E. MicroRNA-10b is overexpressed in malignant glioma and associated with tumor invasive factors, uPAR and RhoC. International Journal of Cancer. 2009;125(6):1407–1413. doi: 10.1002/ijc.24522. [DOI] [PubMed] [Google Scholar]
- 65.Dahiya N, Sherman-Baust CA, Wang TL, et al. MicroRNA expression and identification of putative miRNA targets in ovarian cancer. PLoS One. 2008;3(6) doi: 10.1371/journal.pone.0002436. Article ID e2436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Li X, Zhang Y, Zhang Y, Ding J, Wu K, Fan D. Survival prediction of gastric cancer by a seven-microRNA signature. Gut. 2010;59(5):579–585. doi: 10.1136/gut.2008.175497. [DOI] [PubMed] [Google Scholar]
- 67.Prueitt RL, Yi M, Hudson RS, et al. Expression of microRNAs and protein-coding genes associated with perineural invasion in prostate cancer. Prostate. 2008;68(11):1152–1164. doi: 10.1002/pros.20786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Ma L, Reinhardt F, Pan E, et al. Therapeutic silencing of miR-10b inhibits metastasis in a mouse mammary tumor model. Nature Biotechnology. 2010;28(4):341–347. doi: 10.1038/nbt.1618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Bai H, Madabushi A, Guan X, Lu AL. Interaction between human mismatch repair recognition proteins and checkpoint sensor Rad9-Rad1-Hus1. DNA Repair. 2010;9(5):478–487. doi: 10.1016/j.dnarep.2010.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Han L, Hu Z, Liu Y, et al. Mouse Rad1 deletion enhances susceptibility for skin tumor development. Molecular Cancer. 2010;9, article 67 doi: 10.1186/1476-4598-9-67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Olaussen KA, Dunant A, Fouret P, et al. DNA repair by ERCC1 in non-small-cell lung cancer and cisplatin-based adjuvant chemotherapy. The New England Journal of Medicine. 2006;355(10):983–991. doi: 10.1056/NEJMoa060570. [DOI] [PubMed] [Google Scholar]
- 72.Sabatino MA, Marabese M, Ganzinelli M, Caiola E, Geroni C, Broggini M. Down-regulation of the nucleotide excision repair gene XPG as a new mechanism of drug resistance in human and murine cancer cells. Molecular Cancer. 2010;9, article 259 doi: 10.1186/1476-4598-9-259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Ding Z, Zhang J, Shao J. ERCC1 expression as a predictor of survival after operation in stage I non-small cell lung cancer patients. Zhongguo Fei Ai Za Zhi. 2010;13(5):522–525. doi: 10.3779/j.issn.1009-3419.2010.05.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Avissar M, Christensen BC, Kelsey KT, Marsit CJ. MicroRNA expression ratio is predictive of head and neck squamous cell carcinoma. Clinical Cancer Research. 2009;15(8):2850–2855. doi: 10.1158/1078-0432.CCR-08-3131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Worley LA, Long MD, Onken MD, Harbour JW. Micro-RNAs associated with metastasis in uveal melanoma identified by multiplexed microarray profiling. Melanoma Research. 2008;18(3):184–190. doi: 10.1097/CMR.0b013e3282feeac6. [DOI] [PubMed] [Google Scholar]
- 76.Li XF, Yan PJ, Shao ZM. Downregulation of miR-193b contributes to enhance urokinase-type plasminogen activator (uPA) expression and tumor progression and invasion in human breast cancer. Oncogene. 2009;28(44):3937–3948. doi: 10.1038/onc.2009.245. [DOI] [PubMed] [Google Scholar]
- 77.Rauhala HE, Jalava SE, Isotalo J, et al. miR-193b is an epigenetically regulated putative tumor suppressor in prostate cancer. International Journal of Cancer. 2010;127(6):1363–1372. doi: 10.1002/ijc.25162. [DOI] [PubMed] [Google Scholar]
- 78.Caramuta S, Egyházi S, Rodolfo M, et al. MicroRNA expression profiles associated with mutational status and survival in malignant melanoma. Journal of Investigative Dermatology. 2010;130(8):2062–2070. doi: 10.1038/jid.2010.63. [DOI] [PubMed] [Google Scholar]
- 79.Chen J, Feilotter HE, Paré GC, et al. MicroRNA-193b represses cell proliferation and regulates cyclin D1 in melanoma. American Journal of Pathology. 2010;176(5):2520–2529. doi: 10.2353/ajpath.2010.091061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Kim J, Sif S, Jones B, et al. Ikaros DNA-binding proteins direct formation of chromatin remodeling complexes in lymphocytes. Immunity. 1999;10(3):345–355. doi: 10.1016/s1074-7613(00)80034-5. [DOI] [PubMed] [Google Scholar]
- 81.Kuiper RP, Waanders E, van der Velden VHJ, et al. IKZF1 deletions predict relapse in uniformly treated pediatric precursor B-ALL. Leukemia. 2010;24(7):1258–1264. doi: 10.1038/leu.2010.87. [DOI] [PubMed] [Google Scholar]
- 82.Joha S, Dauphin V, Leprêtre F, et al. Genomic characterization of Imatinib resistance in CD34+ cell populations from chronic myeloid leukaemia patients. doi: 10.1016/j.leukres.2010.07.012. Leukemia Research. In press. [DOI] [PubMed] [Google Scholar]
- 83.Lacobucci I, Lonetti A, Messa F, et al. Expression of spliced oncogenic Ikaros isoforms in Philadelphia-positive acute lymphoblastic leukemia patients treated with tyrosine kinase inhibitors: implications for a new mechanism of resistance. Blood. 2008;112(9):3847–3855. doi: 10.1182/blood-2007-09-112631. [DOI] [PubMed] [Google Scholar]
- 84.Li W-Q, Li Y-M, Tao B-B, et al. Downregulation of ABCG2 expression in glioblastoma cancer stem cells with miRNA-328 may decrease their chemoresistance. Case Reports and Clinical Practice Review. 2010;16(10):HY27–HY30. [PubMed] [Google Scholar]
- 85.Malzkorn B, Wolter M, Liesenberg F, et al. Identification and functional characterization of microRNAs involved in the malignant progression of gliomas. Brain Pathology. 2010;20(3):539–550. doi: 10.1111/j.1750-3639.2009.00328.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Pan YZ, Morris ME, Yu AM. MicroRNA-328 negatively regulates the expression of breast cancer resistance protein (BCRP/ABCG2) in human cancer cells. Molecular Pharmacology. 2009;75(6):1374–1379. doi: 10.1124/mol.108.054163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Krishnan AV, Feldman D. Mechanisms of the anti-cancer and anti-inflammatory actions of vitamin D. Annual Review of Pharmacology and Toxicology. 2011;51:311–336. doi: 10.1146/annurev-pharmtox-010510-100611. [DOI] [PubMed] [Google Scholar]
- 88.Srinivasan M, Parwani AV, Hershberger PA, Lenzner DE, Weissfeld JL. Nuclear vitamin D receptor expression is associated with improved survival in non-small cell lung cancer. Journal of Steroid Biochemistry and Molecular Biology. 2011;123(1-2):30–36. doi: 10.1016/j.jsbmb.2010.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Sonveaux P, Végran F, Schroeder T, et al. Targeting lactate-fueled respiration selectively kills hypoxic tumor cells in mice. Journal of Clinical Investigation. 2008;118(12):3930–3942. doi: 10.1172/JCI36843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Schulte JH, Marschall T, Martin M, et al. Deep sequencing reveals differential expression of microRNAs in favorable versus unfavorable neuroblastoma. Nucleic Acids Research. 2010;38(17):5919–5928. doi: 10.1093/nar/gkq342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Nicholson DW, Ali A, Thornberry NA, et al. Identification and inhibition of the ICE/CED-3 protease necessary for mammalian apoptosis. Nature. 1995;376(6535):37–43. doi: 10.1038/376037a0. [DOI] [PubMed] [Google Scholar]
- 92.Zou HY, Li Q, Lee JH, et al. An orally available small-molecule inhibitor of c-Met, PF-2341066, exhibits cytoreductive antitumor efficacy through antiproliferative and antiangiogenic mechanisms. Cancer Research. 2007;67(9):4408–4417. doi: 10.1158/0008-5472.CAN-06-4443. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Table 1 - List of 18 drugs analyzed in this study.
Supplemental Table 2 - List of lung cancer cell lines with either SNP 6.0 copy number, miRNA expression, or IC50 response.
Supplemental Table 3 - Summary of copy number alteration comparisons for all miRNAs across all drugs.
Supplemental Table 4 - miRNAs significantly differentially expressed across 18 drugs.
Supplemental Table 5 - mRNA targets found by TargetScan and TargetSpy for the four identified miRNAs.