Abstract
Background
A problem has remained unresolved regarding the exceptions to the unilateral inheritance of chloroplast DNA (cpDNA) from MT+/female in Chlamydomonas and other volvocaleans demonstrated by the previous genetic analyses. For identification of the parental types of cpDNA, these studies used parents that have differences in restriction fragment length polymorphisms and exhibit partial sexual incompatibility.
Methodology/Principal Findings
In the present study, we used sexually compatible parents of the isogamous colonial volvocalean Gonium maiaprilis that seemed an ideal species to identify the pattern of cpDNA inheritance based on the length difference in the putative group I intron interrupted in the Rubisco large subunit gene and objective identification of mating types by the presence or absence of the minus-dominance (MID) gene. We examined patterns of inheritance of cpDNA and presence/absence of a MID ortholog (GmMID) in 107 F1 progeny of G. maiaprilis that were obtained by inducing germination of separated single zygotes. The results demonstrated no exception of the uniparental inheritance of cpDNA from the MT+ parent (lacking GmMID) in sexually compatible or genetically less divergent strains of G. maiaprilis.
Conclusions/Significance
The present data suggest that the uniparental inheritance of cpDNA is likely more strict in crossings of less diverged strains or sexually compatible parental volvocaleans, and some genetic inconsistency between the parents may cause exceptional uniparental inheritance of cpDNA.
Introduction
Chloroplast DNA (cpDNA) in the volvocalean algae is predominantly transmitted from only one of the two parental mating types to the progeny; from mating type plus (MT+) in the isogamous species Chlamydomonas reinhardtii [1] and Gonium pectorale [2] or from female in the oogamous Volvox carteri [3]. However, these studies showed that 2–8% of the F1 progeny have an exceptional pattern of uniparental inheritance of cpDNA (cpDNA) [1]–[3], i.e. they inherit cpDNA from the MT−/male. For identification of the parental types of cpDNA, these studies used strains of complementary mating types (sexes) that have differences in restriction fragment length polymorphisms (RFLPs) and exhibit partial sexual incompatibility [2]–[4].
Studies of intra/interspecific crossings in mouse demonstrated that paternal mitochondrial DNA (mtDNA) is selectively eliminated during early embryogenesis in intraspecific crossings, whereas 50% of paternal mtDNA are transmitted to progeny in interspecifc crossings [5], [6]. Thus, crossings between pairs with partial sexual isolation or between genetically differentiated entities in the volvocaleans may also increase the exceptional rate of uniparental inheritance of organelle DNA when compared with intraspecific crossings.
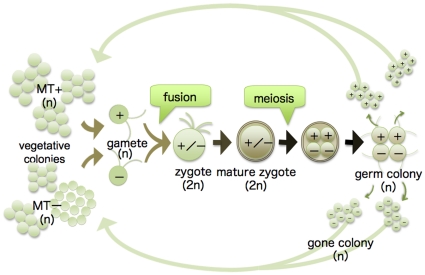
Gonium maiaprilis is an isogamous colonial volvocalean that exhibits heterothallic sexuality [7] (Figure 1). The mating type (MT−)-determining minus dominance gene, MID [8], was recently identified in the closely related species G. pectorale [2]. In addition, our preliminary comparison of cpDNA sequences including a putative group I intron in the Rubisco large subunit (rbcL) genes indicated a difference in length of the introns among the G. maiaprilis strains. Thus, G. maiaprilis seems an ideal species to identify the pattern of cpDNA inheritance based on the difference in the group I intron and objective identification of mating types by the presence or absence of the MID gene [2].
Figure 1. Diagram of sexual reproduction in heterothallic Gonium maiaprilis.
Based on Hayama et al. [7].
In this study, we examined patterns of inheritance of cpDNA in 107 F1 progeny of G. maiaprilis. The results demonstrated no exception of the uniparental inheritance of cpDNA from the MT+ parent in sexually compatible strains of G. maiaprilis.
Results
One hundred and thirty-three gone colonies, each representing a separate meiotic product, were isolated from 44 germinating zygotes of G. maiaprilis Asa041901×Asa041903 to establish F1 strains (Figure 1). Ultimately, 77% (103/133) of the gone colonies became actively growing cultures. Based on backcrossing, 58 of the 103 exhibit a minus mating phenotype and the remaining 45 a plus mating phenotype (Table 1).
Table 1. Mating phenotypes, presence/absence of GmMID and inheritance of cpDNA in F1 progeny of G. maiaprilis Asa041901×Asa041903.
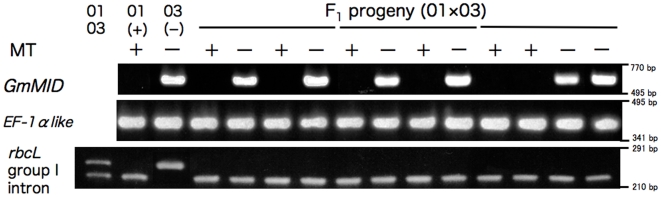
To determine the presence or absence of the MID gene, the MID orthologue (GmMID) was isolated from G. maiaprilis and characterized (Figures S1, S2, S3). Genomic PCR using GmMID-specific primers demonstrated that all 60 F1 strains (including additional two F1 strains previously established [7]) with minus mating phenotype have GmMID whereas all 47 F1 strains (including additional two F1 strains previously established [7]) with plus mating phenotype lack this gene (Figures 2 and S4). On the other hand, all 107 F1 strains had cpDNA of the Asa041901 (MT+) type based on genomic PCR using rbcL group I intron-specific primers (Figures 2, 3, S4 and Table 1).
Figure 2. Mating phenotypes (MT) and results of genomic PCR for parental strains (Asa041901[01] and Asa041903 [03]) and 12 representative F1 strains of Gonium maiaprilis.
Presence/absence of GmMID and the length polymorphism within the cpDNArbcL group I intron are assessed by gel electrophoreses. The nuclear gene EF-1alpha like serves as a control. The horizontal line over the F1 progeny indicates F1 strains originating from the same zygote.
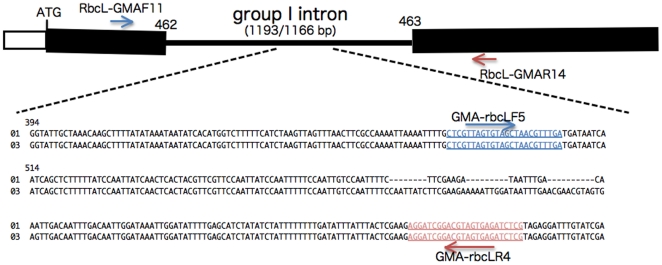
Figure 3. Diagram showing intron/exon structure and positions of specific primers (Table S2) in the rbcL genes from Gonium maiaprilis Asa041901 (01) and Asa041903 (03) (GenBank/EMBL/DDBJ accession nos. AB520743 and AB520744).
Thick bars represent exons interrupted by a putative group I intron between basepairs 462 and 463 of the sequence of the rbcL gene of Chlorella vulgaris [accession no. AB001684]. Numbers above the alignment indicate the nucleotide position within the intron.
The secondary structures of the nuclear ribosomal DNA internal transcribed spaces 1 and 2 (ITS-1 and ITS-2) contain single base substitutions in four positions between G. maiaprilis Asa041901 and Asa041903 (Figure S5). These substitutions did not correspond to compensatory base change (CBC), suggesting that the strains fall within a range of an interfertile entity or a biological species [7], [9]. In G. pectorale Mongolia1 and Mongolia4, seven single base substitutions were detected in the ITS secondary structures (Table 2) although no CBC was recognized (Figure S6). Furthermore, the nucleotide sequences of the rbcL coding region (1128 bp) of the G. maiaprilis parents are exactly the same (GenBank/EMBL/DDBJ accession nos. AB520743-5, [7]) whereas one nucleotide substitution is present between the parents of G. pectorale (Table 2).
Table 2. Comparison of Gonium maiaprilis and G. pectorale crosses.
| Source of zygotes | No. of nt change in ITSa (rbcL b) | No. of F1 strains examined | Survival rate of F1 strains | cpDNA from MT+ parent | cpDNA from MT− parent | Percentage of exceptions | Reference | |
| Mating type + | Mating type − | |||||||
| G. pectorale Mongolia1 | G. pectorale Mongolia4 | 7 (1) | 78 | Poor | 73 | 5 | 6.4% | Hamaji et al. [2] |
| G. maiaprilis Asa041901 | G. maiaprilis Asa041903 | 4 (0) | 107 | 78% | 107 | 0 | 0% | This paper |
Total number in ITS-1 and ITS-2 of nuclear ribosomal DNA (GenBank/EMBL/DDBJ accession nos. AB520746 and AB623040-2) (Figures S5 and S6).
Discussion
In 78 F1 strains of G. pectorale Mongolia1×Mongolia4, five exceptions of the uniparental inheritance of cpDNA were reported [2] (Table 2). In contrast, there were no exceptions of the uniparental inheritance of from the MT+ parent (lacking GmMID) among the 107 G. maiaprilis F1 strains (Table 2 and Figure S4). This difference in the rate of exceptional uniparental inheritance of cpDNA between G. maiaprilis and G. pectorale is significant (P = 0.0014<0.05) by Fisher's exact test [10]. On the other hand, the survival rate of F1 progeny (77%) in G. maiaprilis is high as in intraspecific crossings of other volvocaleans (Table S1) [3], [11], [12]. In G. pectorale Mongolia1×Mongolia4, however, the survival of F1 progeny was poor thus obviating tetrad analysis [2]. In addition, genetic difference between G. maiaprilis Asa041901 and Asa041903 is smaller than that between G. pectorale Mongolia1 and Mongolia4 (Table 2). Therefore, reproductive/genetic isolation between G. maiaprilis Asa041901 and Asa041903 is apparently less than that between G. pectorale Mongolia1 and Mongolia4.
These results suggest that the uniparental inheritance of cpDNA may be more strict in crossings of less diverged strains or sexually compatible parental volvocalean parents, and some genetic inconsistency between the parents may cause exceptional uniparental inheritance of cpDNA. The difference in the rate of exceptional uniparental inheritance of cpDNA (Table 2) could be considered to result from the difference in maturation of zygotes prior to germination between G. maiaprilis and G. pectorale. The zygotes of G. maiaprilis were induce to germinate after six-week dark treatment while immature zygotes were used for germination in G. pectorale Mongolia1×Mongolia4 [2]. However, determination of the uniparental inheritance or complete digestion of cpDNA from MT− occurs in the early stage of zygote formation or quadriflagellate zygotes in Chlamydomonas reinhardtii [13]. Thus, the uniparental inheritance of cpDNA in the volvocaleans may be based on a precision molecular system that requires interactions of alleles from both parental cells, of sex-related genes that may be evolving rapidly [14], although details of the molecular mechanism for uniparental inheritance in the Volvocales remain unresolved [15].
Exceptional cases of the uniparental inheritance of mutations to streptomycin resistance in Chlamydomonas reinhardtii [16], [17] and the colonial volvocalean Eudorina elegans [18] were reported in classic genetic studies. However, these studies are based on crossings of UV-induced mutant strains that might have been affected by additional mutations causing confusion of the consortium of the parental cells for uniparental inheritance of the organelle DNAs.
Materials and Methods
Cultures and induction of sexual reproduction in Gonium maiaprilis
Two G. maiaprilis strains of complementary mating types (Asa041901 and Asa041903) were used in this study. These two strains are available from the Microbial Culture Collection at the National Institute for Environmental Studies (NIES-Collection [19]). The cultures were grown in screw-cap tubes (18×150 mm) containing about 10 mL VTAC or AF-6 medium modified by elimination of CaCO3 and addition of 400 mg L−1 MES [19]–[21]. Cultures were grown at 20°C, on a 14:10 h light-dark cycle, under cool-white fluorescent lamps at 165–175 µmol m−2 s−1 intensity.
For induction of sexual reproduction, approximately 10 ml of a 14-day-old culture in VTAC medium were reduced to 1 mL by centrifugation. The concentrated cultures of the two complementary mating types were mixed in Petri dishes (60-mm diameter) with 5.0 ml mating medium [20]. These dishes were cultured at 25°C on a 14:10 h light-dark cycle, under cool-white fluorescent lamps at 165–175 µmol m−2 s−1 intensity. After 10–14 days under these conditions, zygotes were pipetted onto the surface of AF-6 medium solidified with 1% agar in Petri dishes (90-mm diameter). The dishes were placed in the dark by wrapping in a double layer of aluminum foil and maintained in darkness at 20°C for about 6 weeks, after which the matured, clumped zygotes were separated using the pressure between a cover slip and slide. The separated zygotes were individually isolated and placed in 500 µL AF-6 liquid medium in a glass depression (20-mm diameter) in Petri dishes. In order to avoid evaporation from the medium containing a single zygote, 5 mL water solidified with 1% agar were placed in the bottom of the Petri dishes. The separated zygotes were then grown at 20°C on 14:10 h light-dark cycle. After the zygote had given rise to one to four gone colonies originating from a four-celled germ colon (Figure 1), each gone colony was transferred into a separate tube of AF-6 medium by a micropipette to establish an F1 strain. Mating phenotypes of F1 strains were determined by backcrossing.
Isolation and characterization of GmMID
The cDNA sequence of GmMID (GenBank/EMBL/DDBJ accession no. AB623043)was obtained from Gonium maiaprilis Asa041903 (MT−) as described previously [2], [22]. The genomic sequence (GenBank/EMBL/DDBJ accession no. AB623044) was determined using GmMID-specific primers (Table S2) that were designed based on the cDNA sequence and using the methods described previously [22].
For phylogenetic analysis, 47 amino acids from the RWP-RK domain of GmMID (Figure S1) were aligned with five other MID proteins (Figure S2) and 25 RWP-RK domain-containing sequences from Chlamydomonas reinhardtii and Volvox carteri genome data [2], [23]. From this alignment, a maximum likelihood analysis using the WAG model [24] was conducted using RAxML [25] with a bootstrap analysis [26] based on 100 replicates. Bootstrap analyses of the maximum parsimony method (based on the full heuristic search with the tree bisection reconnection branch-swapping algorithm) and neighbor joining methods (using p-distances) were also carried out based on 1,000 replications, using PAUP 4.0b10 [27] and Clustal X [28], respectively.
Determination of the length polymorphism within the cpDNA rbcL group I intron of Gonium maiaprilis
The nucleotide sequence of the putative rbcL group I intron from G. maiaprilis Asa041901 and Asa041903 (GenBank/EMBL/DDBJ accession nos. AB520743 and AB520744) was determined by direct sequencing [7] using two specific primers located in the adjoining rbcL coding regions (Table S2) and showed a 27 bp difference in sequence length between the two strains (Figure 3).
Genomic PCR for parental and F1 strains of Gonium maiaprilis
Presence/absence of GmMID and the length polymorphism within the cpDNA rbcL group I intron are assessed by gel electrophoreses. The nuclear gene EF-1alpha like of G. maiaprilis (GenBank/EMBL/DDBJ accession nos. AB623051 and AB623052) was determined by direct sequencing [7] using specific primers (Table 2) and genomic DNA, and serves as a control. PCR was performed with two specific primers for each gene (Table S2) and TaKaRa LA Taq (Takara bio inc., Shiga, Japan), under the following conditions: 30 cycles of 94°C for 30 seconds, 55°C for 30 seconds, and 72°C for 40 seconds, followed by 72°C for 7 minutes.
ITS-1 and ITS-2 secondary structures
The ITS-1 and ITS-2 sequences (GenBank/EMBL/DDBJ accession nos. AB623040-2) were directly determined by the methods described in Hayama et al. [7] with primers for ITS regions (Table S2). The secondary structures of ITS-1 and ITS-2 were predicted using CentroidFold [29], [30] and revise the secondary structure models of ITS-1 and ITS-2 from earlier studies [7], [9], [31]–[33].
Supporting Information
Comparison of exon-intron structure between GmMID and five other MID homologs.
(TIF)
Alignment of six MID proteins from Volvox carteri (VcMID), Pleodorina starrii (PlestMID), Gonium pectorale (GpMID), G. maiaprilis (GmMID), Chlamydomonas reinhardtii (CrMID), and C. globosa (previously misidentified as C. incersta [34] ) (CiMID). Solid and shaded backgrounds indicate identity in 100% or in over 60% of the sequences aligned, respectively. Five amino acids composing a leucine zipper are marked with asterisks. A line above the alignment marks the RWP-RK domain of 47 amino acids used for the phylogenetic analyses (Figure S3).
(TIF)
Maximum likelihood (ML) phylogenetic tree showing MID proteins from Volvox carteri (VcMID), Pleodorina starrii (PlestMID), Gonium maiaprilis (GmMID), G. pectorale (GpMID), Chlamydomonas reinhardtii (CrMID) and C. globosa (previously misidentified as C. incersta [34] ) (CiMID). Other members of the RWP-RK family from Chlamydomonas and Volvox are included as outgroup. Numbers next to branch points are bootstrap values for ML/neighbor joining/maximum parsimony methods.
(TIF)
Summary of mating phenotypes (MT), presence (gray)/absence (white) of GmMID and types of cpDNA ( rbcL group I intron) from parental strains (Asa041901[01] and Asao41903 [03]) and their 107 F1 strains of Gonium maiaprilis . White or gray box represents the same character as that of Asa041901 or Asa041903, respectively, for each of the three attributes. Each horizontal line indicates those F1 strains originating from the same germinating zygote. Isolation of progeny representing both mating types in the 3 and 4-membered tetrads indicate that these are meiotic products.
(TIF)
Secondary structures of the ITS-1 and ITS-2 RNA transcript of Gonium maiaprilis Asa041901 and Asa041902 (GenBank/EMBL/DDBJ accession nos. AB520746 and AB623042). Arrows mark the four single base substitutions between Asa041901 and Asa041903. The number between the two characters indicates the nucleotide position where the single base substitution occurred; the left character is the base of Asa041901 whereas the right character is the base of Asa041903.
(TIF)
Secondary structures of the ITS-1 and ITS-2 RNA transcript of Gonium pectorale Mongolia1 and Mongolia4 (GenBank/EMBL/DDBJ accession nos. AB623040 and AB623041). Arrows mark the seven single base substitutions between Mongolia1 and Mongolia4. The number between the two characters indicates the nucleotide position where the single base substitution occurred; the left character is the base of Mongolia1 whereas the right character is the base of Mongolia4.
(TIF)
Survival rates of F1 progeny from intra and interspecific crossing in various colonial volvocaceans.
(DOC)
Primers used for amplifications and sequencing of four DNA regions in the present study.
(DOC)
Acknowledgments
We thank Dr. Patrick Ferris for helpful comments, discussion and critical reading and correction of the manuscript.
Footnotes
Competing Interests: The authors have declared that no competing interests exist.
Funding: This work was supported by a Grant-in-Aid for Scientific Research (no. 22112505 to HN) from the Ministry of Education, Culture, Sports, Science and Technology, Japan (http://www.mext.go.jp/english/). The funder had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Boynton JE, Harris EH, Burkhart BD, Lamerson PM, Gillham NW. Transmission of mitochondrial and chloroplast genomes in crosses of Chlamydomonas. Proc Natl Acad Sci U S A. 1987;84:2391–2395. doi: 10.1073/pnas.84.8.2391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hamaji T, Ferris PJ, Coleman AW, Waffenschmidt S, Takahashi F, et al. Identification of the minus-dominance gene ortholog in the mating-type locus of Gonium pectorale. Genetics. 2008;178:283–294. doi: 10.1534/genetics.107.078618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Adams CR, Stamer KA, Miller JK, Mcnally JK, Kirk MM, et al. Patterns of organellar and nuclear inheritance among progeny of two geographically isolated strains of Volvox carteri. Curr Genet. 1990;18:141–153. doi: 10.1007/BF00312602. [DOI] [PubMed] [Google Scholar]
- 4.Bell RA, Cain JR. Sexual reproduction and hybridization in Chlamydomonas smithii and C. reinhardtii (Chlorophyceae, Volvocales). Phycologia. 1983;22:243–227. [Google Scholar]
- 5.Kaneda H, Hayashi J, Takahama S, Taya C, Lindahl KF, et al. Elimination of paternal mitochondrial DNA in intraspecific crosses during early mouse embryogenesis. Proc Natl Acad Sci U S A. 1995;92:4542–4546. doi: 10.1073/pnas.92.10.4542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shitara H, Hayashi J, Takahama S, Kaneda H, Yonekawa H. Maternal inheritance of mouse mtDNA in interspecific hybrids: segregation of the leaked paternal mtDNA followed by the prevention of subsequent paternal leakage. Genetics. 1998;148:851–858. doi: 10.1093/genetics/148.2.851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hayama M, Nakada T, Hamaji T, Nozaki H. Morphology, molecular phylogeny and taxonomy of Gonium maiaprilis sp. nov. (Goniaceae, Chlorophyta) from Japan. Phycologia. 2010;49:221–234. [Google Scholar]
- 8.Ferris PJ, Goodenough UW. Mating type in Chlamydomonas is specified by mid, the minus-dominance gene. Genetics. 1997;146:859–869. doi: 10.1093/genetics/146.3.859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Coleman AW. Is there a molecular key to the level of “biological species” in eukaryotes? A DNA guide. Mol Phylogenet Evol. 2009;50:197–203. doi: 10.1016/j.ympev.2008.10.008. [DOI] [PubMed] [Google Scholar]
- 10.Conover WJ. Practical Nonparametric Statistics, 3rd ed. John Wiley & Sons, Inc., Hoboken, NJ; 1998. 592 [Google Scholar]
- 11.Goldstein M. 1964 Speciation and mating behavior in Eudorina. J Protozool. 1964;11:317–344. [Google Scholar]
- 12.Nozaki H. Morphology, sexual reproduction and taxonomy of Volvox carteri f. kawasakiensis f. nov. (Chlorophyta) from Japan. Phycologia. 1988;27:209–220. [Google Scholar]
- 13.Nishimura Y, Misumi O, Matsunaga S, Higashiyama T, Yokota A, et al. The active digestion of uniparental chloroplast DNA in a single zygote of Chlamydomonas reinhardtii is revealed by using the optical tweezer. Proc Natl Acad Sci U S A. 1999;96:12577–12582. doi: 10.1073/pnas.96.22.12577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ferris PJ, Pavlovic C, Fabry S, Goodenough UW. Rapid evolution of sex related genes in Chlamydomonas. Proc Natl Acad Sci U S A. 1997;94:8634–8639. doi: 10.1073/pnas.94.16.8634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nishimura Y. Uniparental inheritance of cpDNA and the genetic control of sexual differentiation in Chlamydomonas reinhardtii. J Plant Res. 2010;123:149–162. doi: 10.1007/s10265-009-0292-y. [DOI] [PubMed] [Google Scholar]
- 16.Sager R. Mendelian and non-Mendelian inheritance of streptomycin resistance in Chlamydomonas reinhardi. Proc Natl Acad Sci U S A. 1954;40:356–363. doi: 10.1073/pnas.40.5.356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gillham NW, Levine RP. Studies on the origin of streptomycin resistant mutants in Chlamydomonas reinhardi. Genetics. 1962;47:1463–1474. doi: 10.1093/genetics/47.11.1463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mishra NC, Threlkeld SFH. Genetic studies in Eudorina. Genet Res Camb. 1968;11:21–31. doi: 10.1017/s0016672300011162. [DOI] [PubMed] [Google Scholar]
- 19.Kasai F, Kawachi M, Erata M, Mori F, Yumoto K, et al. NIES-Collection. List of Strains. 8th Edition. Jpn J Phycol. 2009;57(supplement):1–350, pls1–7. [Google Scholar]
- 20.Nozaki H, Kuroiwa H, Mita T, Kuroiwa T. Pleodorina japonica sp. nov. (Volvocales, Chlorophyta) with bacteria-like endosymbionts. Phycologia. 1989;28:252–267. [Google Scholar]
- 21.Kato S. Laboratory culture and morphology of Colacium vesiculosum Ehrb. (Euglenophyceae). Jpn J Phycol. 1982;30:63–67. (in Japanese with English abstract) [Google Scholar]
- 22.Nozaki H, Mori T, Misumi O, Matsunaga S, Kuroiwa T. Males evolved from the dominant isogametic mating type. Curr Biol. 2006;16:R1018–R1020. doi: 10.1016/j.cub.2006.11.019. [DOI] [PubMed] [Google Scholar]
- 23.Ferris P, Olson BJ, De Hoff PL, Douglass S, Casero D, et al. Evolution of an expanded sex-determining locus in Volvox. Science. 2010;328:351–354. doi: 10.1126/science.1186222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Whelan S, Goldman N. A general empirical model of protein evolution derived from multiple protein families using a maximum-likelihood approach. Mol Biol Evol. 2001;18:691–699. doi: 10.1093/oxfordjournals.molbev.a003851. [DOI] [PubMed] [Google Scholar]
- 25.Stamatakis A, Hoover P, Rougemont J. A rapid bootstrap algorithm for the RAxML web-servers. Syst Biol. 2008;75:758–771. doi: 10.1080/10635150802429642. [DOI] [PubMed] [Google Scholar]
- 26.Felsenstein J. Confidence limits on phylogenesis: an approach using the bootstrap. Evolution. 1985;39:783–791. doi: 10.1111/j.1558-5646.1985.tb00420.x. [DOI] [PubMed] [Google Scholar]
- 27.Swofford DL. PAUP*: Phylogenetic Analysis Using Parsimony (* and Other Methods). Version 4.0b10. Sinauer, Sunderland, Massachusetts; 2002. [Google Scholar]
- 28.Thompson JD, Gibson TJ, Plewnia F, Jeanmougin F, Higgins DG. The ClustalX windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucl Acids Res. 1997;25:4876–4882. doi: 10.1093/nar/25.24.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sato K, Hamada M, Asai K, Mituyama T. CentroidFold: a web server for RNA secondary structure prediction. Nuc Acids Res. 2009;37(No. suppl_2):W277–W280. doi: 10.1093/nar/gkp367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hamada M, Kiryu H, Sato K, Mituyama T, Asai K. Prediction of RNA secondary structure using generalized centroid estimators. Bioinformatics. 2009;25:465–473. doi: 10.1093/bioinformatics/btn601. [DOI] [PubMed] [Google Scholar]
- 31.Mai JC, Coleman AW. The internal transcribed spacer 2 exhibits a common secondary structure in green algae and flowering plants. J Mol Evol. 1997;44:258–271. doi: 10.1007/pl00006143. [DOI] [PubMed] [Google Scholar]
- 32.Pröschold T, Harris EH, Coleman AW. Portrait of a species: Chlamydomonas reinhardtii. Genetics. 2005;170:1601–1610. doi: 10.1534/genetics.105.044503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Coleman AW. Is there a molecular key to the level of ‘biological species’ in eukaryotes? A DNA guide. Mol Phylogenet Evol. 2009;50:197–203. doi: 10.1016/j.ympev.2008.10.008. [DOI] [PubMed] [Google Scholar]
- 34.Nakada T, Shinkawa H, Ito T, Tomita M. Recharacterization of Chlamydomonas reinhardtii and its relatives with new isolates from Japan. J Plant Res. 2010;123:67–78. doi: 10.1007/s10265-009-0266-0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Comparison of exon-intron structure between GmMID and five other MID homologs.
(TIF)
Alignment of six MID proteins from Volvox carteri (VcMID), Pleodorina starrii (PlestMID), Gonium pectorale (GpMID), G. maiaprilis (GmMID), Chlamydomonas reinhardtii (CrMID), and C. globosa (previously misidentified as C. incersta [34] ) (CiMID). Solid and shaded backgrounds indicate identity in 100% or in over 60% of the sequences aligned, respectively. Five amino acids composing a leucine zipper are marked with asterisks. A line above the alignment marks the RWP-RK domain of 47 amino acids used for the phylogenetic analyses (Figure S3).
(TIF)
Maximum likelihood (ML) phylogenetic tree showing MID proteins from Volvox carteri (VcMID), Pleodorina starrii (PlestMID), Gonium maiaprilis (GmMID), G. pectorale (GpMID), Chlamydomonas reinhardtii (CrMID) and C. globosa (previously misidentified as C. incersta [34] ) (CiMID). Other members of the RWP-RK family from Chlamydomonas and Volvox are included as outgroup. Numbers next to branch points are bootstrap values for ML/neighbor joining/maximum parsimony methods.
(TIF)
Summary of mating phenotypes (MT), presence (gray)/absence (white) of GmMID and types of cpDNA ( rbcL group I intron) from parental strains (Asa041901[01] and Asao41903 [03]) and their 107 F1 strains of Gonium maiaprilis . White or gray box represents the same character as that of Asa041901 or Asa041903, respectively, for each of the three attributes. Each horizontal line indicates those F1 strains originating from the same germinating zygote. Isolation of progeny representing both mating types in the 3 and 4-membered tetrads indicate that these are meiotic products.
(TIF)
Secondary structures of the ITS-1 and ITS-2 RNA transcript of Gonium maiaprilis Asa041901 and Asa041902 (GenBank/EMBL/DDBJ accession nos. AB520746 and AB623042). Arrows mark the four single base substitutions between Asa041901 and Asa041903. The number between the two characters indicates the nucleotide position where the single base substitution occurred; the left character is the base of Asa041901 whereas the right character is the base of Asa041903.
(TIF)
Secondary structures of the ITS-1 and ITS-2 RNA transcript of Gonium pectorale Mongolia1 and Mongolia4 (GenBank/EMBL/DDBJ accession nos. AB623040 and AB623041). Arrows mark the seven single base substitutions between Mongolia1 and Mongolia4. The number between the two characters indicates the nucleotide position where the single base substitution occurred; the left character is the base of Mongolia1 whereas the right character is the base of Mongolia4.
(TIF)
Survival rates of F1 progeny from intra and interspecific crossing in various colonial volvocaceans.
(DOC)
Primers used for amplifications and sequencing of four DNA regions in the present study.
(DOC)